Patterns of Genetic and Clonal Diversity in Myriophyllum spicatum in Streams and Reservoirs of Republic of Korea
Abstract
1. Introduction
2. Results
2.1. Variation in Microsatellite Loci
2.2. Genetic and Clonal Diversity
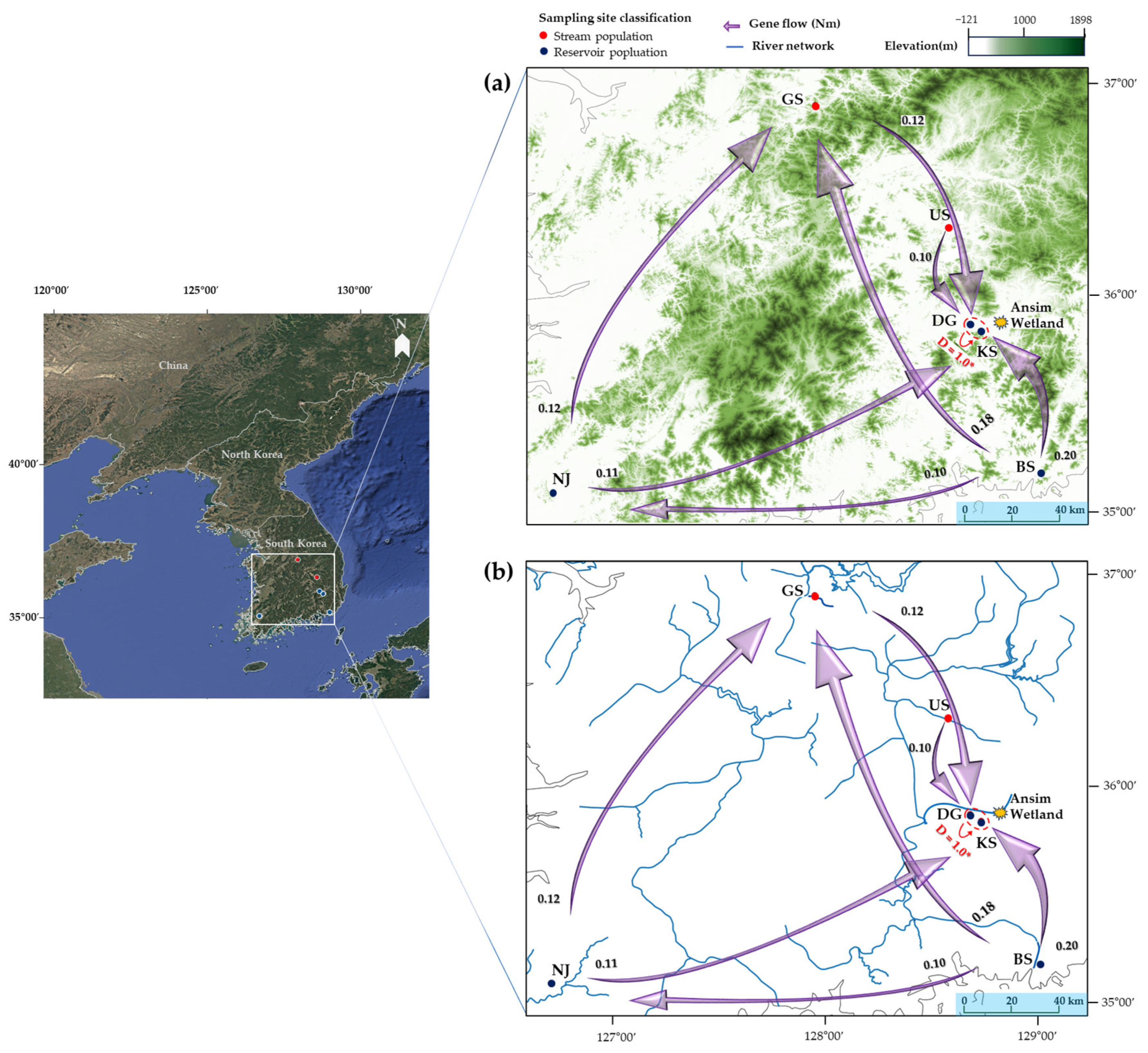
2.3. Genetic Differentiation and Gene Flow
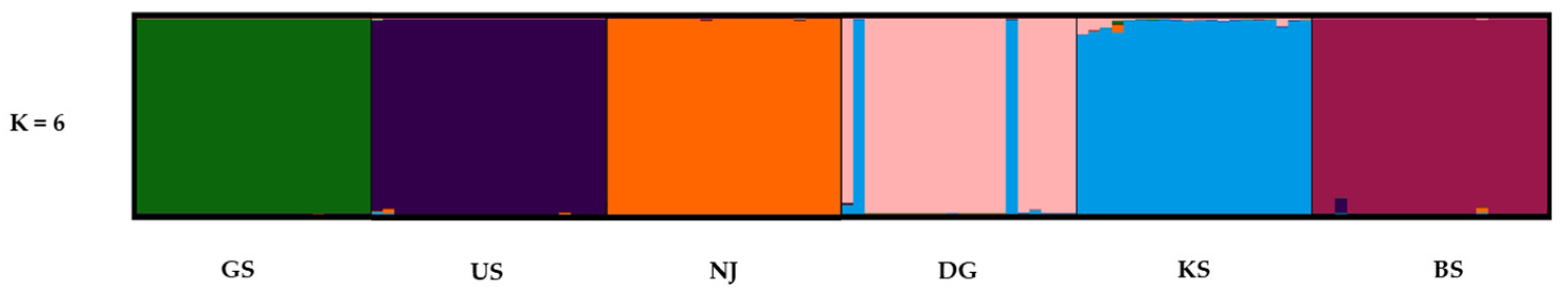
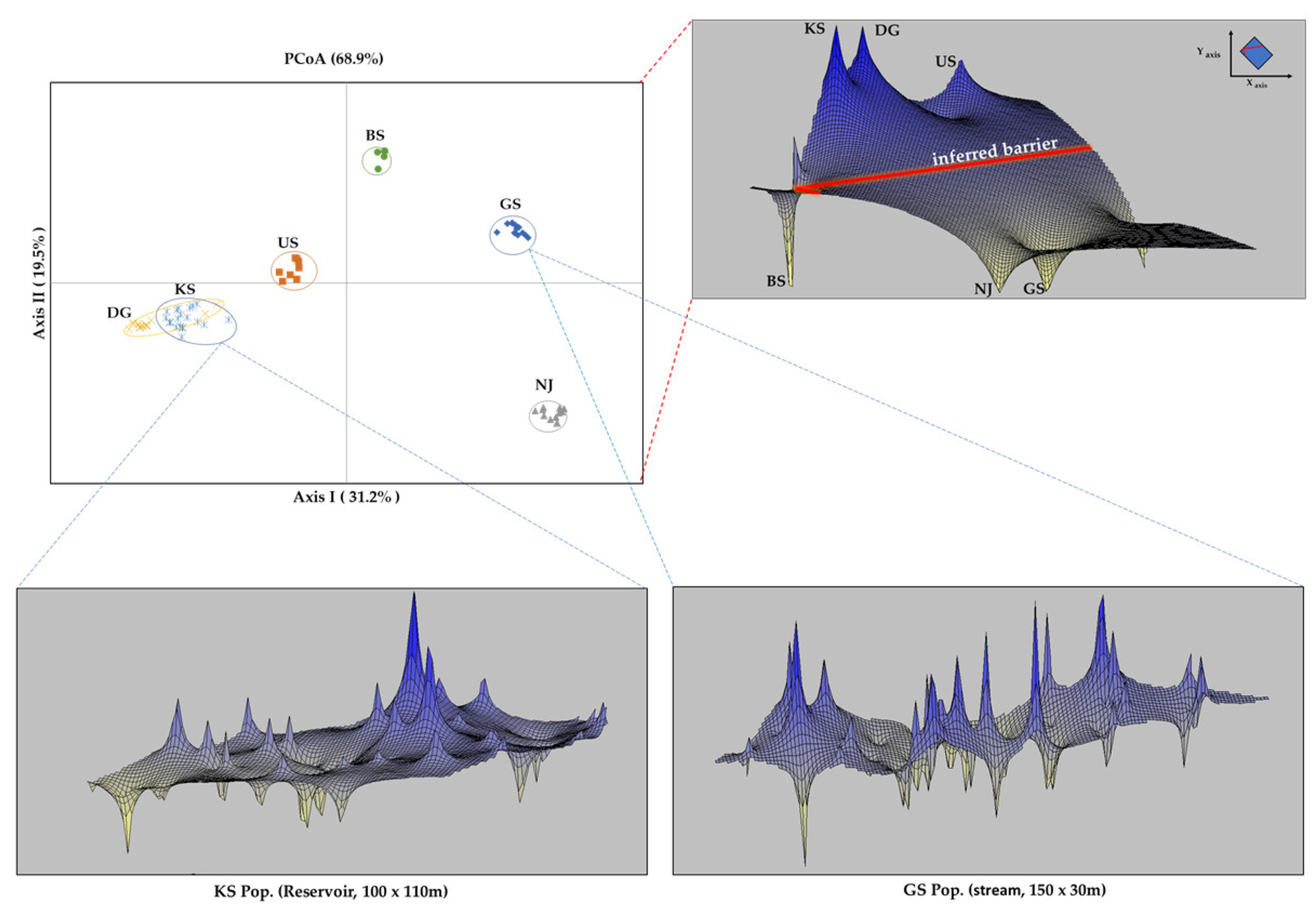
2.4. Spatial and Genetic Structure
3. Discussion
3.1. Variation in Microsatellite Loci
3.2. Genetic and Clonal Diversity
3.3. Genetic Differentiation and Gene Flow
3.4. Spatial and Genetic Structure
3.5. Implications for Conservation and Sustainable Management
4. Materials and Methods
4.1. Sample Preparation
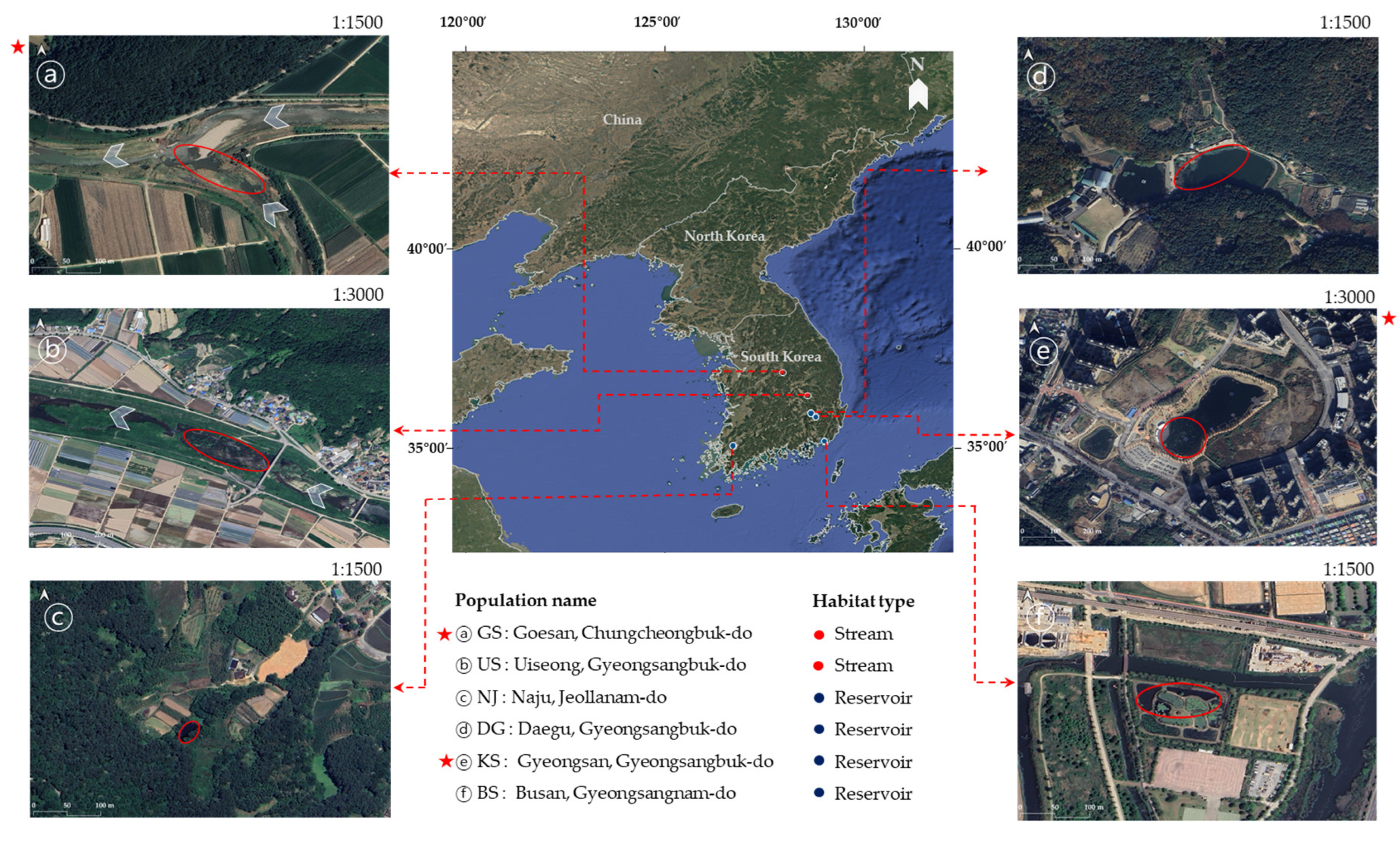
4.2. Microsatellite Marker Development and Polymerase Chain Reaction (PCR) Amplification
4.3. Genetic and Clonal Diversity Analysis
4.4. Genetic Differentiation and Gene Flow
4.5. Spatial and Genetic Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Young, A.; Boyle, T.; Brown, T. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol. 1996, 11, 413–418. [Google Scholar] [CrossRef]
- Booy, G.; Hendriks, R.J.J.; Smulders, M.J.M.; van Groenendael, J.M.; Vosman, B. Genetic diversity and the survival of populations. Plant Biol. 2000, 2, 379–395. [Google Scholar] [CrossRef]
- Hussner, A.; Heidbüchel, P.; Coetzee, J.; Gross, E.M. From introduction to nuisance growth: A review of traits of alien aquatic plants which contribute to their invasiveness. Hydrobiologia 2021, 848, 2119–2151. [Google Scholar] [CrossRef]
- Adomako, M.O.; Zhang, Q.; Yu, F.H. Genotypic differences in response to different patterns of clonal fragmentation in the aquatic macrophyte Pistia stratiotes. J. Plant Ecol. 2022, 15, 1199–1212. [Google Scholar] [CrossRef]
- Zhang, H.; Chase, J.M.; Liao, J. Habitat amount modulates biodiversity responses to fragmentation. Nat. Ecol. Evol. 2024, 8, 1437–1447. [Google Scholar] [CrossRef] [PubMed]
- Sakai, A.K.; Allendorf, F.W.; Holt, J.S.; Lodge, D.M.; Molofsky, J.; With, K.A.; Baughman, S.; Cabin, R.J.; Cohen, J.E.; Ellstrand, N.C.; et al. The population biology of invasive species. Annu. Rev. Ecol. Syst. 2001, 32, 305–332. [Google Scholar] [CrossRef]
- Eckert, C.G.; Lui, K.; Bronson, K.; Corradini, P.; Bruneau, A. Population genetic consequences of extreme variation in sexual and clonal reproduction in an aquatic plant. Mol. Ecol. 2003, 12, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Barrett, L.G.; Thrall, P.H.; Burdon, J.J.; Linde, C.C. Life history determines genetic structure and evolutionary potential of host–parasite interactions. Trends Ecol. Evol. 2008, 23, 678–685. [Google Scholar] [CrossRef]
- Stange, M.; Barrett, R.D.H.; Hendry, A.P. The importance of genomic variation for biodiversity, ecosystems and people. Nat. Rev. Genet. 2021, 22, 89–105. [Google Scholar] [CrossRef]
- Eckert, C.G.; Dorken, M.E.; Barrett, S.C. Ecological and evolutionary consequences of sexual and clonal reproduction in aquatic plants. Aquat. Bot. 2016, 135, 46–61. [Google Scholar] [CrossRef]
- Ashton, P.J.; Mitchell, D.S. Aquatic plants: Patterns and modes of invasion, attributes of invading species and assessment of control programs. In Biological Invasions: A Global Perspective; Drake, J.A., Mooney, H.A., Di Castri, F., Groves, R.H., Kruger, F.J., Rejmânek, M., Williamson, M.H., Eds.; John Wiley and Sons, Ltd.: London, UK, 1989; pp. 111–154. [Google Scholar]
- Starfinger, U.; Stöcklin, J. Seed, pollen, and clonal dispersal and their role in structuring plants populations. Prog. Bot. 1996, 57, 336–355. [Google Scholar]
- Piquot, Y.; Petit, D.; Valero, M.; Cuguen, J.; de Laguerie, P.; Vernet, P. Variation in sexual and asexual reproduction among young and old populations of the perennial macrophyte Dendroseris. Oikos 1998, 82, 139–148. [Google Scholar] [CrossRef]
- Silvertown, J. The evolutionary maintenance of sexual reproduction: Evidence from the ecological distribution of asexual reproduction in clonal plants. Int. J. Plant Sci. 2008, 169, 157–168. [Google Scholar] [CrossRef]
- Vallejo–Marín, M.; Dorken, M.E.; Barrett, S.C.H. The ecological and evolutionary consequences of clonality for plant mating. Annu. Rev. Ecol. Evol. Syst. 2010, 41, 193–213. [Google Scholar] [CrossRef]
- da Cunha, N.L.; Fischer, E.; Lorenz–Lemke, A.P.; Barrett, S.C.H. Floral variation and environmental heterogeneity in a tristylous clonal aquatic of the Pantanal wetlands of Brazil. Ann. Bot. 2014, 114, 1637–1649. [Google Scholar] [CrossRef]
- Wetzel, R.G. A comparative study of the primary production of higher aquatic plants, periphyton, and phytoplankton in a large, shallow lake. Int. Rev. Gesamt Hydrobiol. Hydrogr. 1964, 49, 1–61. [Google Scholar] [CrossRef]
- Jiang, H.S.; Zhang, Y.; Yin, L.; Li, W.; Jin, Q.; Fu, W.; Zhang, T.; Huang, W. Diurnal changes in photosynthesis by six submerged macrophytes measured using fluorescence. Aquat. Bot. 2018, 149, 33–39. [Google Scholar] [CrossRef]
- Yu, C.; Yang, L.; Leah, N.; Wei, L.; Ling, X.; Hong, S.J. The analysis of leaf traits of eight Ottelia populations and their potential ecosystem functions in karst freshwaters in China. Front. Plant Sci. 2019, 9, 1938. [Google Scholar] [CrossRef] [PubMed]
- Crawford, D.J.; Stuessy, T.F.; Silva, M. Allozyme divergence and the evolution of Dendroseris (Compositae: Lactuceae) on the Juan Fernandez Islands. Syst. Bot. 1987, 12, 435–443. [Google Scholar] [CrossRef]
- Barrett, S.C.H.; Eckert, C.G.; Husband, B.C. Evolutionary processes in aquatic plant populations. Aquat. Bot. 1993, 44, 105–145. [Google Scholar] [CrossRef]
- Santamaría, L. Why are most aquatic plants widely distributed? Dispersal, clonal growth and small–scale heterogeneity in a stressful environment. Acta Oecol. 2002, 23, 137–154. [Google Scholar] [CrossRef]
- Junk, W.J.; Piedade, M.T.F. Plant life in the floodplain with special reference to herbaceous plants. In The Central Amazon Floodplain: Ecology of a Pulsing System; Springer Nature: Berlin/Heidelberg, Germany, 1997; pp. 147–185. [Google Scholar]
- Sculthorpe, C.D. The Biology of Aquatic Vascular Plants; Edward Arnold: London, UK, 1967; p. 610. [Google Scholar]
- van Groenendael, J.M.; Klimeš, L.; Klimešová, J.; Hendriks, R.J.J. Comparative ecology of clonal plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1996, 351, 1331–1339. [Google Scholar]
- Yakimowski, S.B.; Barrett, S.C.H. Clonal genetic structure and diversity in populations of an aquatic plant with combined vs. separate sexes. Mol. Ecol. 2014, 23, 2914–2928. [Google Scholar] [CrossRef]
- Klimešová, J.; Martínková, J.; Ottaviani, G. Belowground plant functional ecology: Towards an integrated perspective. Funct. Ecol. 2018, 32, 2115–2126. [Google Scholar] [CrossRef]
- Franklin, S.; Alpert, P.; Salguero–Gómez, R.; Janovský, Z.; Herben, T.; Klimešová, J.; Douhovnikoff, V. Next–gen plant clonal ecology. Perspect. Plant Ecol. Evol. Syst. 2021, 49, 125601. [Google Scholar] [CrossRef]
- Bowes, G. Aquatic plant photosynthesis: Strategies that enhance carbon gain. In Plant Life in Aquatic and Amphibious Habitats; Crawford, R.M.M., Ed.; Blackwell Scientific Publications: Oxford, UK, 1987; pp. 79–98. [Google Scholar]
- Gavrilescu, M. Water, Soil, and Plants Interactions in a Threatened Environment. Water 2021, 13, 2746. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, Z.; Xie, D.; Wang, H.; Zhao, A.; Wang, Y.; Hanling, W.; xinwei, X.; Tao, L.; Zhao, J. Effects of highland environments on clonal diversity in aquatic plants: An interspecific comparison study on the Qinghai–Tibetan Plateau. Front. Plant Sci. 2022, 13, 1040282. [Google Scholar] [CrossRef]
- Higgisson, W.; Broadhurst, L.; Shams, F.; Gruber, B.; Dyer, F. Reproductive strategies and population genetic structure in two dryland river floodplain plants, Marsilea drummondii and Eleocharis acuta. Genes 2022, 13, 1506. [Google Scholar] [CrossRef]
- Korea National Arboretum. Myriophyllum spicatum L. Available online: http://www.nature.go.kr/kbi/plant/pilbk/selectPlantPilbkDtl.do?plantPilbkNo=26182 (accessed on 1 April 2025).
- Patten, B.C., Jr. Notes on the biology of Myriophyllum spicatum L. in a New Jersey lake. Bull. Torrey Bot. Club. 1956, 83, 5–18. [Google Scholar] [CrossRef]
- Aiken, S.G.; Newroth, P.R.; Wile, I. The biology of Canadian Weeds.: 34. Myriophyllum spicatum L. Can. J. Plant Sci. 1979, 59, 201–215. [Google Scholar] [CrossRef]
- Ortiz, J.E.; Marcarelli, A.M.; Juneau, K.J.; Huckins, C.J. Invasive Myriophyllum spicatum and nutrients interact to influence algal assemblages. Aquat. Bot. 2019, 156, 1–9. [Google Scholar] [CrossRef]
- Lin, Z.; Zhong, C.; Yu, G.; Fu, Y.; Guan, B.; Liu, Z.; Yu, J. Effects of sediments phosphorus inactivation on the life strategies of Myriophyllum spicatum: Implications for lake restoration. Water 2021, 13, 2112. [Google Scholar] [CrossRef]
- Glisson, W.J.; Larkin, D.J. Hybrid watermilfoil (Myriophyllum spicatum × Myriophyllum sibiricum) exhibits traits associated with greater invasiveness than its introduced and native parental taxa. Biol. Invasions 2021, 23, 2417–2433. [Google Scholar] [CrossRef]
- Gassmann, A.J.; Levy, A.; Tran, T.; Futuyma, D.J. Adaptations of an insect to a novel host plant: A phylogenetic approach. Funct. Ecol. 2006, 20, 478–485. [Google Scholar] [CrossRef]
- Beck, K.G.; Zimmerman, K.; Schardt, J.D.; Stone, J.; Lukens, R.R.; Reichard, S.; Randall, J.; Cangelosi, A.A.; Cooper, D.; Thompson, J.P. Invasive species defined in a policy context: Recommendations from the Federal Invasive Species Advisory Committee. Invasive Plant Sci. Manag. 2008, 1, 414–421. [Google Scholar] [CrossRef]
- Hussner, A.; Stiers, I.; Verhofstad, M.J.J.M.; Bakker, E.S.; Grutters, B.M.C.; Haury, J.; van Valkenburg, J.L.C.H.; Brundu, G.; Newman, J.; Clayton, J.S.; et al. Management and control methods of invasive alien freshwater aquatic plants: A review. Aquat. Bot. 2017, 136, 112–137. [Google Scholar] [CrossRef]
- Li, F.; Qin, Y.; Zhu, L.; Xie, Y.; Liang, S.; Hu, C.; Chen, X.; Deng, Z. Effects of fragment size and sediment heterogeneity on the colonization and growth of Myriophyllum spicatum. Ecol. Eng. 2016, 95, 457–462. [Google Scholar] [CrossRef]
- Fan, X.R.; Njeri, H.K.; Pu, Y.H.; La, Q.; Li, W.; Li, X.L.; Chen, Y.Y. Contrasting relationships between genetic diversity and species diversity in conserved and disturbed submerged macrophyte communities of Honghu Lake, a typical freshwater lake of Yangtze River Basin. Glob. Ecol. Conserv. 2021, 31, 01873. [Google Scholar] [CrossRef]
- Ahn, K.; Lim, J.; Lee, Y.; Choi, T.; Lee, K.; Im, M.; Go, Y.; Suh, J.; Shin, Y.; Kim, M. Vegetation classification and distributional pattern in Damyang riverine wetland. J. Environ. Impact Assess. 2016, 25, 89–102. [Google Scholar] [CrossRef]
- Xiao, C.; Wang, X.F.; Xia, J.; Li, G.H. The effect of temperature, water level and burial depth on seed germination of Myriophyllum spicatum and Potamogeton malaianus. Aquat. Bot. 2010, 92, 28–32. [Google Scholar] [CrossRef]
- Lillie, R.A.; Budd, J. Habititat architecture of Myriophyllum spicatum L. as an index to habitat quality for fish and macroinvertebrates. J. Freshw. Ecol. 1992, 7, 113–125. [Google Scholar] [CrossRef]
- Madsen, J.D. Methods for Management of Nonindigenous Aquatic Plants. In Assessment and Management of Plant Invasions; Luken, J.O., Thieret, J.W., Eds.; Springer Series on Environmental Management; Springer: New York, NY, USA, 1997; pp. 145–171. [Google Scholar]
- National Institute of Biological Resources. Available online: https://species.nibr.go.kr (accessed on 14 February 2025).
- Jeong, S.; Yang, D.; Joo, S.; Park, S. Allelopathic inhibition effects of Myriophyllum spicatum on growths of bloom–forming cyanobacteria and other phytoplankton species in coexistence experiments. J. Plant Biol. 2021, 64, 501–510. [Google Scholar] [CrossRef]
- Kim, C.H.; Lee, Y.K.; Kim, M.J.; Choi, J.S.; Hwang, B.S.; Cho, P.Y.; Kim, Y.J.; Jeong, Y.T. Evaluation antioxidant and anti–inflammatory activity of ethanolic extracts of Myriophyllum spicatum L. in lipopolysaccharide–stimulated RAW 264.7 cells. Korean J. Plant Res. 2023, 36, 15–25. [Google Scholar]
- Pichersky, E.; Gang, D.R. Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends Plant Sci. 2000, 5, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Wink, M. Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 2003, 64, 3–19. [Google Scholar] [CrossRef]
- Christensen, K.B.; Petersen, R.K.; Petersen, S.; Kristiansen, K.; Christensen, L.P. Activation of PPARγ by metabolites from the flowers of purple coneflower (Echinacea purpurea). J. Nat. Prod. 2009, 72, 933–937. [Google Scholar] [CrossRef]
- Kwon, S.T.; Kamiya, J. Genetic variation of cytochrome P450 genes in garlic cultivars. Korean J. Plant Res. 2011, 24, 584–590. [Google Scholar] [CrossRef]
- Govindaraj, M.; Vetriventhan, M.; Srinivasan, M. Importance of genetic diversity assessment in crop plants and its recent advances: An overview of its analytical perspectives. Genet. Res. Int. 2015, 431–487. [Google Scholar] [CrossRef]
- Bhandari, H.R.; Bhanu, A.N.; Srivastava, K.; Singh, M.N.; Shreya, H.A. Assessment of genetic diversity in crop plants—An overview. Adv. Plants Agric. Res. 2017, 7, 279–286. [Google Scholar]
- Ribeiro, M.M.; Diamantino, T.; Domingues, J.; Montanari, Í., Jr.; Alves, M.N.; Gonçalves, J.C. Stevia rebaudiana germplasm characterization using microsatellite markers and steviol glycosides quantification by HPLC. Mol. Biol. Rep. 2021, 48, 2573–2582. [Google Scholar] [CrossRef]
- LaRue, E.A.; Grimm, D.; Thum, R.A. Laboratory crosses and genetic analysis of natural populations demonstrate sexual viability of invasive hybrid watermilfoils (Myriophyllum spicatum × M. sibiricum). Aquat. Bot. 2013, 109, 49–53. [Google Scholar] [CrossRef]
- Wu, Z.; Yu, D.; Wang, Z.; Li, X.; Xu, X. Great influence of geographic isolation on the genetic differentiation of Myriophyllum spicatum under a steep environmental gradient. Sci. Rep. 2015, 5, 15618. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yu, D.; Li, X.; Xu, X. Influence of geography and environment on patterns of genetic differentiation in a widespread submerged macrophyte, Eurasian watermilfoil (Myriophyllum spicatum L., Haloragaceae). Ecol. Evol. 2016, 6, 460–468. [Google Scholar] [CrossRef]
- Tóth, V.R.; Endre, G.; Kovács, S.; Présing, M.; Horváth, H. Morphological and genetic variability of Myriophyllum spicatum in different shallow water bodies of Hungary. Wetlands 2017, 37, 351–362. [Google Scholar] [CrossRef]
- Cao, Q.; Hu, F.; Liu, N. Local–scale patterns of genetic variation in coexisting floating–leaved Nymphoides peltata and submerged Myriophyllum spicatum in Donghu Lake. J. Oceanol. Limnol. 2020, 38, 1825–1834. [Google Scholar] [CrossRef]
- Thum, R.A.; Chorak, G.M.; Newman, R.M.; Eltawely, J.A.; Latimore, J.; Elgin, E.; Parks, S. Genetic diversity and differentiation in populations of invasive Eurasian (Myriophyllum spicatum) and hybrid (Myriophyllum spicatum × Myriophyllum sibiricum) watermilfoil. Invasive Plant Sci. Manag. 2020, 13, 59–67. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, Z.; Xie, D.; Zhang, J.; Cai, P.; Li, X.; Zhao, J. Extensive Sympatry and Frequent Hybridization of Ecologically Divergent Aquatic Plants on the Qinghai–Tibetan Plateau. Front. Plant Sci. 2022, 13, 851151. [Google Scholar] [CrossRef]
- Schlötterer, C. Evolutionary dynamics of microsatellite DNA. Chromosoma 2000, 109, 365–371. [Google Scholar] [CrossRef]
- Hearne, C.M.; Ghosh, S.; Todd, J.A. Microsatellites for linkage analysis of genetic traits. Trends Genet. 1992, 8, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Morgante, M.; Olivieri, A.M. PCR-amplified microsatellites as markers in plant genetics. Plant J. 1993, 3, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Bonin, A.; Bellemain, E.; Bronken Eidesen, P.; Pompanon, F.; Brochmann, C.; Taberlet, P. How to track and assess genotyping errors in population genetics studies. Mol. Ecol. 2004, 13, 3261–3273. [Google Scholar] [CrossRef]
- Ellstrand, N.C.; Roose, M.L. Patterns of genotypic diversity in clonal plant species. Am. J. Bot. 1987, 74, 123–131. [Google Scholar] [CrossRef]
- Hidding, B.; Meirmans, P.G.; Klaassen, M.; de Boer, T.; Ouborg, N.J.; Wagemaker, C.A.M.; Nolet, B.A. The effect of herbivores on genotypic diversity in a clonal aquatic plant. Oikos 2014, 123, 1112–1120. [Google Scholar] [CrossRef]
- Barrat–Segretain, M.H. Strategies of reproduction, dispersion, and competition in river plants: A review. Vegetatio 1996, 123, 13–37. [Google Scholar] [CrossRef]
- Dawson, H.F. Water flow and the vegetation of running waters. In Vegetation of Inland Waters, 1st ed.; Symoens, J.J., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1988; pp. 283–309. [Google Scholar]
- Cao, Q.J.; Mei, F.F.; Wang, L. Population genetic structure in six sympatric and widespread aquatic plants inhabiting diverse lake environments in China. Ecol. Evol. 2017, 7, 5713–5723. [Google Scholar] [CrossRef]
- Wani, A.A.; Arshid, S. Assessment of seed quality parameters and effect of physical and chemical treatments on seed germination of Myriophyllum spicatum L. Comunic. Sci. 2013, 4, 1–11. [Google Scholar]
- Ministry of Environment, Republic of Korea, National Institute of Environmental Research. Water Environment Information System. Available online: https://water.nier.go.kr/web/waterMeasure?pMENU_NO=2 (accessed on 1 April 2025).
- Ministry of Environment, Korea. Flood Hazard Map Information. Available online: https://floodmap.go.kr/fldara/fldaraList.do (accessed on 10 August 2025).
- Korea Meteorological Administration (KMA). Korea Meteorological Administration (KMA). Rainy Season Data and Typhoon Data. Available online: https://data.kma.go.kr/climate/rainySeason/selectRainySeasonList.do (accessed on 10 August 2025).
- Son, D.; Cho, K.H.; Lee, E.J. The potential habitats of two submerged macrophytes, Myriophyllum spicatum and Hydrilla verticillata in the river ecosystems, South Korea. Knowl. Manag. Aquat. Ecosyst. 2017, 418, 58. [Google Scholar] [CrossRef]
- Obeso, J.R. The costs of reproduction in plants. New Phytol. 2002, 155, 321–348. [Google Scholar] [CrossRef]
- Kleunen, M.V.; Fischer, M.; Schmid, B. Effects of intraspecific competition on size variation and reproductive allocation in a clonal plant. Oikos 2001, 94, 515–524. [Google Scholar] [CrossRef]
- Tobiessen, P.; Snow, P.D. Temperature and light effects on the growth of Potamogeton crispus in Collins Lake, New York State. Can. J. Bot. 1984, 62, 2822–2826. [Google Scholar] [CrossRef]
- Strand, J.A.; Weisner, S.E. Morphological plastic responses to water depth and wave exposure in an aquatic plant (Myriophyllum spicatum). J. Ecol. 2001, 89, 166–175. [Google Scholar] [CrossRef]
- Eriksson, O. Dynamics of genets in clonal plants. Trends Ecol. Evol. 1993, 8, 313–316. [Google Scholar] [CrossRef]
- Bazzaz, F.A. Plants in Changing Environments: Linking Physiological, Population, and Community Ecology, 1st ed.; Cambridge University Press: Cambridge, UK, 1996. [Google Scholar]
- Stueffer, J.F.; de Kroon, H.; During, H.J. Exploitation of environmental heterogeneity by spatial division of labour in a clonal plant. Funct. Ecol. 1996, 10, 328–334. [Google Scholar] [CrossRef]
- Klimešová, J.; Dolezal, J.; Prach, K.; Kosnar, J. Clonal growth forms in Arctic plants and their habitat preferences: A study from Petuniabukta, Spitsbergen. Polish Polar Res. 2012, 33, 421–442. [Google Scholar] [CrossRef]
- Klimešová, J.; Ottaviani, G.; Charles–Dominique, T.; Campetella, G.; Canullo, R.; Chelli, S.; Janovský, Z.; Lubbe, F.C.; Martínková, J.; Herben, T. Incorporating clonality into the plant ecology research agenda. Trends Plant Sci. 2021, 26, 1236–1247. [Google Scholar] [CrossRef]
- Demetrio, G.R.; de Freitas Coelho, F. What are the consequences of clonal integration for floral traits and reproductive investment of a broadly distributed aquatic plant? Flora 2023, 303, 152292. [Google Scholar] [CrossRef]
- Hutchings, M.J.; Wijesinghe, D.K. Patchy habitats, division of labour and growth dividends in clonal plants. Trends Ecol. Evol. 1997, 12, 390–394. [Google Scholar] [CrossRef] [PubMed]
- De Kroon, H.; Huber, H.; Stuefer, J.F.; Van Groenendael, J.M. A modular concept of phenotypic plasticity in plants. New Phytol. 2005, 166, 73–82. [Google Scholar] [CrossRef]
- Fazlioglu, F.; Bonser, S.P. Phenotypic plasticity and specialization in clonal versus non–clonal plants: A data synthesis. Acta Oecol. 2016, 77, 193–200. [Google Scholar] [CrossRef]
- Timpane–Padgham, B.L.; Beechie, T.; Klinger, T. A systematic review of ecological attributes that confer resilience to climate change in environmental restoration. PLoS ONE 2017, 12, e0173812. [Google Scholar] [CrossRef]
- Muller, H.J. The relation of recombination to mutational advance. Mutat. Res.—Fund. Mol. M. 1964, 1, 2–9. [Google Scholar] [CrossRef]
- Felsenstein, J. The evolutionary advantage of recombination. Genetics 1974, 78, 737–756. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Latta, L.; Hicks, J.; Giorgianni, M. Mutation, selection, and the maintenance of life-history variation in a natural population. Evolution 1998, 52, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Ivey, C.T.; Richards, J.H. Genotypic diversity and clonal structure of everglades sawgrass, Cladium jamaicense (Cyperaceae). Int. J. Pl. Sci. 2001, 162, 1327–1335. [Google Scholar] [CrossRef]
- Cousens, R.D.; Hill, J.; French, K.; Bishop, I.D. Towards Better Prediction of Seed Dispersal by Animals. Funct. Ecol. 2010, 24, 1163–1170. [Google Scholar] [CrossRef]
- Figuerola, J.; Green, A.J. Dispersal of aquatic organisms by waterbirds: A review of past research and priorities for future studies. Freshw. Biol. 2002, 47, 483–494. [Google Scholar] [CrossRef]
- Green, A.J.; Figuerola, J.; Sánchez, M.I. Implications of waterbird ecology for the dispersal of aquatic organisms. Acta Oecol. 2002, 23, 177–189. [Google Scholar] [CrossRef]
- Clausen, P.; Nolet, B.A.; Fox, A.D.; Klaassen, M. Long–distance endozoochorous dispersal of submerged macrophyte seeds by migratory waterbirds in northern Europe—A critical review of possibilities and limitations. Acta Oecol. 2002, 23, 191–203. [Google Scholar] [CrossRef]
- Reynolds, C.; Cumming, G.S. Seed traits and bird species influence the dispersal parameters of wetland plants. Freshw. Biol. 2016, 61, 1157–1170. [Google Scholar] [CrossRef]
- Yoon, J.; Park, H.C.; Park, C. Distribution and diversity of birds in the Ansim Wetland area of Daegu, a wildlife protection area. Korean J. Ornithol. 2023, 30, 125–135. [Google Scholar] [CrossRef]
- Willson, M.F.; Traveset, A. The Ecology of Seed Dispersal. In Seeds: The Ecology of Regeneration in Plant Communities; CABI: Wallingford, UK, 2000; pp. 85–110. [Google Scholar]
- Guastello, P.R.; Thum, R.A. Mesocosm and field evaluation of Eurasian and hybrid watermilfoil response to endothall in Jefferson Slough, Montana. J. Aquat. Plant Manag. 2018, 56, 63–67. [Google Scholar]
- Engloner, A.I.; Németh, K.; Kós, P.B.; Meglécz, E.; Bereczki, J. Genetic Diversity of the Submerged Macrophyte Ceratophyllum demersum Depends on Habitat Hydrology and Habitat Fragmentation. Front. Plant Sci. 2023, 14, 1277916. [Google Scholar] [CrossRef]
- Nazareno, A.G.; Knowles, L.L.; Dick, C.W.; Lohmann, L.G. By Animal, Water, or Wind: Can Dispersal Mode Predict Genetic Connectivity in Riverine Plant Species? Front. Plant Sci. 2021, 12, 626405. [Google Scholar] [CrossRef] [PubMed]
- Madsen, J.D.; Smith, D.H. Vegetative spread of eurasian watermilfoil colonies. J. Aquatic Plant Manag. 1997, 35, 63–68. [Google Scholar]
- De Meester, L.; Gómez, A.; Okamura, B.; Schwenk, K. The Monopolization Hypothesis and the Dispersal–Gene Flow Paradox in Aquatic Organisms. Acta Oecol. 2002, 23, 121–135. [Google Scholar] [CrossRef]
- Hussner, A. Long–Term Macrophyte Mapping Documents a Continuously Shift from Native to Non–Native Aquatic Plant Dominance in the Thermally Abnormal River Erft (North Rhine–Westphalia, Germany). Limnologica 2014, 48, 39–45. [Google Scholar] [CrossRef]
- Heidbüchel, P.; Hussner, A. Falling into Pieces: In Situ Fragmentation Rates of Submerged Aquatic Plants and the Influence of Discharge in Lowland Streams. Aquat. Bot. 2020, 160, 103164. [Google Scholar] [CrossRef]
- Thum, R.A. Genetic variation and aquatic plant management: Key concepts and practical implications. J. Aquat. Plant Manag. 2018, 56, 101–106. [Google Scholar]
- You, W.H.; Yu, D.; Liu, C.H.; Xie, D.; Xiong, W. Clonal integration facilitates invasiveness of the alien aquatic plant Myriophyllum aquaticum L. under heterogeneous water availability. Hydrobiologia 2013, 718, 27–39. [Google Scholar] [CrossRef]
- Bernos, T.A.; Jeffries, K.M.; Mandrak, N.E. Aquatic invasive species specialists’ perceptions on the importance of genetic tools and concepts to inform management. Biol. Invasions 2022, 24, 1863–1879. [Google Scholar] [CrossRef]
- National Assembly Budget Office (NABO). Evaluation of the Ecological Stream Restoration Project; NABO: Seoul, Republic of Korea, 2015; Available online: https://www.nabo.go.kr (accessed on 1 August 2025).
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Steeves, T.E.; Hale, M.L.; Gemmell, N.J. Development of polymorphic microsatellite markers for the New Zealand black stilt (Himantopus novaezelandiae) and cross-amplification in the pied stilt (Himantopus himantopus leucocephalus). Mol. Ecol. Res. 2008, 8, 1105–1107. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. FSTAT (version 1.2): A computer program to calculate F–statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.C.; Boyle, T. POPGENE Version 1.32: Microsoft Windows–Based Freeware for Population Genetic Analysis, Quick User Guide; Center for International Forestry Research, University of Alberta: Edmonton, AB, Canada, 1999; pp. 1–29. [Google Scholar]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Pielou, E.C. An Introduction to Mathematical Ecology; Wiley–Interscience: New York, NY, USA, 1969. [Google Scholar]
- Cristescu, R.; Sherwin, W.B.; Handasyde, K.; Cahill, V.; Cooper, D.W. Detecting bottlenecks using BOTTLENECK 1.2. 02 in wild populations: The importance of the microsatellite structure. Conserv. Genet. 2010, 11, 1043–1049. [Google Scholar] [CrossRef]
- Wright, S. The interpretation of population structure by F–statistics with special regard to systems of mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar] [PubMed]
- Bohonak, A.J. IBD (isolation by distance): A program for analyses of isolation by distance. J. Hered. 2002, 93, 153–154. [Google Scholar] [CrossRef] [PubMed]
- Sundqvist, L.; Keenan, K.; Zackrisson, M.; Prodöhl, P.; Kleinhans, D. Directional genetic differentiation and relative migration. Ecol. Evol. 2016, 6, 3461–3475. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Wen, W.; Falush, D. Documentation for STRUCTURE Software: Version 2; University of Chicago: Chicago, IL, USA, 2003. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Li, Y.L.; Liu, J.X. Structure Selector: A Web-Based Software to Select and Visualize the Optimal Number of Clusters Using Multiple Methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M. The Vegan Package. Community Ecol. Package 2007, 10, 719. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.r-project.org/ (accessed on 20 January 2025).
- Miller, M.P. Alleles in Space (AIS): Computer Software for the Joint Analysis of Interindividual Spatial and Genetic Information. J. Hered. 2005, 96, 722–724. [Google Scholar] [CrossRef]
- Ministry of Environment. 2025 Budget and Fund Management Plan. 2024. Available online: https://www.me.go.kr (accessed on 4 April 2025). (In Korean).
- Lee, M.; Kim, H.; Lee, J.Y.; Yang, J.E.; Lim, C. A shift towards integrated and adaptive water management in South Korea: Building resilience against climate change. Water Resour. Manag. 2022, 36, 1611–1625. [Google Scholar] [CrossRef]
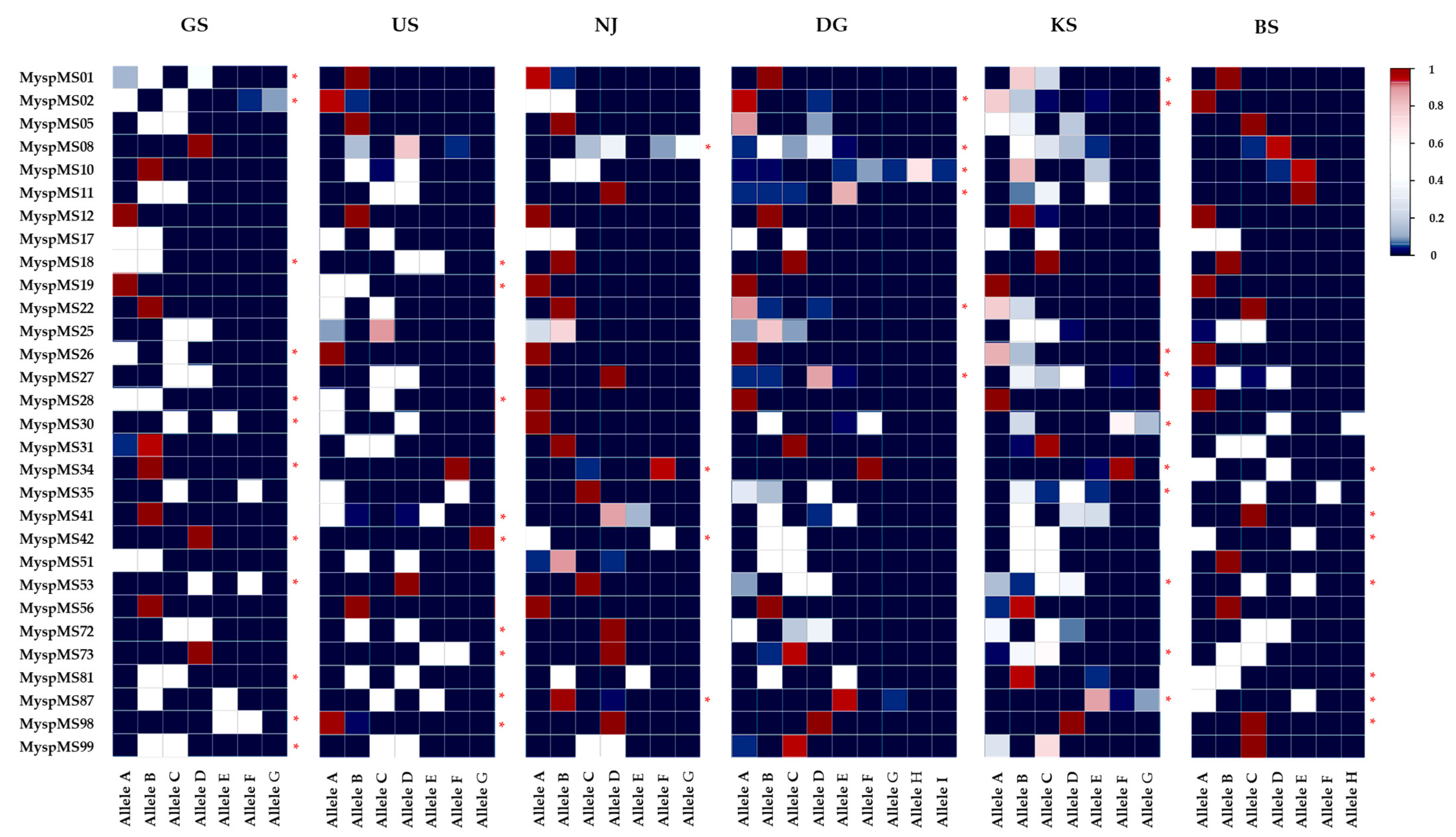
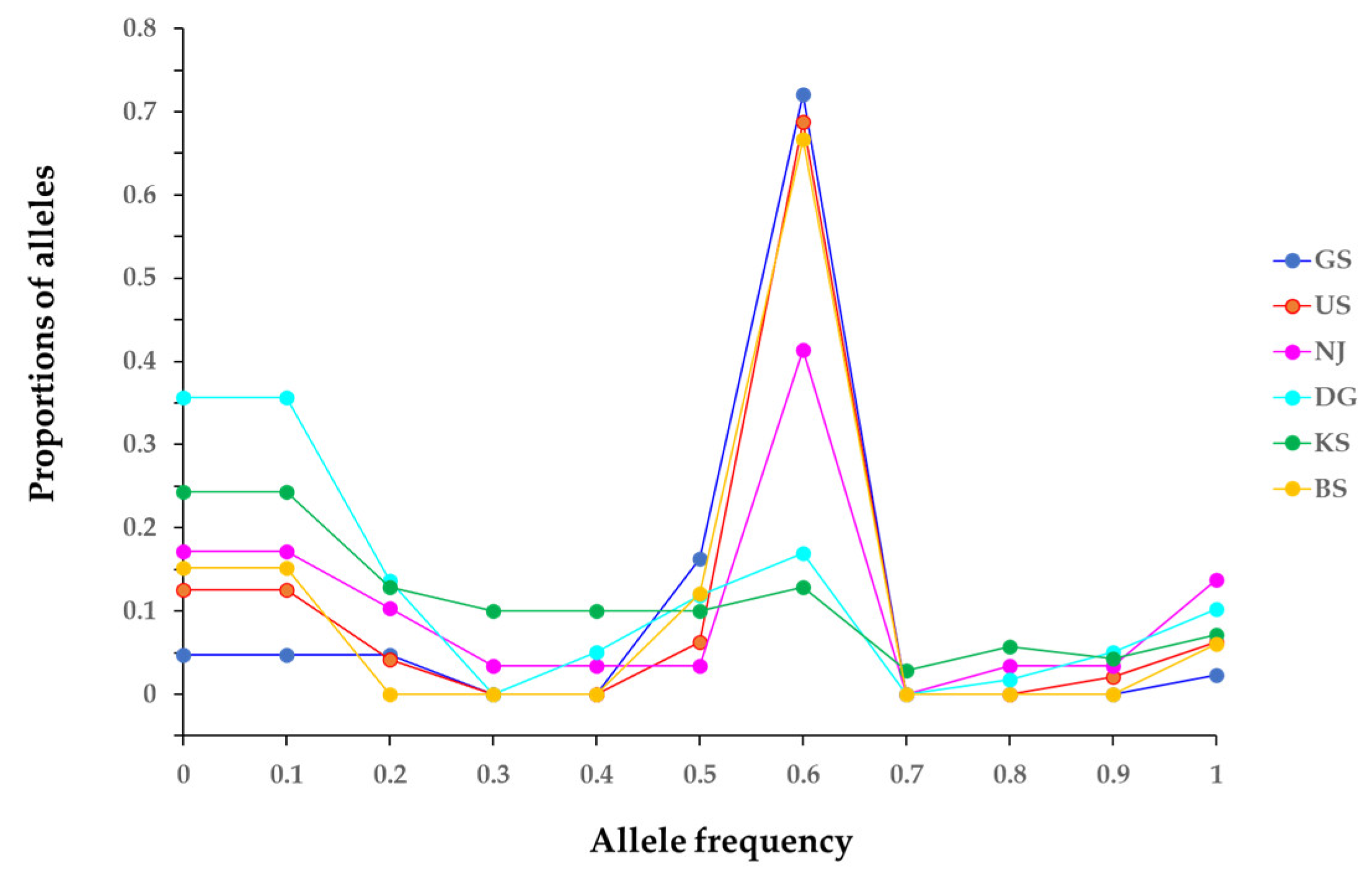
| GeneBank Accession No. | Locus | PIC | A | Ae/L | Unique Allele | Ho | He | FIS | FST | |
|---|---|---|---|---|---|---|---|---|---|---|
| Common | Rare | |||||||||
| PV231646 | MyspMS01 | 0.409 | 4 | 1.96 | 2 | 0 | 0.250 | 0.449 | −0.418 | 0.650 |
| PV231647 | MyspMS02 | 0.367 | 7 | 1.88 | 3 | 1 | 0.433 | 0.392 | −0.526 | 0.314 |
| PV231648 | MyspMS05 | 0.592 | 4 | 2.75 | 0 | 0 | 0.225 | 0.652 | −0.021 | 0.702 |
| PV231649 | MyspMS08 | 0.548 | 7 | 3.73 | 2 | 0 | 0.292 | 0.588 | 0.289 | 0.341 |
| PV231650 | MyspMS10 | 0.673 | 9 | 2.77 | 4 | 1 | 0.392 | 0.708 | −0.213 | 0.589 |
| PV231651 | MyspMS11 | 0.658 | 5 | 3.66 | 1 | 0 | 0.492 | 0.709 | −0.613 | 0.614 |
| PV231652 | MyspMS12 | 0.381 | 3 | 1.96 | 0 | 1 | 0.008 | 0.504 | 0 | 0.986 |
| PV231653 | MyspMS17 | 0.555 | 3 | 2.63 | 0 | 0 | 1.000 | 0.625 | −1 | 0.231 |
| PV231654 | MyspMS18 | 0.644 | 5 | 2.90 | 3 | 0 | 0.333 | 0.694 | −1 | 0.792 |
| PV231655 | MyspMS19 | 0.141 | 2 | 1.13 | 1 | 0 | 0.167 | 0.153 | −1 | 0.500 |
| PV231656 | MyspMS22 | 0.590 | 4 | 2.57 | 1 | 0 | 0.258 | 0.663 | −0.480 | 0.770 |
| PV231657 | MyspMS25 | 0.569 | 4 | 3.02 | 0 | 0 | 0.492 | 0.640 | −0.184 | 0.394 |
| PV231658 | MyspMS26 | 0.185 | 3 | 1.30 | 2 | 0 | 0.217 | 0.198 | −0.709 | 0.401 |
| PV231659 | MyspMS27 | 0.486 | 6 | 2.25 | 0 | 2 | 0.658 | 0.538 | −0.617 | 0.278 |
| PV231660 | MyspMS28 | 0.272 | 3 | 1.34 | 2 | 0 | 0.333 | 0.292 | −1 | 0.474 |
| PV231661 | MyspMS30 | 0.820 | 8 | 5.57 | 3 | 0 | 0.725 | 0.840 | −0.686 | 0.534 |
| PV231662 | MyspMS31 | 0.387 | 3 | 2.02 | 1 | 0 | 0.342 | 0.508 | −0.783 | 0.665 |
| PV231663 | MyspMS34 | 0.494 | 6 | 1.67 | 4 | 1 | 0.192 | 0.530 | −0.776 | 0.825 |
| PV231664 | MyspMS35 | 0.725 | 6 | 4.46 | 1 | 0 | 0.700 | 0.763 | −0.565 | 0.459 |
| PV231665 | MyspMS41 | 0.728 | 5 | 3.46 | 2 | 0 | 0.492 | 0.766 | −0.499 | 0.616 |
| PV231666 | MyspMS42 | 0.828 | 7 | 5.60 | 4 | 0 | 0.625 | 0.847 | −0.87 | 0.648 |
| PV231667 | MyspMS51 | 0.484 | 4 | 2.29 | 0 | 0 | 0.625 | 0.521 | −0.713 | 0.339 |
| PV231668 | MyspMS53 | 0.617 | 6 | 2.97 | 3 | 0 | 0.600 | 0.676 | −0.593 | 0.488 |
| PV231669 | MyspMS56 | 0.247 | 2 | 1.46 | 0 | 0 | 0.000 | 0.289 | 1 | 0.951 |
| PV231670 | MyspMS72 | 0.592 | 4 | 3.00 | 1 | 0 | 0.775 | 0.648 | −0.736 | 0.351 |
| PV231671 | MyspMS73 | 0.691 | 6 | 3.46 | 2 | 1 | 0.483 | 0.735 | −0.811 | 0.678 |
| PV231672 | MyspMS81 | 0.577 | 5 | 2.15 | 3 | 0 | 0.833 | 0.611 | −0.927 | 0.332 |
| PV231673 | MyspMS87 | 0.566 | 7 | 2.29 | 2 | 2 | 0.558 | 0.614 | −0.785 | 0.536 |
| PV231674 | MyspMS98 | 0.645 | 6 | 2.17 | 4 | 1 | 0.175 | 0.682 | −0.909 | 0.885 |
| PV231675 | MyspMS99 | 0.441 | 4 | 2.04 | 1 | 0 | 0.608 | 0.478 | −0.823 | 0.342 |
| Mean | 0.530 | 4.9 | 2.68 | 1.73 | 0.33 | 0.443 | 0.577 | −0.566 | 0.556 | |
| Overall | 0.554 | 148 | 80.46 | 52 | 10 | 0.443 | 0.577 | −0.610 | 0.569 | |
| Population | N | Genetic | Clonal | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P (0.95) | A | Ae/L | Ho | He | G(R) | G/N | D | E | ||
| GS | 20 | 66.7 | 53 | 1.67 | 0.618 | 0.327 | 11 (2–9) | 0.550 | 0.763 | 0.839 |
| US | 20 | 70.0 | 56 | 1.64 | 0.608 | 0.324 | 8 (2–12) | 0.400 | 0.567 | 0.648 |
| NJ | 20 | 40.0 | 46 | 1.32 | 0.220 | 0.157 | 12 (2–8) | 0.600 | 0.901 | 0.982 |
| DG | 20 | 66.7 | 69 | 1.52 | 0.352 | 0.252 | 12 (2–8) | 0.600 | 0.948 | 1.000 |
| KS | 20 | 76.7 | 74 | 1.75 | 0.427 | 0.348 | 20 (0) | 1.000 | 1.000 | 1.000 |
| BS | 20 | 50.0 | 48 | 1.45 | 0.432 | 0.225 | 4 (2–17) | 0.200 | 0.284 | 0.379 |
| Mean | 20 | 61.7 | 57.7 | 1.56 | 0.443 | 0.272 | 11.2 | 0.558 | 0.752 | 0.818 |
| Total | 120 | 100 | 148 | 2.80 | 0.443 | 0.577 | 67 (53) | 0.558 | 0.959 | 0.987 |
| Population | He | IAM | SMM | TPM | FIS |
|---|---|---|---|---|---|
| GS | 0.327 | 0.250 *** | 0.278 *** | 0.299 *** | −0.888 |
| US | 0.324 | 0.256 *** | 0.285 *** | 0.306 *** | −0.870 |
| NJ | 0.157 | 0.261 * | 0.289 | 0.314 | −0.382 |
| DG | 0.252 | 0.353 | 0.396 | 0.431 | −0.372 |
| KS | 0.348 | 0.331 * | 0.369 | 0.406 | −0.200 |
| BS | 0.225 | 0.258 *** | 0.284 *** | 0.308 *** | −0.911 |
| Locus | GS | US | NJ | DG | KS | BS | |
|---|---|---|---|---|---|---|---|
| MyspMS01 | −0.570 *** | - | −0.027 | - | −0.267 | - | |
| MyspMS02 | −0.498 ** | −0.027 *** | −1.000 *** | −0.027 | −0.200 | - | |
| MyspMS05 | −0.727 ** | - | - | 1.000 | 0.220 | - | |
| MyspMS08 | - | 0.714 | 0.861 | −0.376 ** | −0.016 | 1.000 | |
| MyspMS10 | - | −0.905 *** | −1.000 *** | 0.411 | 0.835 | 1.000 | |
| MyspMS11 | −1.000 *** | −1.000 *** | - | 0.645 | −0.553 ** | - | |
| MyspMS12 | - | - | - | - | 0.000 | - | |
| MyspMS17 | −1.000 *** | −1.000 *** | −1.000 *** | −1.000 *** | −1.000 *** | −1.000 *** | |
| MyspMS18 | −1.000 *** | −1.000 *** | - | - | - | - | |
| MyspMS19 | - | −1.000 *** | - | - | - | - | |
| MyspMS22 | - | −1.000 *** | - | 0.479 | −0.267 | - | |
| MyspMS25 | −1.000 *** | 1.000 | 1.000 | 0.719 | −0.615 ** | −0.905 *** | |
| MyspMS26 | −1.000 *** | - | - | - | −0.152 | - | |
| MyspMS27 | −1.000 *** | −1.000 *** | - | 0.791 | −0.376 * | −0.818 *** | |
| MyspMS28 | −1.000 *** | −1.000 *** | - | - | - | - | |
| MyspMS30 | −1.000 *** | −1.000 *** | - | −0.905 *** | 0.370 | −1.000 *** | |
| MyspMS31 | 1.000 | −1.000 *** | - | - | 0.000 | −1.000 *** | |
| MyspMS34 | - | - | −0.027 | - | 0.000 | −1.000 *** | |
| MyspMS35 | −1.000 *** | −1.000 *** | - | −0.520 *** | 0.493 | −1.000 *** | |
| MyspMS41 | - | −0.818 *** | 0.782 | −0.744 *** | −0.473 ** | - | |
| MyspMS42 | - | - | −1.000 *** | −1.000 *** | −0.484 * | −1.000 *** | |
| MyspMS51 | −1.000 *** | −1.000 *** | −0.056 | −1.000 ** | −0.100 | - | |
| MyspMS53 | −0.900 *** | - | - | −0.520 | −0.122 | −1.000 *** | |
| MyspMS56 | - | - | - | - | 1.000 | - | |
| MyspMS72 | −0.727 ** | −1.000 *** | - | −0.510 | −0.524 ** | −1.000 *** | |
| MyspMS73 | - | −1.000 *** | - | −0.027 *** | −0.587 ** | −1.000 *** | |
| MyspMS81 | −1.000 *** | −1.000 *** | −1.000 *** | −0.810 | −0.027 | −1.000 *** | Overall |
| MyspMS87 | −1.000 *** | −1.000 *** | 0.000 | −0.027 | −0.092 | −0.900 *** | Nm = 0.190 |
| MyspMS98 | −1.000 *** | 0.000 | - | - | - | - | FIS = −0.610 |
| MyspMS99 | −1.000 *** | −1.000 *** | −1.000 *** | −0.027 | −0.357 | - | FIT = 0.305 |
| All | −0.888 *** | −0.870 *** | −0.382 *** | −0.372 *** | −0.200 *** | −0.911 *** | FST = 0.569 |
| Pop. | GS | US | NJ | DG | KS | BS |
|---|---|---|---|---|---|---|
| GS | - | 0.219 | 0.167 | 0.161 | 0.229 | 0.189 |
| US | 0.533 | - | 0.134 | 0.111 | 0.153 | 0.110 |
| NJ | 0.600 | 0.652 | - | 0.229 | 0.315 | 0.175 |
| DG | 0.608 | 0.692 | 0.522 | - | 1.300 | 0.169 |
| KS | 0.521 | 0.620 | 0.442 | 0.161 | - | 0.215 |
| BS | 0.569 | 0.694 | 0.588 | 0.597 | 0.538 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, E.-H.; Kim, K.-R.; Lee, M.-H.; Goh, J.; Yu, J.-N. Patterns of Genetic and Clonal Diversity in Myriophyllum spicatum in Streams and Reservoirs of Republic of Korea. Plants 2025, 14, 2648. https://doi.org/10.3390/plants14172648
Kim E-H, Kim K-R, Lee M-H, Goh J, Yu J-N. Patterns of Genetic and Clonal Diversity in Myriophyllum spicatum in Streams and Reservoirs of Republic of Korea. Plants. 2025; 14(17):2648. https://doi.org/10.3390/plants14172648
Chicago/Turabian StyleKim, Eun-Hye, Kang-Rae Kim, Mi-Hwa Lee, Jaeduk Goh, and Jeong-Nam Yu. 2025. "Patterns of Genetic and Clonal Diversity in Myriophyllum spicatum in Streams and Reservoirs of Republic of Korea" Plants 14, no. 17: 2648. https://doi.org/10.3390/plants14172648
APA StyleKim, E.-H., Kim, K.-R., Lee, M.-H., Goh, J., & Yu, J.-N. (2025). Patterns of Genetic and Clonal Diversity in Myriophyllum spicatum in Streams and Reservoirs of Republic of Korea. Plants, 14(17), 2648. https://doi.org/10.3390/plants14172648







