GhSTZ-Mediated Suppression of Metabolic–Immune Coordination Compromises Cotton Defense Against Verticillium Wilt
Abstract
1. Introduction
2. Results
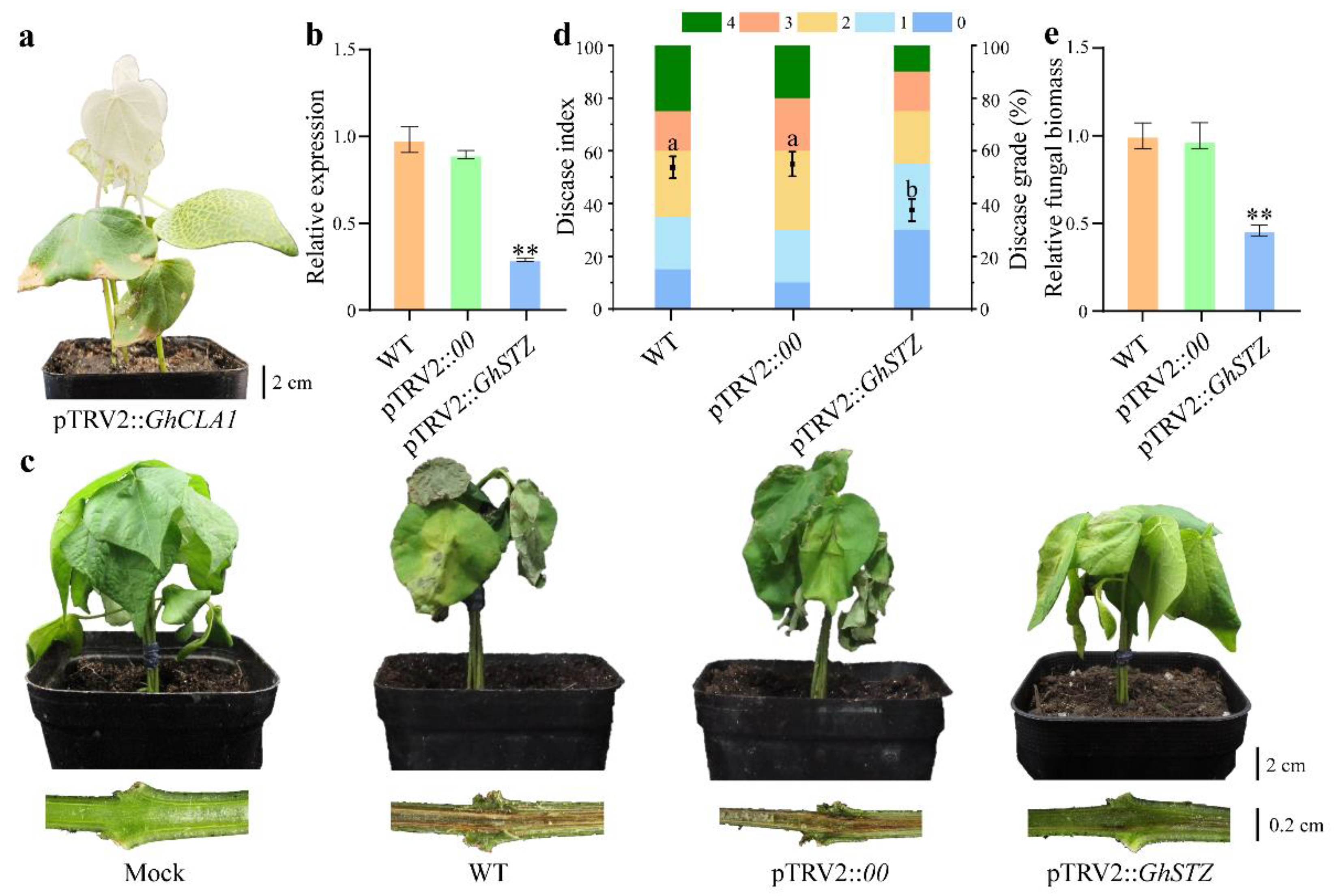
2.1. GhSTZ Serves a Critical Role in Cotton Resistance to VW
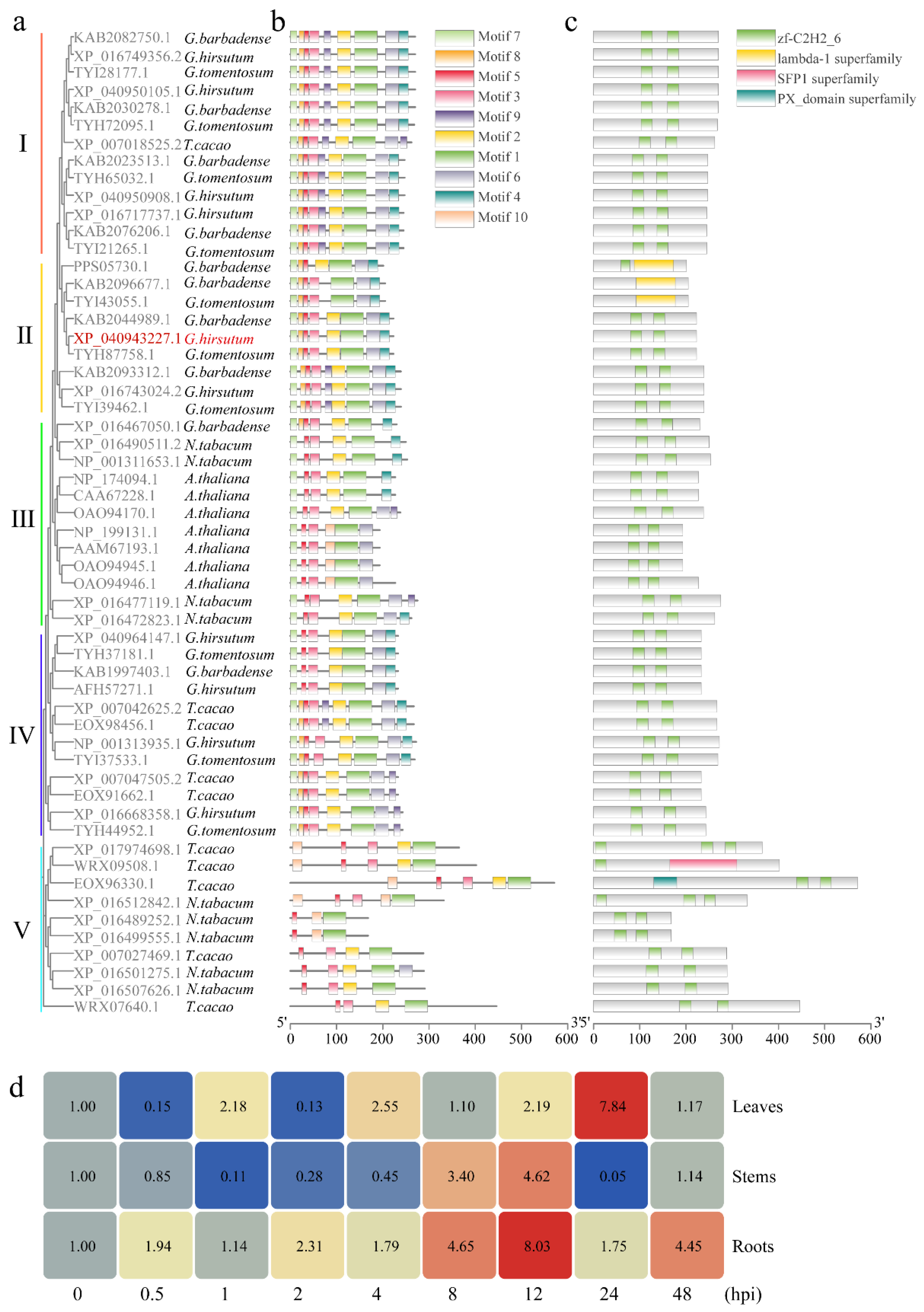
2.2. Bioinformatic Characterization and Expression Profiling of GhSTZ
2.3. Silencing GhSTZ Enhances Cotton Resistance to VW
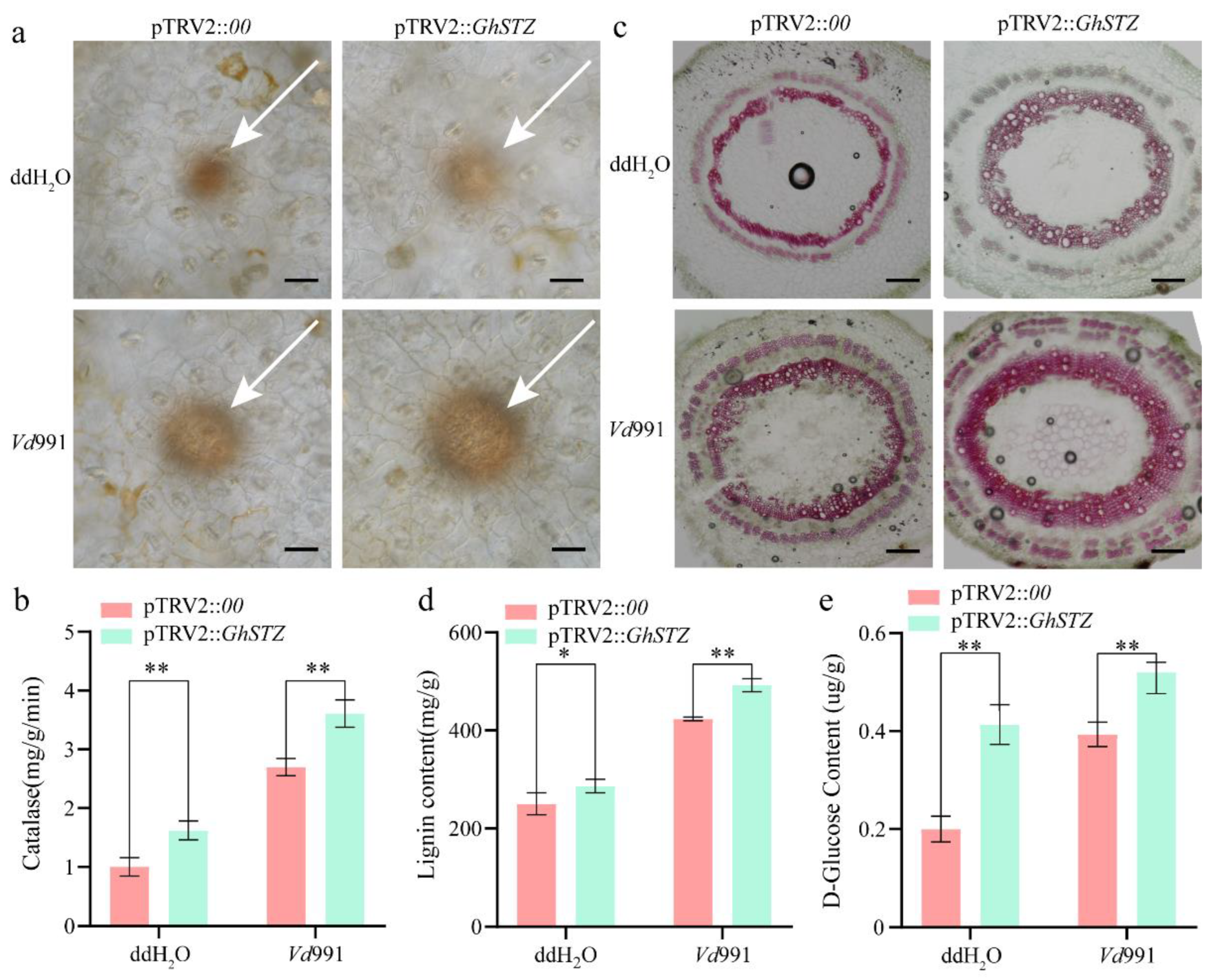
2.4. Silencing GhSTZ Promotes Accumulation of Defense Compounds
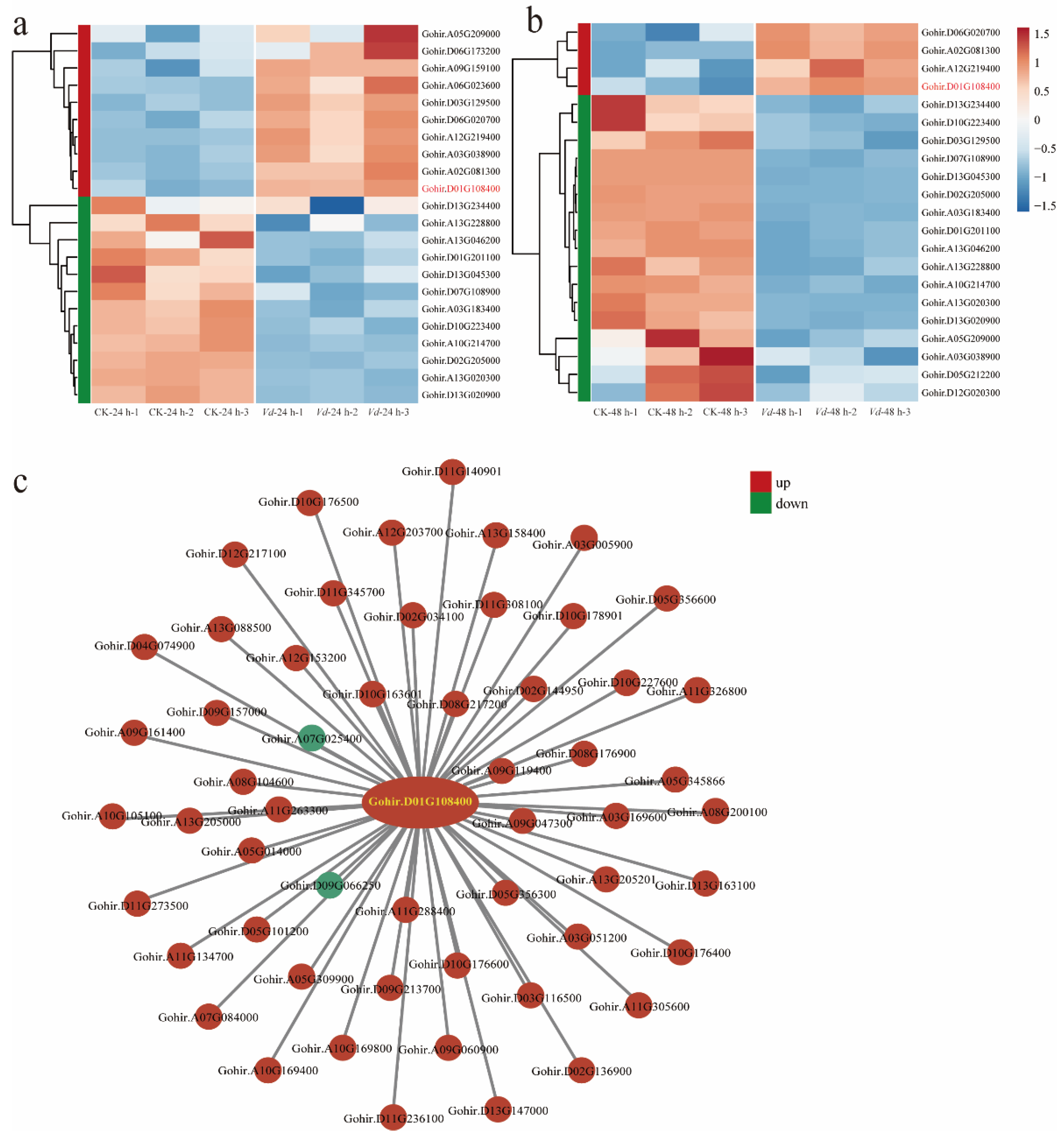
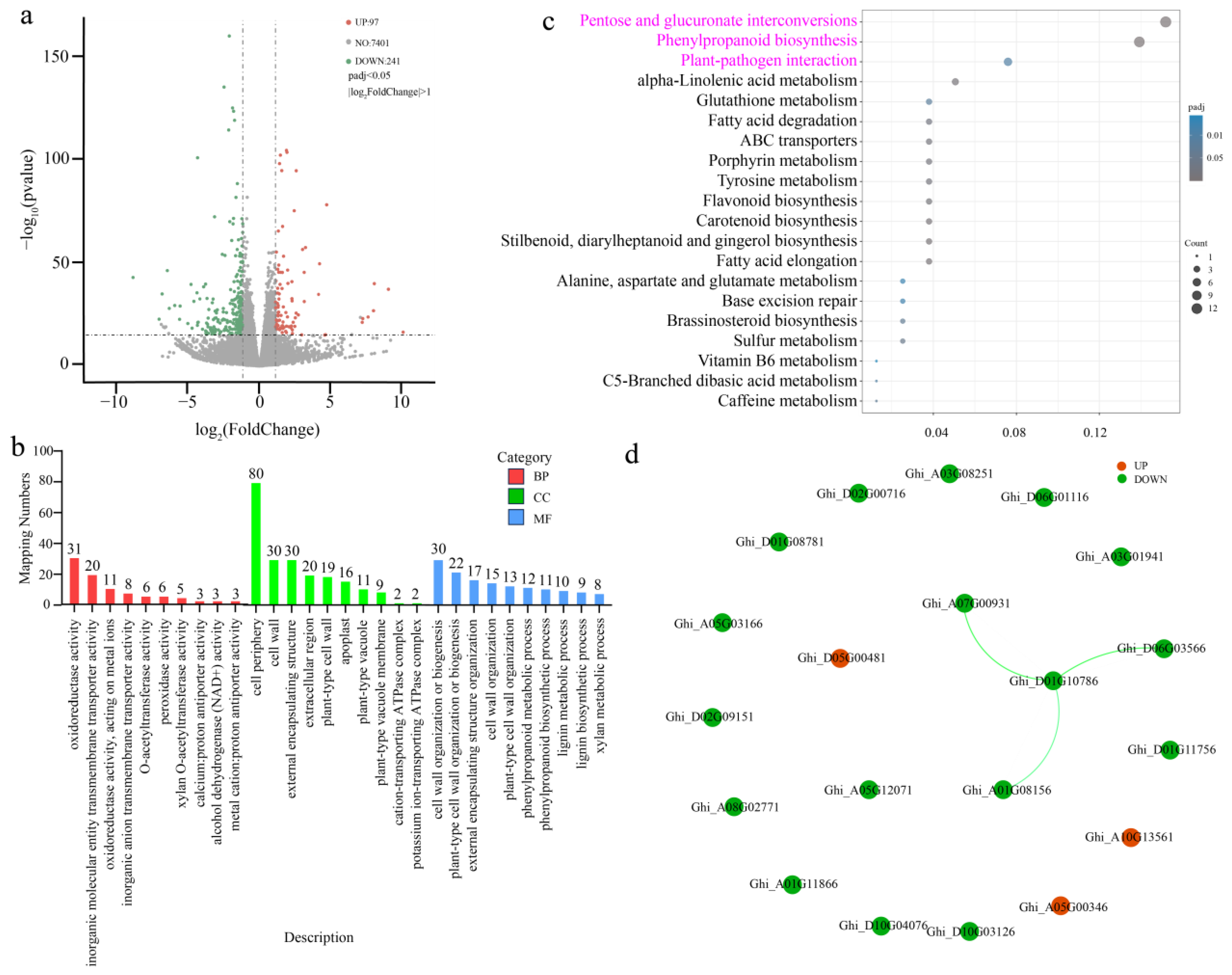
2.5. Differential Gene Expression Analysis and Validation
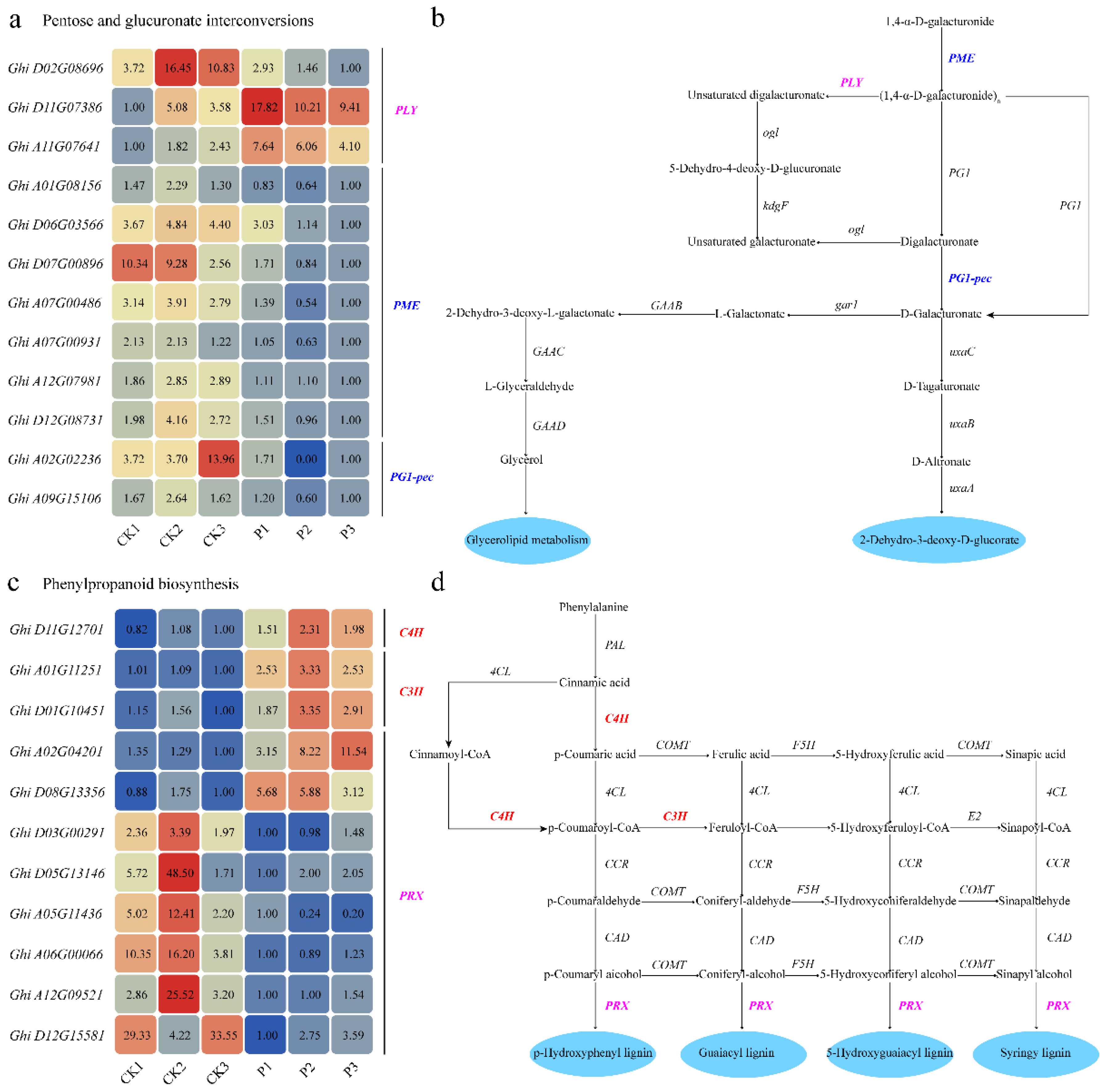
2.6. GhSTZ Regulates Expression of Key Genes in Defense-Related Metabolic Pathways
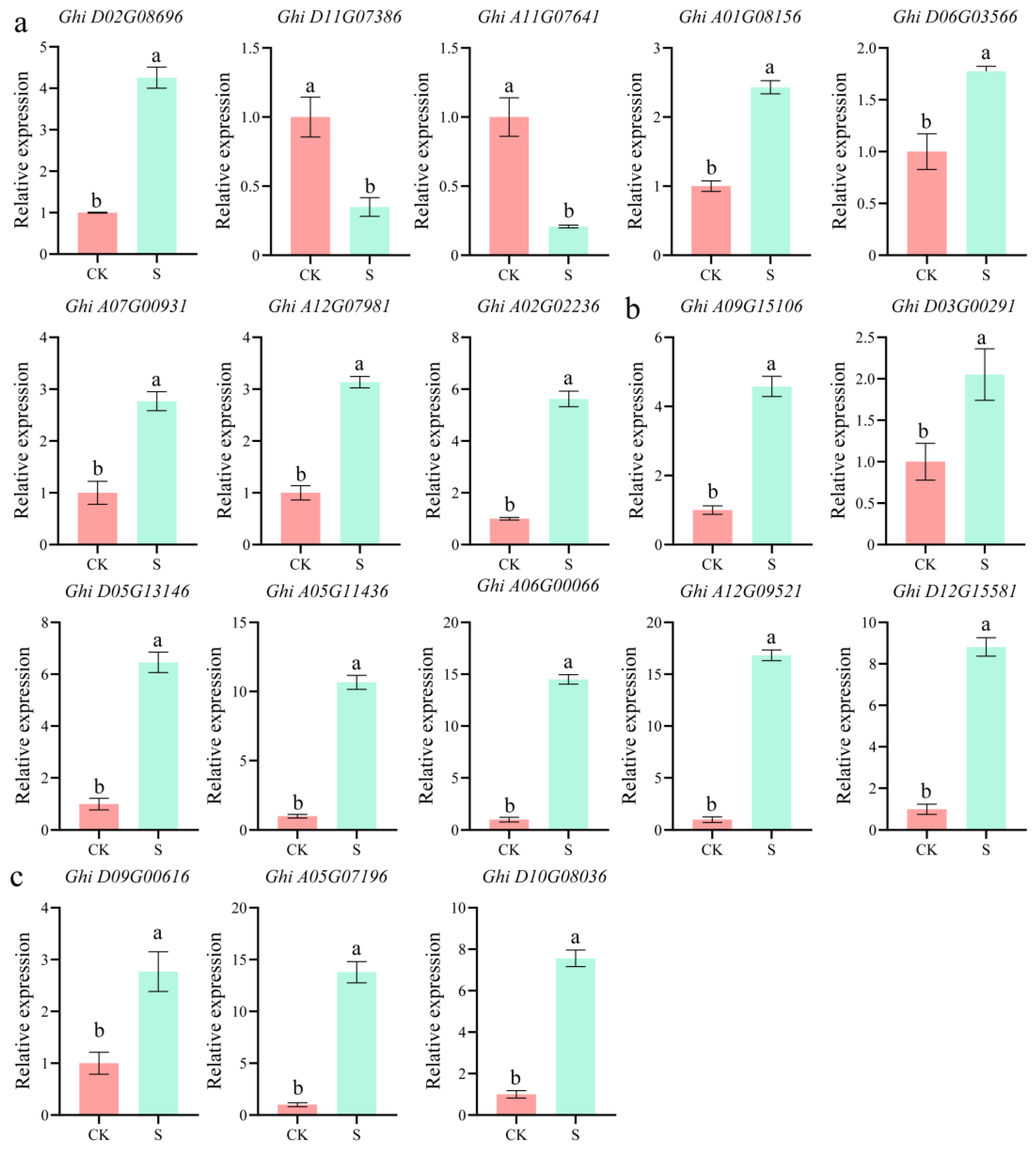
2.7. Validation of Transcriptome Data via RT-qPCR
3. Discussion
4. Materials and Methods
4.1. Plant and Fungal Materials
4.2. Identification of Key Genes and Co-Expression Network Analysis
4.3. Bioinformatics Analysis
4.4. Gene Expression Analysis
4.5. VIGS in Cotton
4.6. Disease Resistance Phenotyping and Pathogen Quantification
4.7. Physiological and Biochemical Parameter Assays
4.8. Transcriptome Sequencing and Analysis
4.9. RT-qPCR Validation of Disease Resistance-Related Genes
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y.J. Flagellin FLiS improves the resistance of cotton to Verticillium wilt through the signaling pathways of salicylic acid and jasmonic acid. Front. Plant Sci. 2025, 16, 1595529. [Google Scholar] [CrossRef]
- Hu, Z.J.; Yuan, J.J.; Zou, R.; Wang, Y.L.; Peng, X.; Yang, X.Y.; Xie, C.J. Identification and Functional Analysis of BAG Gene Family Contributing to Verticillium Wilt Resistance in Upland Cotton. Plant Sci. Int. J. Exp. Plant Biol. 2025, 356, 112501. [Google Scholar] [CrossRef]
- Zhao, Z.Q.; Zhu, Z.C.; Jiao, Y.; Zhang, G.L. Pan-genome analysis of GT64 gene family and expression response to Verticillium wilt in cotton. BMC Plant Biol. 2024, 24, 893. [Google Scholar] [CrossRef]
- Wang, W.Y.; Sun, Y.D.; Han, L.B.; Su, L.; Xia, G.X.; Wang, H.Y. Overexpression of GhPFN2 enhances protection against Verticillium dahliae invasion in cotton. Sci. China Life Sci. 2017, 60, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Mo, S.J.; Zhang, Y.; Wang, X.F.; Yang, J.; Sun, Z.W.; Zhang, D.M.; Chen, B.; Wang, G.N.; Ke, H.F.; Liu, Z.W.; et al. Cotton GhSSI2 isoforms from the stearoyl acyl carrier protein fatty acid desaturase family regulate Verticillium wilt resistance. Mol. Plant Pathol. 2021, 22, 1041–1056. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, B.; Sun, Z.W.; Liu, Z.W.; Cui, Y.R.; Ke, H.F.; Wang, Z.C.; Wu, L.Q.; Zhang, G.Y.; Wang, G.N.; et al. A large-scale genomic association analysis identifies a fragment in Dt11 chromosome conferring cotton Verticillium wilt resistance. Plant Biotechnol. J. 2021, 19, 2126–2138. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Cao, H.H.; Wang, J.H.; Zhang, H.B. Recent advances in functional assays of WRKY transcription factors in plant immunity against pathogens. Front. Plant Sci. 2025, 15, 1517595. [Google Scholar] [CrossRef]
- Dai, X.Y.; Wang, Z.W.; Bao, Y.F.; Jia, C.C.; Bai, F.F.; Hasi, A.; Che, G. Identification and functional characterization of the C2H2 ZFP transcription factor CmSUP7 in regulating melon plant growth and fruit development. Plant Physiol. Biochem. 2025, 220, 109513. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Xie, R.; Hu, Y.; Du, L.; Wang, F.; Zhao, X.; Huang, Y.; Chen, X.; Hao, M.; Xu, Q.; et al. A C2H2-type zinc finger protein TaZFP8-5B negatively regulates disease resistance. BMC Plant Biol. 2024, 24, 1116. [Google Scholar] [CrossRef]
- Evans, S.E.; Franks, A.E.; Bergman, M.E.; Sethna, N.S.; Currie, M.A.; Phillips, M.A. Plastid ancestors lacked a complete Entner-Doudoroff pathway, limiting plants to glycolysis and the pentose phosphate pathway. Nat. Commun. 2024, 15, 1102. [Google Scholar] [CrossRef]
- Hu, H.C.; Liu, Y.H.; He, B.B.; Chen, X.; Ma, L.; Luo, Y.L.; Fei, X.T.; Wei, A.Z. Integrative physiological, transcriptome, and metabolome analysis uncovers the drought responses of two Zanthoxylum bungeanum cultivars. Ind. Crops Prod. 2022, 189, 115812. [Google Scholar] [CrossRef]
- Tao, S.K.; Zhu, Y.; Pan, Y.G.; Zhang, Z.K.; Huang, L.J. Enhancement of respiratory metabolism of the pentose phosphate pathway (PPP) strengthens the chilling tolerance of postharvest papaya fruit stored at 1 °C. Postharvest Biol. Technol. 2022, 191, 111988. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Z.-X.; Tian, H.-Y.; Zeng, Y.-L.; Xue, H.; Mao, W.-T.; Zhang, L.-Y.; Chen, J.-N.; Lu, X.; Zhu, Y.; et al. The miR172a-SNB module orchestrates induced and adult resistance to multiple diseases via MYB30-mediated lignin accumulation in rice. Mol. Plant 2024, 18, 59–75. [Google Scholar] [CrossRef]
- Wang, Y.J.; Qin, J.; Wei, M.M.; Liao, X.W.; Shang, W.J.; Chen, J.Y.; Subbarao, K.V.; Hu, X.P. Verticillium dahliae Elicitor VdSP8 Enhances Disease Resistance Through Increasing Lignin Biosynthesis in Cotton. Plant Cell Environ. 2024, 48, 728–745. [Google Scholar] [CrossRef] [PubMed]
- Ranade, S.S.; Seipel, G.; Gorzsás, A.; GarcíaGil, M.R. Enhanced lignin synthesis and ecotypic variation in defense-related gene expression in response to shade in Norway spruce. Plant Cell Environ. 2022, 45, 2671–2681. [Google Scholar] [CrossRef]
- Ma, Q.H. Lignin Biosynthesis and Its Diversified Roles in Disease Resistance. Genes 2024, 15, 295. [Google Scholar] [CrossRef]
- Luo, X.C.; Hu, Z.W.; Chu, L.Y.; Li, J.P.; Tang, Z.-R.; Sun, X.X.; An, H.L.; Wan, P.; Wang, X.P.; Yang, Y.Z.; et al. GhRac9 improves cotton resistance to Verticillium dahliae via regulating ROS production and lignin content. Physiol. Plant. 2025, 177, e70091. [Google Scholar] [CrossRef]
- Yi, F.F.; Li, Y.Z.; Song, A.S.; Shi, X.Y.; Hu, S.C.; Wu, S.; Shao, L.L.; Chu, Z.Y.; Xu, K.; Li, L.L.; et al. Positive roles of the Ca2+ sensors GbCML45 and GbCML50 in improving cotton Verticillium wilt resistance. Mol. Plant Pathol. 2024, 25, e13483. [Google Scholar] [CrossRef]
- Pan, X.; Xu, S.P.; Cao, G.H.; Chen, S.P.; Zhang, T.; Yang, B.B.; Zhou, G.H.; Yang, X. A novel peptide encoded by a rice circular RNA confers broad-spectrum disease resistance in rice plants. New Phytol. 2025, 246, 689–701. [Google Scholar] [CrossRef]
- Zhang, L.; Xi, D.M.; Luo, L.; Meng, F.; Li, Y.Z.; Wu, C.A.; Guo, X.Q. Cotton GhMPK2 is involved in multiple signaling pathways and mediates defense responses to pathogen infection and oxidative stress. FEBS J. 2011, 278, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhao, Y.; Zhang, Y.Q.; Niu, L.H.; Li, W.Y.; Lu, W.Q.; Li, J.F.; Schäfer, P.; Meng, Y.L.; Shan, W.X. A mitochondrial RNA processing protein mediates plant immunity to a broad spectrum of pathogens by modulating the mitochondrial oxidative burst. Plant Cell 2022, 34, 2343–2363. [Google Scholar] [CrossRef]
- Zhao, N.; Guo, A.H.; Wang, W.R.; Li, B.; Wang, M.; Zhou, Z.X.; Jiang, K.Y.; Aierxi, A.; Wang, B.L.; Adjibolosoo, D.; et al. GbPP2C80 Interacts with GbWAKL14 to Negatively Co-Regulate Resistance to Fusarium and Verticillium wilt via MPK3 and ROS Signaling in Sea Island Cotton. Adv. Sci. 2024, 11, e2309785. [Google Scholar] [CrossRef]
- Zhao, Y.N.; Yu, H.; Zhou, J.M.; Smith, S.M.; Li, J.Y. Malate Circulation: Linking Chloroplast Metabolism to Mitochondrial ROS. Trends Plant Sci. 2020, 25, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, A.R.; Luo, Q.; Hu, Z.J.; Ma, Q.M.; Li, Y.M.; Lin, T.; Liang, X.; Yu, J.Q.; Foyer, C.H.; et al. Glucose sensing by regulator of G protein signaling 1 (RGS1) plays a crucial role in coordinating defense in response to environmental variation in tomato. New Phytol. 2022, 236, 561–575. [Google Scholar] [CrossRef]
- Zhang, M.L.; Wei, F.; Guo, K.; Hu, Z.; Li, Y.Y.; Xie, G.S.; Wang, Y.T.; Cai, X.W.; Peng, L.C.; Wang, L.Q. A Novel FC116/BC10 Mutation Distinctively Causes Alteration in the Expression of the Genes for Cell Wall Polymer Synthesis in Rice. Front. Plant Sci. 2016, 7, 1366. [Google Scholar] [CrossRef]
- Zhang, B.C.; Liu, X.L.; Qian, Q.; Liu, L.F.; Dong, G.J.; Xiong, G.Y.; Zeng, D.L.; Zhou, Y.H. Golgi nucleotide sugar transporter modulates cell wall biosynthesis and plant growth in rice. Proc. Natl. Acad. Sci. USA 2011, 108, 5110–5115. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Si, Z.F.; Wang, G.P.; Shi, Z.L.; Chen, J.W.; Qi, G.A.; Jin, S.K.; Han, Z.G.; Gao, W.H.; Tian, Y.; et al. Genomic Insights into the Genetic Basis of Cotton Breeding in China. Mol. Plant 2023, 16, 662–677. [Google Scholar] [CrossRef]
- Liu, H.Q.; Zhang, W.S.; Zeng, J.G.; Zheng, Q.H.; Guo, Z.; Ruan, C.; Li, W.X.; Wang, G.L.; Wang, X.Y.; Guo, W.Z. A Golgi vesicle-membrane-localized cytochrome B561 regulates ascorbic acid regeneration and confers Verticillium wilt resistance in cotton. Plant J. 2025, 121, e17162. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Cui, L.F.; Liu, R.B.; Feng, Z.L.; Feng, H.J.; Zhou, J.L.; Zhao, L.H.; Wei, F.; Zhu, H.Q. In the coevolution of cotton and pathogenic fungi, resistant cotton varieties lead to an escalation in the virulence of Verticillium dahliae. Ecotoxicol. Environ. Saf. 2025, 290, 117730. [Google Scholar] [CrossRef]
- Li, C.Z.; Zhang, G.S.; Cheng, G.F.; Wang, Q. Phenylalanine Ammonia-Lyase GhPAL9 Confers Resistance to Verticillium Wilt in Cotton. Int. J. Mol. Sci. 2025, 26, 4983. [Google Scholar] [CrossRef]
- Wang, L.; Yang, Y.L.; Qin, J.H.; Ma, Q.F.; Qiao, K.K.; Fan, S.L.; Qu, Y.Y. Integrative GWAS and transcriptomics reveal GhAMT2 as a key regulator of cotton resistance to Verticillium wilt. Front. Plant Sci. 2025, 16, 1563466. [Google Scholar] [CrossRef]
- Chai, Q.C.; Zheng, M.N.; Li, Y.L.; Gao, M.W.; Wang, Y.C.; Wang, X.L.; Zhang, C.; Jiang, H.; Chen, Y.; Wang, J.B.; et al. GhWRKY75 positively regulates GhPR6-5b via binding to a W-box TTGAC (C/T) to orchestrate cotton resistance to Verticillium dahliae. J. Integr. Agric. 2024, 23, 3343–3357. [Google Scholar] [CrossRef]
- Zhu, Y.T.; Hu, X.Q.; Wang, P.; Wang, H.W.; Ge, X.Y.; Li, F.G.; Hou, Y.X. GhODO1, an R2R3-type MYB transcription factor, positively regulates cotton resistance to Verticillium dahliae via the lignin biosynthesis and jasmonic acid signaling pathway. Int. J. Biol. Macromol. 2022, 201, 580–591. [Google Scholar] [CrossRef]
- Sánchez-López, Á.M.; Bahaji, A.; Gámez-Arcas, S.; De Diego, N.; Vrobel, O.; Tarkowski, P.; Baroja-Fernández, E.; Muñoz, F.J.; Almagro, G.; Seguí-Simarro, J.M.; et al. PGI1-mediated vascular oxidative pentose phosphate pathway modulates photosynthesis via long-distance cytokinin signaling. Plant Physiol. Biochem. 2024, 209, 108520. [Google Scholar] [CrossRef]
- Wei, Q.H.; Zhang, F.; Sun, F.S.; Luo, Q.C.; Wang, R.B.; Hu, R.; Chen, M.J.; Chang, J.L.; Yang, G.X.; He, G.Y. A wheat MYB transcriptional repressor TaMyb1D regulates phenylpropanoid metabolism and enhances tolerance to drought and oxidative stresses in transgenic tobacco plants. Plant Sci. 2017, 265, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, K.; Qu, X.Y.; Kretschmer, M.; Kronstad, J.W. The phosphate language of fungi. Trends Microbiol. 2021, 30, 338–349. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.P.; Zhao, M.; Gao, X.Y.; Shan, Y.; Li, F.J.; Tian, X.L.; Li, Z.H. GhWRKY1bD improves drought tolerance by co-regulation of ABA, ROS, and proline homeostasis in cotton (Gossypium hirsutum). Ind. Crops Prod. 2024, 220, 119179. [Google Scholar] [CrossRef]
- Sun, Y.R.; Tian, Z.L.; Zuo, D.Y.; Wang, Q.L.; Song, G.L. GhUBC10-2 mediates GhGSTU17 degradation to regulate salt tolerance in cotton (Gossypium hirsutum). Plant Cell Environ. 2024, 47, 1606–1624. [Google Scholar] [CrossRef]
- Mehdi, F.; Galani, S.; Wickramasinghe, K.P.; Zhao, P.f.; Lu, X.; Lin, X.Q.; Xu, C.H.; Liu, H.B.; Li, X.J.; Liu, X.L. Current perspectives on the regulatory mechanisms of sucrose accumulation in sugarcane. Heliyon 2024, 10, e27277. [Google Scholar] [CrossRef]
- Khan, Q.; Qin, Y.; Guo, D.J.; Chen, J.Y.; Zeng, X.P.; Mahmood, A.; Yang, L.T.; Liang, Q.; Song, X.P.; Xing, Y.X.; et al. Sucrose metabolism analysis in a high sucrose sugarcane mutant clone at a mature stage in contrast to low sucrose parental clone through the transcriptomic approach. Chem. Biol. Technol. Agric. 2023, 10, 39. [Google Scholar] [CrossRef]
- Wang, L.; Ruan, Y.L. New insights into roles of cell wall invertase in early seed development revealed by comprehensive spatial and temporal expression patterns of GhCWIN1 in cotton. Plant Physiol. 2012, 160, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Xie, Y.J.; Jiang, Y.; Nadeem, H.; Wang, Y.; Yang, N.; Zhu, H.Q.; Tang, C.M. GhTLP1, a thaumatin-like protein 1, improves Verticillium wilt resistance in cotton via JA, ABA and MAPK signaling pathway-plant pathways. Int. J. Biol. Macromol. 2023, 253, 127388. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.D.; Zhong, M.M.; Li, Y.B.; Zhang, R.H.; Su, L.; Xia, G.X.; Wang, H.Y. GhADF6-mediated actin reorganization is associated with defence against Verticillium dahliae infection in cotton. Mol. Plant Pathol. 2021, 22, 1656–1667. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, G.; Zhu, X.; Bi, Y.; Fernando, W.G.D.; Lv, X.; Lei, J.; Dai, P.; Li, Y. GhSTZ-Mediated Suppression of Metabolic–Immune Coordination Compromises Cotton Defense Against Verticillium Wilt. Plants 2025, 14, 2638. https://doi.org/10.3390/plants14172638
Zhang G, Zhu X, Bi Y, Fernando WGD, Lv X, Lei J, Dai P, Li Y. GhSTZ-Mediated Suppression of Metabolic–Immune Coordination Compromises Cotton Defense Against Verticillium Wilt. Plants. 2025; 14(17):2638. https://doi.org/10.3390/plants14172638
Chicago/Turabian StyleZhang, Guoshuai, Xinyu Zhu, Yanqing Bi, W. G. Dilantha Fernando, Xiaodi Lv, Jianfeng Lei, Peihong Dai, and Yue Li. 2025. "GhSTZ-Mediated Suppression of Metabolic–Immune Coordination Compromises Cotton Defense Against Verticillium Wilt" Plants 14, no. 17: 2638. https://doi.org/10.3390/plants14172638
APA StyleZhang, G., Zhu, X., Bi, Y., Fernando, W. G. D., Lv, X., Lei, J., Dai, P., & Li, Y. (2025). GhSTZ-Mediated Suppression of Metabolic–Immune Coordination Compromises Cotton Defense Against Verticillium Wilt. Plants, 14(17), 2638. https://doi.org/10.3390/plants14172638






