1. Introduction
The genus
Liriodendron, belonging to the family Magnoliaceae, comprises two extant species:
Liriodendron chinense, native to East Asia, and
Liriodendron tulipifera, found in eastern North America.
Liriodendron species are large deciduous trees that can grow up to 40 m tall and are characterized by their distinctive leaf shapes, showy flowers, and straight, upright trunks—traits that contribute to their ecological, ornamental, and timber value. The wood is lightweight, fine-textured, and naturally resistant to pests, while the trees themselves demonstrate strong environmental adaptability, including tolerance to air pollution [
1]. Owing to their ecological resilience and economic utility,
Liriodendron species hold significant potential for research on phylogeny, genetic diversity, and species conservation, as well as for breeding and practical applications in forestry and landscaping [
2].
The comprehensive collection, preservation, and assessment of
Liriodendron germplasm resources serve dual critical purposes of safeguarding genetic diversity and ecological functionality while establishing the essential foundation for breeding superior cultivars through contemporary forest tree improvement programs. Phenotypic characterization represents the most immediate and reliable methodology for germplasm evaluation, enabling thorough documentation of accession performance and revealing underlying genetic diversity and adaptive potential [
3]. Recent investigations have yielded systematic advances in understanding key phenotypic attributes of
L. tulipifera,
L. chinense, and their interspecific hybrids. Notably, Zong et al. identified three AP2/ERF transcription factors exhibiting shoot apical meristem-specific expression patterns in
Liriodendron through genome-wide analysis, potentially governing early leaf morphogenesis [
4]. Significant progress has also been made in flowering trait research, with Sheng et al. elucidating floral transition regulatory mechanisms via comparative transcriptomic profiling [
5]. Furthermore, Liu et al. characterized spatiotemporal expression dynamics among MADS-box transcription factors during floral development, providing mechanistic insights into floral architecture variation within the genus [
6]. These meticulous phenotypic analyses have significantly advanced our comprehension of phenotypic plasticity and adaptive evolutionary processes in
Liriodendron, while simultaneously informing practical applications in germplasm classification, conservation management, and genetic enhancement initiatives. Although many researchers have conducted extensive studies on phenotypic traits and genetic mechanisms in
Liriodendron [
7,
8,
9], the integrated evaluation systems combining phenotypic traits with SNP markers remains underdeveloped.
Evaluating and characterizing the genetic diversity of germplasm resources is essential for constructing core germplasm collections. To date, a variety of molecular markers have been utilized in plant genetic research, including conventional markers such as restriction fragment length polymorphism (RFLP), random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), inter-simple sequence repeat (ISSR), and simple sequence repeat (SSR), as well as sequencing-based markers such as single nucleotide polymorphisms (SNPs) and insertion/deletion polymorphisms (InDels) [
10]. Recent advances in high-throughput sequencing technologies have significantly enhanced association analyses between molecular markers and plant phenotypic traits, establishing this approach as a powerful methodology for elucidating genetic diversity and developing comprehensive germplasm resource maps. Representative studies demonstrate the effectiveness of this strategy. Wang et al. successfully identified multiple SCoT marker loci significantly associated with 12 ornamental traits through marker-trait association analysis of 65 chrysanthemum germplasm accessions [
11]. In parallel research, Donkpegan et al. performed genome-wide association analysis on 23 fruit quality traits across 116 sweet cherry germplasm resources, pinpointing SNP markers strongly correlated with critical agronomic characteristics including fruit size and firmness [
12]. These investigations collectively provide molecular-level insights into phenotypic expression patterns. In phenotypic–molecular association studies, SNPs have become the predominant marker class owing to their abundance, genome-wide coverage, and high information density [
13]. Their effectiveness has been demonstrated across diverse taxa, including
Acorus tatarinowii [
14],
Zea mays [
15], and
Dioscorea rotundata [
16], in which these markers have proved useful for assessing genetic diversity and constructing DNA fingerprints.
With the advancement of DNA molecular marker technology, the evaluation of germplasm resources in an increasing number of species has shifted from phenotypic characterization to high-resolution genotyping, providing a foundation for the development of standardized DNA fingerprinting systems. In recent years, SNP-based and SSR-based fingerprinting platforms have been established in most tree species [
17]. For example, Yan et al. [
18] analyzed the genetic diversity and genetic structure of 161 clonal lines of
Pinus koraiensis using SSR markers and successfully constructed a robust DNA fingerprinting system. Similar studies have also been conducted in
Ailanthus altissima [
19] and
Camellia sinensis [
20]. This technique has been widely applied across various taxa, including vegetables (e.g.,
Raphanus sativus [
21],
Brassica oleracea var.
botrytis [
22],
Ipomoea batatas [
23]) and fruits (e.g.,
Vaccinium corymbosum [
24],
Morus alba [
25],
Prunus avium [
26]). However, to date, there is no efficient and high-resolution molecular identification system available for
Liriodendron germplasm resources, which presents challenges for germplasm management, breeding, and intellectual property protection in this genus [
27]. Therefore, establishing a comprehensive DNA fingerprinting platform for
Liriodendron using high-throughput molecular markers has become a pressing research priority. As a fundamental tool for germplasm characterization, DNA fingerprinting enables the generation of unique molecular identifiers by detecting genomic variations with high specificity [
19]. Compared with traditional phenotypic assessments, this approach offers significant advantages in accuracy, reproducibility, and scalability. The development of SNP-based DNA fingerprints for
Liriodendron is thus of great significance for both theoretical research and practical applications in germplasm conservation and breeding programs.
This study utilized 297 Liriodendron germplasm accessions as experimental materials and performed systematic assessments of 34 phenotypic traits. Comprehensive analysis demonstrated considerable phenotypic variation in most evaluated traits, along with statistically significant inter-trait correlations. Based on high-throughput sequencing of 197 representative samples, a set of high-quality single-nucleotide polymorphism (SNP) markers was identified, from which core SNP markers were selected for downstream analysis. The resulting genome-wide SNP marker system enabled precise assessments of genetic diversity and genetic structure within the Liriodendron germplasm collection. Furthermore, a robust DNA fingerprinting platform with high discriminatory power was developed to support accurate resource authentication, digital archiving, and traceability. These methodological advances substantially improve the precision, efficiency, and reliability of Liriodendron germplasm conservation and utilization.
3. Discussion
Recent advances in forest genetic improvement have underscored the need for systematic conservation and utilization of rare tree germplasm resources. This paradigm shift reflects both ecological imperatives and the demands of modern breeding programs, particularly for relict species with narrow natural distributions [
29,
30,
31]. As a representative genus within the Magnoliaceae family,
Liriodendron possesses considerable ecological, ornamental, and economic value, rendering genetic diversity assessment and germplasm identification key research priorities. Genetic diversity, a fundamental indicator of a species’ adaptive capacity, is shaped by factors such as genetic drift, natural selection, and gene flow [
32]. To support the development of effective breeding strategies for
Liriodendron, we first evaluated phenotypic variation among 297 accessions. Phenotypic traits, which reflect morphological-level genetic diversity, were assessed using coefficients of variation (CV), with higher values indicating greater variability in germplasm resources [
33]. Analysis of key growth traits including annual mean diameter at breast height increment, annual height increment, and crown spread revealed consistently high coefficients of variation, demonstrating substantial genetic differentiation within the
Liriodendron genus. This pronounced phenotypic variation not only establishes critical selection criteria for superior germplasm identification but also underscores the remarkable phenotypic plasticity of
Liriodendron species in specific growth characteristics, thereby enhancing breeding potential. Notably, germplasm exhibiting greater crown dimensions shows particular suitability for landscape applications, while accessions with accelerated growth rates are ideally suited for timber plantation development [
34]. Of particular significance is the exceptional variation observed in stem form and branching architecture traits, which directly determine crown structure and wood properties [
35]. These findings provide valuable insights for optimizing silvicultural practices and informing strategic breeding programs for varietal improvement.
As a fundamental determinant of evolutionary resilience and adaptive capacity, genetic diversity provides an essential baseline for both germplasm conservation and breeding applications [
22]. In this study, we conducted a genome-wide SNP analysis across 297
Liriodendron accessions representing three taxonomically distinct groups:
L. chinense,
L. tulipifera, and their interspecific hybrids. As the most abundant form of genomic variation, SNPs offer several advantages—including codominance, amplification stability, and high reproducibility—which make them particularly suitable for assessing genetic diversity [
36]. In recent years, SNP-based approaches have been widely applied in plant genetics research, significantly advancing germplasm management and varietal conservation efforts [
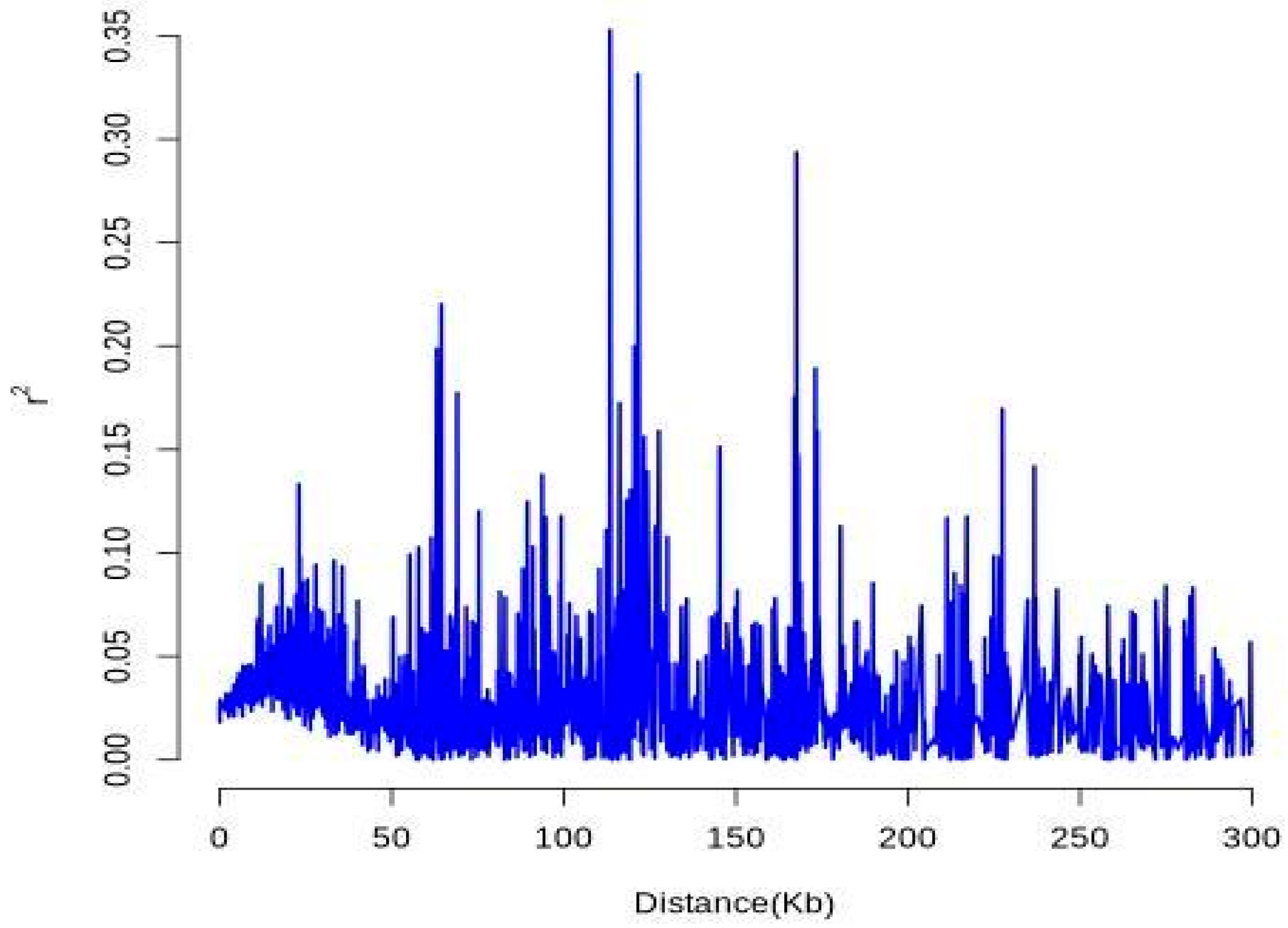
37]. From an initial pool of variants, 4204 high-quality SNPs were stringently selected, showing a non-uniform distribution across the genome with evident regional variation. These markers exhibited an average PIC of 0.159, a critical index for evaluating inter-accession polymorphism and supporting gene pool development and breeding acceleration [
38]. Compared with those in other forest species, such as
Picea abies (PIC ≈ 0.12) [
39] and
Betula platyphylla (He ≈ 0.141) [
40], the polymorphism levels observed in
Liriodendron were representative and suitable for germplasm evaluation. The overall observed heterozygosity (Ho = 0.203), expected heterozygosity (He = 0.154), and Shannon’s diversity index (H′ = 0.364) collectively indicated a substantial degree of genetic diversity within the sampled population. Notably, the
Liriodendron sino-americanum group exhibited the highest diversity across all indices, likely resulting from the incorporation of biparental allelic variation through interspecific hybridization. This pattern is consistent with prior studies on interspecific heterosis in
Liriodendron sino-americanum and may represent a broader evolutionary trend, as similar diversity-enhancing effects have been reported in other woody taxa, such as poplar hybrid systems [
41].
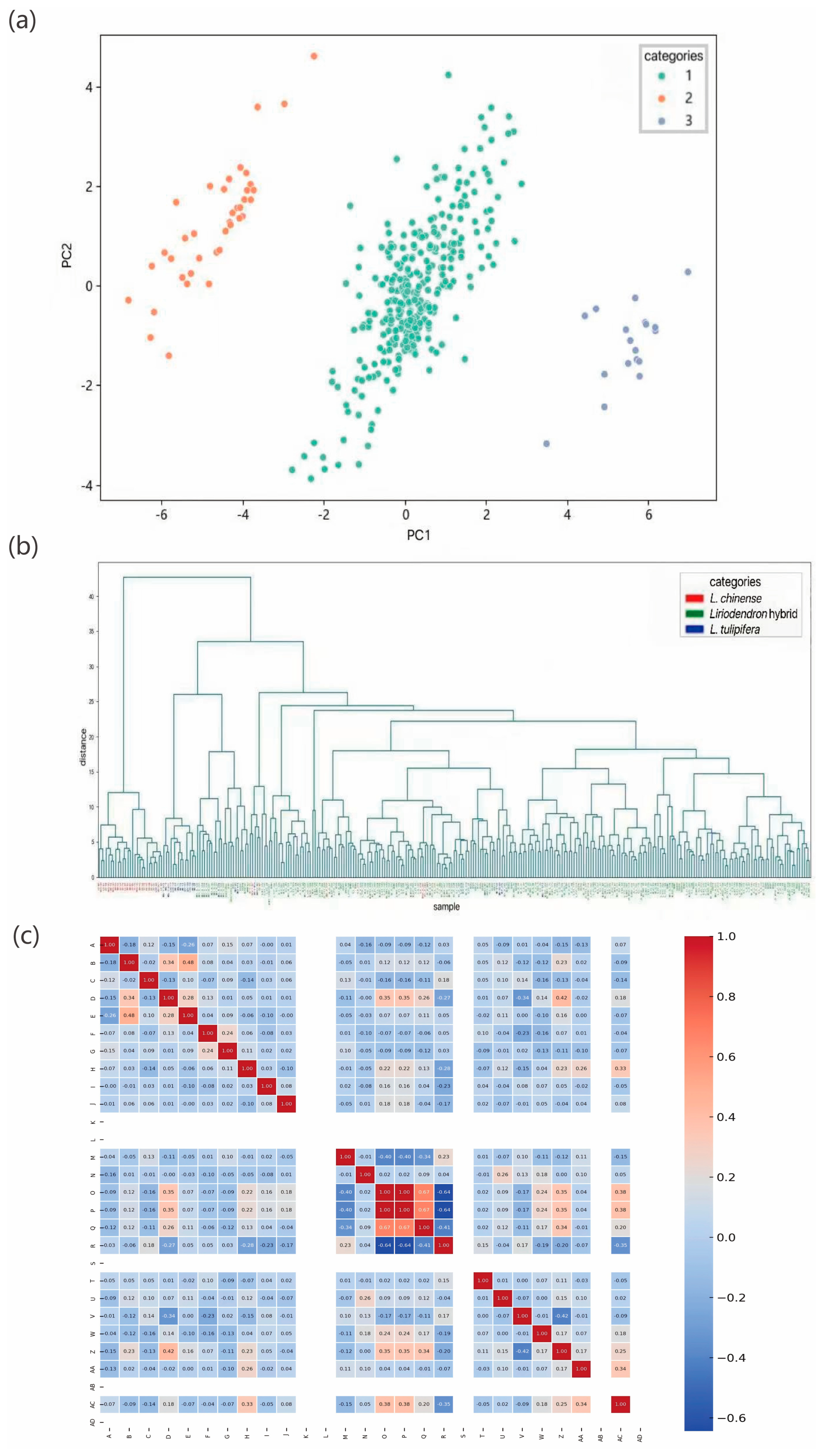
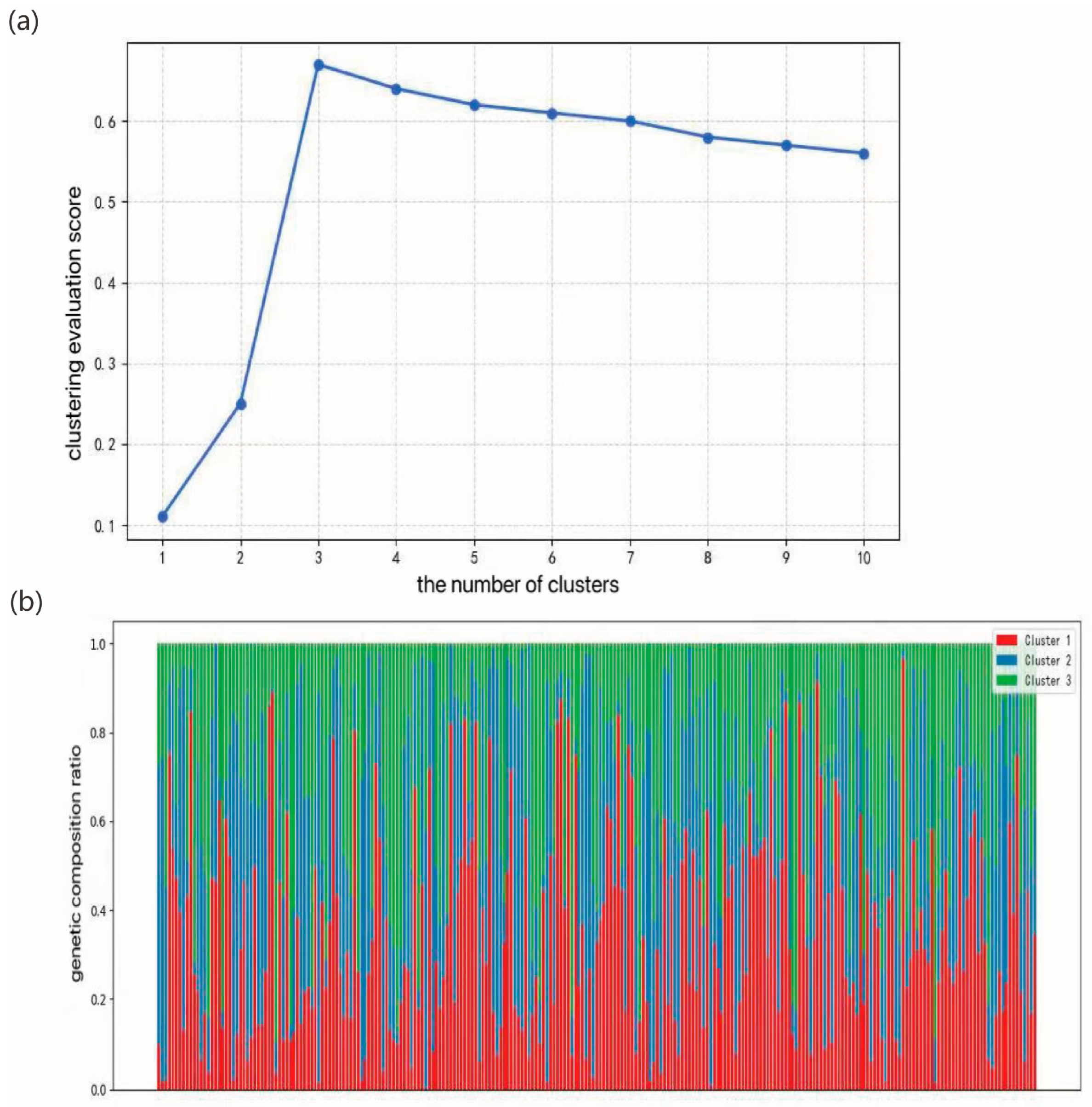
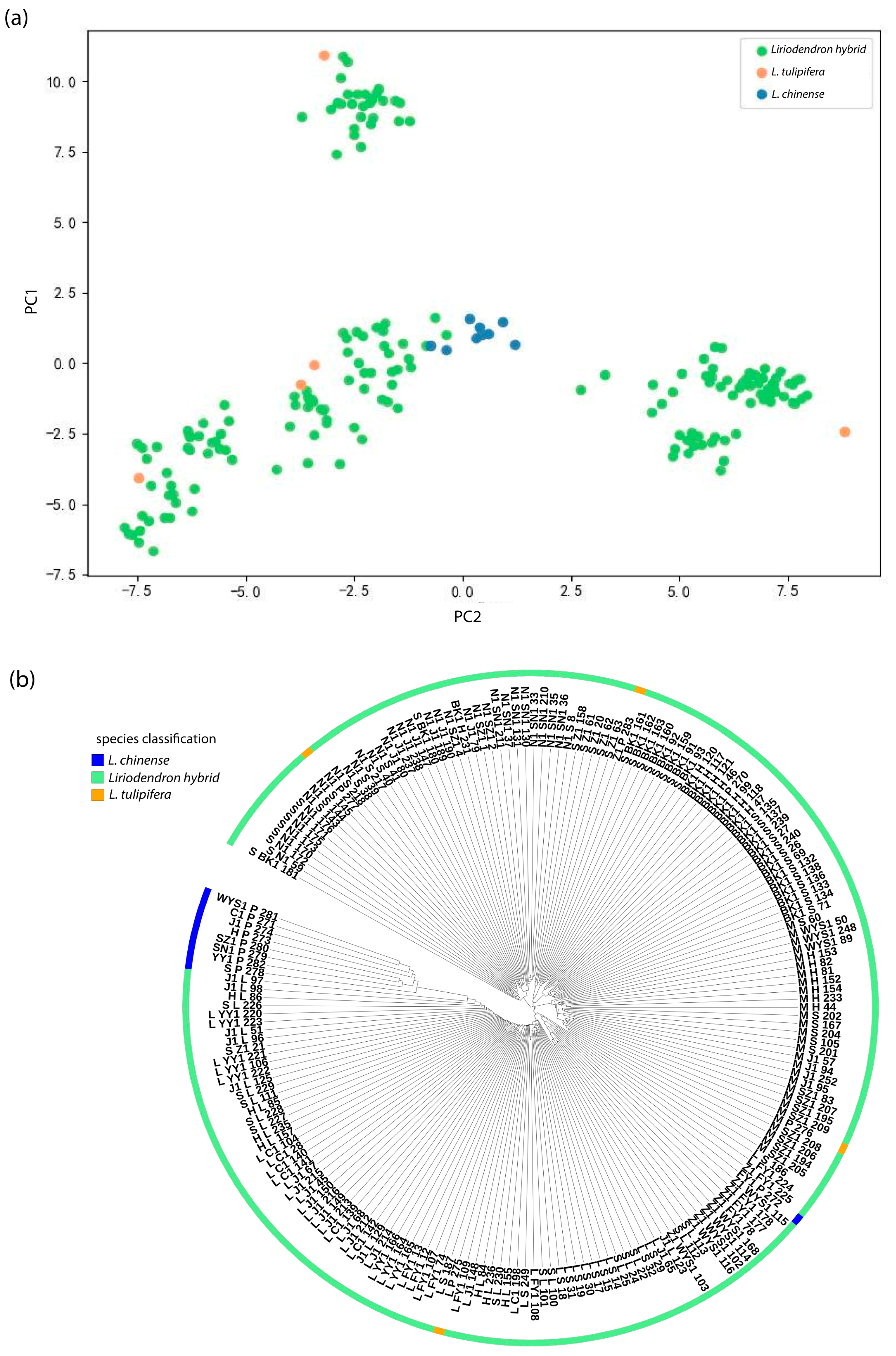
Genetic structure reveals the distribution patterns of genetic diversity within and among populations, serving as an important indicator of a species’ adaptive potential to its environment [
42]. To characterize the genetic structure of
Liriodendron germplasm resources, we conducted a series of genetic variation analyses. Population structure analysis indicated that the optimal clustering occurred at K = 3, dividing the 297 accessions into three genetic groups. Both principal component analysis and hierarchical clustering produced consistent results, which aligned with our expectations. Notably, North American
L. tulipifera exhibited a broader genetic distribution than
L. chinense and
Liriodendron sino-americanum, potentially due to higher genetic heterogeneity or more complex evolutionary lineages within its populations. Long et al. similarly reported that
L. tulipifera harbors approximately 1.8 times the genetic diversity of
L. chinense, [
43] likely resulting from multiple contributing factors such as geographic isolation, restricted gene flow [
44], and historical domestication bottlenecks [
45]. In the K = 3 population structure simulation, most accessions displayed mixed ancestry components, indicating frequent gene flow among groups and leading to partial differentiation without complete population separation. This observation is consistent with findings from red-fruited
Ailanthus altissima varieties [
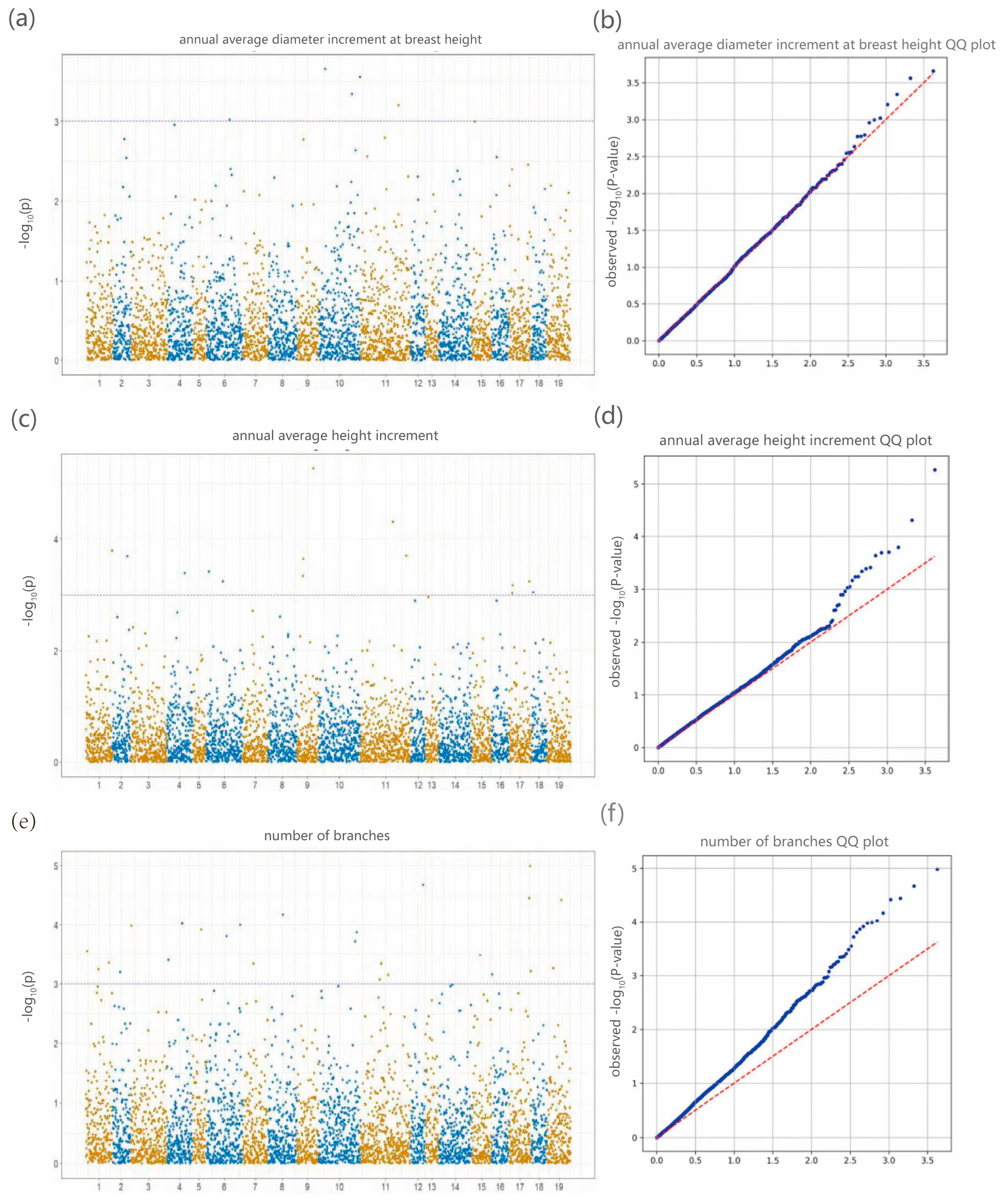
19]. Genome-wide association analysis integrating genomic and phenotypic datasets revealed 25 significantly associated SNPs corresponding to DBH, tree height, and branch number traits, with primary genomic distributions on chromosomes 10, 11, and 17. Notably, the most strongly associated SNPs for these respective traits were 10_31605746, 9_55280744, and 17_69375264. These findings not only elucidate the molecular basis of key phenotypic characteristics in
Liriodendron but also provide reliable target loci for molecular marker-assisted selection. The identified SNPs facilitate early trait prediction at the seedling stage through genotyping, potentially shortening the breeding cycle and accelerating the development of superior cultivars [
46]. This dual “structure-function” approach has demonstrated practical utility, as reported by Resende et al. in their landmark study on
Eucalyptus, this dual structure–function approach has already proven its practical value in tree breeding [
47].
Building upon our previous systematic evaluations of phenotypic traits, genetic diversity, population structure, and genome-wide association analyses of key trait-associated SNPs, we established a robust theoretical and data-driven foundation for DNA fingerprint development in
Liriodendron. Although DNA fingerprinting has been widely applied in crops and woody shrubs [
48,
49], no such database previously existed for
Liriodendron species. The selection of appropriate SNP markers is essential for developing a DNA fingerprint database. Using a stepwise additive algorithm, we identified 13 core SNP markers demonstrating high polymorphism (PIC = 0.34), balanced genotype distribution, broad genomic coverage (spanning 8 chromosomes), and excellent discriminative ability. After validation, these markers were used to establish a DNA fingerprint database for 297 Populus germplasm samples. This database enables precise germplasm identification, promotes the shift from conventional to precision breeding methods, and offers molecular tools for Populus germplasm evaluation and breeding. Moreover, these core SNP markers serve as key links between molecular assays and breeding applications, valuable tools for germplasm identification, genetic relationship studies, marker-assisted breeding, and genetic map development [
50,
51,
52]. As the first comprehensive effort to characterize
Liriodendron germplasm at both the phenotypic and molecular levels, our findings offer critical scientific support for the conservation, precise identification, and innovative utilization of these valuable genetic resources.