Arabidopsis Ubiquitin E3 Ligase AtCHYR1 Promotes ROS Production in Plant Responses to Sugar Availability
Abstract
1. Introduction
2. Results
2.1. Expression of AtCHYR1 Is Repressed by High Concentration of Exogenous Sugar
2.2. AtCHYR1 Positively Promotes High Glc-Mediated Inhibition in Seed Germination and Post-Germination Growth
2.3. AtCHYR1 Is a Glc Starvation-Response Gene and Aggravates Plant Starvation Response
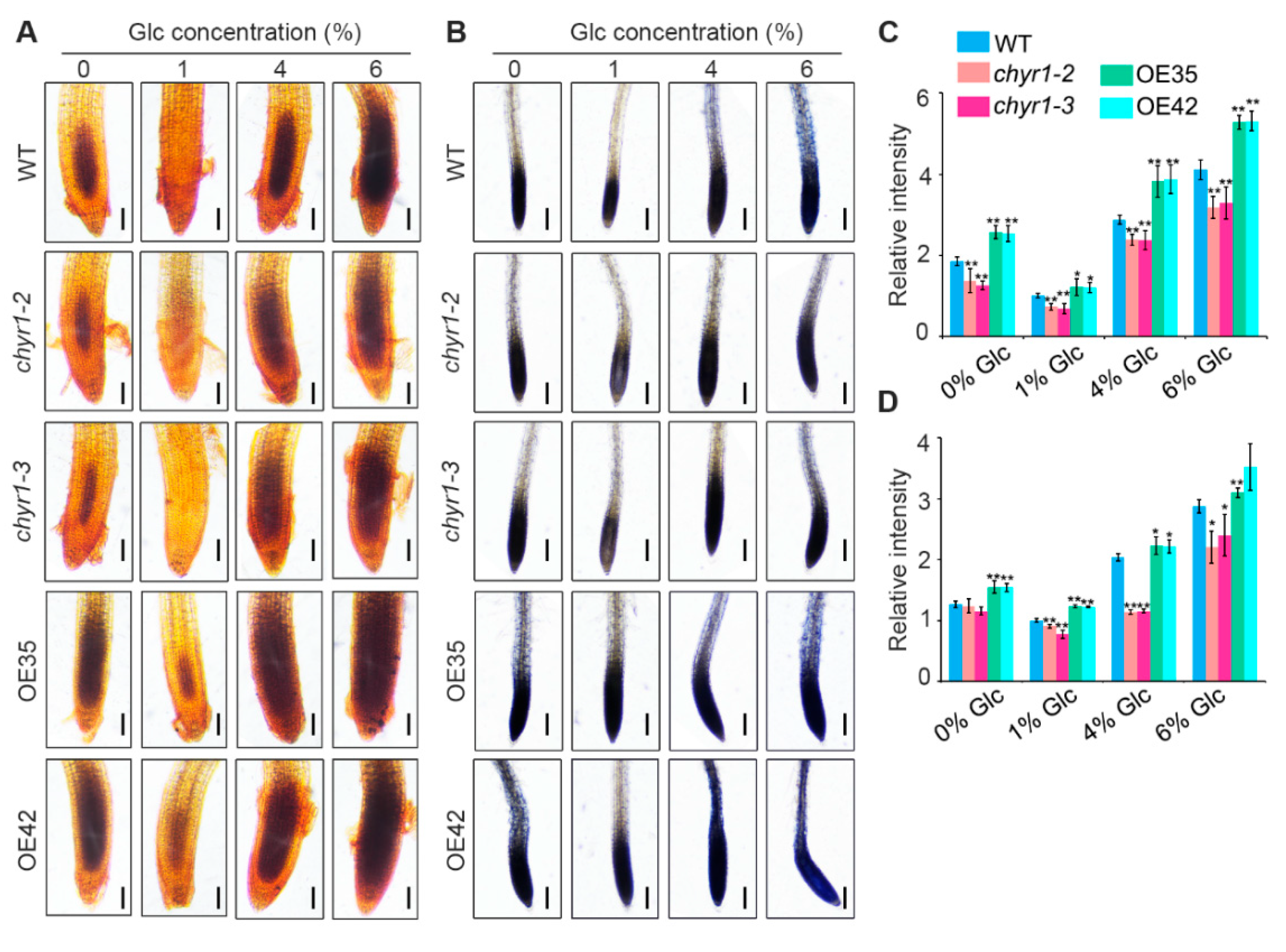
2.4. AtCHYR1 Enhances ROS Accumulation in Roots Under High-Glucose Conditions and Sugar-Starvation
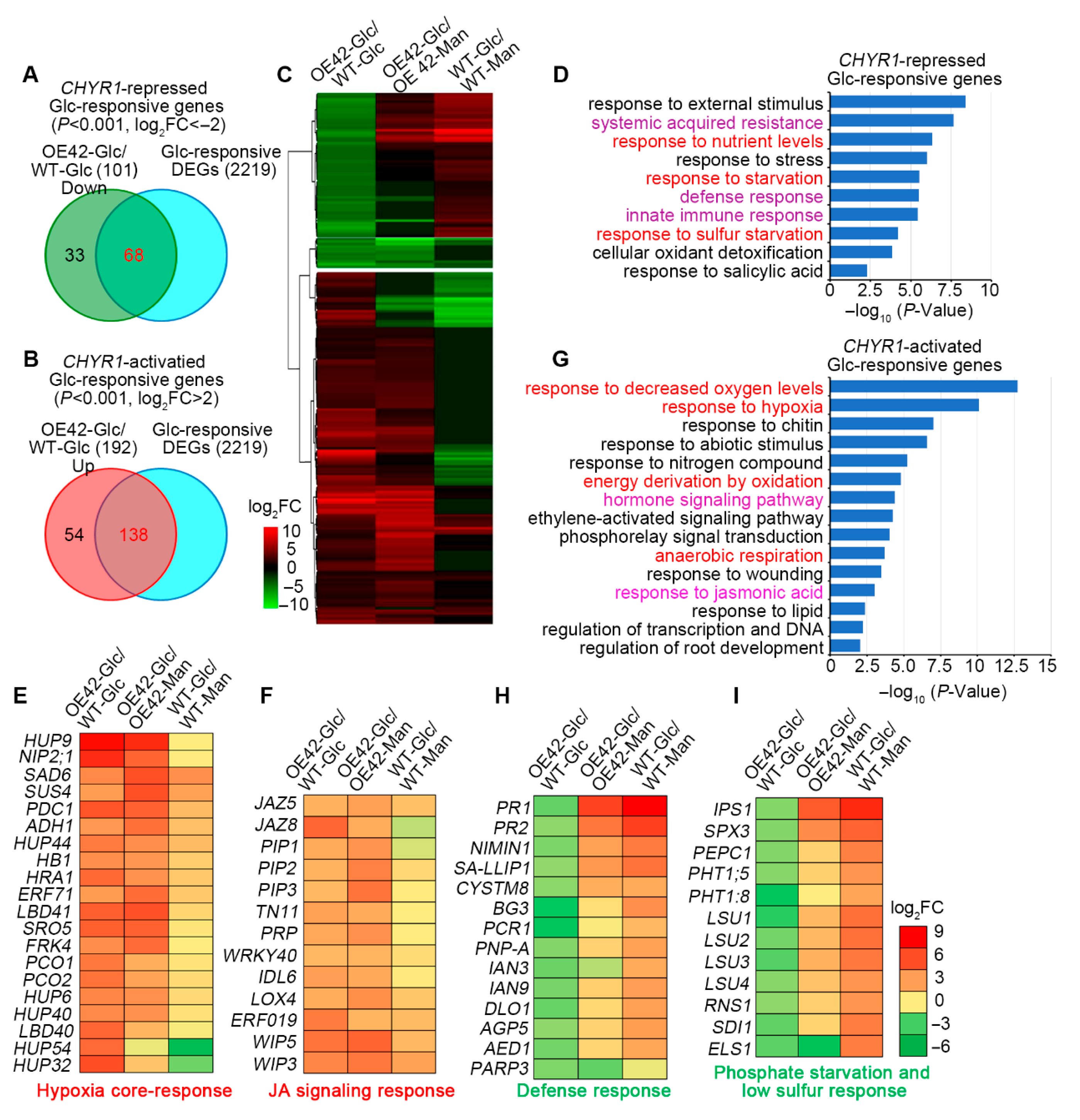
2.5. Transcriptomic Analysis Reveals That Glc-Inducible Genes Are Regulated in AtCHYR1-Overexpressing Plants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Germination Assay and the Response of the Root to Glc
4.3. Histochemical GUS Staining
4.4. ROS Analyses
4.5. RT-qPCR Analysis
4.6. RNA-Seq Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sami, F.; Yusuf, M.; Faizan, M.; Faraz, A.; Hayat, S. Role of sugars under abiotic stress. Plant Physiol. Biochem. 2016, 109, 54–61. [Google Scholar] [CrossRef]
- Cai, J.; Li, D.; Aharoni, A. The role of long-distance mobile metabolites in the plant stress response and signaling. Plant J. 2023, 114, 1115–1131. [Google Scholar] [CrossRef]
- Eom, S.H.; Kim, E.; Hyun, T.K. HXK, SnRK1, and TOR signaling in plants: Unraveling mechanisms of stress response and secondary metabolism. Sci. Prog. 2024, 107, 368504241301533. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.; Zhou, L.; Rolland, F.; Hall, Q.; Cheng, W.H.; Liu, Y.X.; Hwang, I.; Jones, T.; Sheen, J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 2003, 300, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Sheen, J.; Jang, J.C. The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol. Biol. 2000, 44, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Sheen, J. Dynamic and diverse sugar signaling. Curr. Opin. Plant Biol. 2016, 33, 116–125. [Google Scholar] [CrossRef]
- Broeckx, T.; Hulsmans, S.; Rolland, F. The plant energy sensor: Evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J. Exp. Bot. 2016, 67, 6215–6252. [Google Scholar] [CrossRef]
- Rodriguez, M.; Parola, R.; Andreola, S. TOR and SnRK1 signaling pathways in plant response to abiotic stresses: Do they always act according to the “yin-yang” model? Plant Sci. 2019, 288, 110220. [Google Scholar] [CrossRef]
- Li, L.; Liu, K.H.; Sheen, J. Dynamic nutrient signaling networks in plants. Annu. Rev. Cell Dev. Biol. 2021, 37, 341–367. [Google Scholar] [CrossRef]
- Sami, F.; Siddiqui, H.; Hayat, S. Interaction of glucose and phytohormone signaling in plants. Plant Physiol. Biochem. 2019, 135, 119–126. [Google Scholar] [CrossRef]
- Yuan, K.; Wysocka-Diller, J. Phytohormone signalling pathways interact with sugars during seed germination and seedling development. J. Exp. Bot. 2006, 57, 3359–3367. [Google Scholar] [CrossRef]
- Rellán-Álvarez, R.; Lobet, G.; Dinneny, J.R. Environmental control of root system biology. Annu. Rev. Plant Biol. 2016, 67, 619–642. [Google Scholar] [CrossRef]
- Yuan, T.T.; Xu, H.H.; Zhang, K.X.; Guo, T.T.; Lu, Y.T. Glucose inhibits root meristem growth via ABA INSENSITIVE 5, which represses PIN1 accumulation and auxin activity in Arabidopsis. Plant Cell Environ. 2014, 37, 1338–1350. [Google Scholar] [CrossRef]
- Liszkay, A.; van der Zalm, E.; Schopfer, P. Production of reactive oxygen intermediates (O2−, H2O2, and OH) by maize roots and their role in wall loosening and elongation growth. Plant Physiol. 2004, 136, 3114–3123. [Google Scholar] [CrossRef]
- Huang, L.; Yu, L.J.; Zhang, X.; Fan, B.; Wang, F.Z.; Dai, Y.S.; Qi, H.; Zhou, Y.; Xie, L.J.; Xiao, S. Autophagy regulates glucose-mediated root meristem activity by modulating ROS production in Arabidopsis. Autophagy 2019, 15, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Kourie, J.I. Interaction of reactive oxygen species with ion transport mechanisms. Am. J. Physiol. 1998, 275, C1–C24. [Google Scholar] [CrossRef] [PubMed]
- Kiselyov, K.; Muallem, S. ROS and intracellular ion channels. Cell Calcium. 2016, 60, 108–114. [Google Scholar] [CrossRef]
- Leymarie, J.; Vitkauskaité, G.; Hoang, H.H.; Gendreau, E.; Chazoule, V.; Meimoun, P.; Corbineau, F.; El-Maarouf-Bouteau, H.; Bailly, C. Role of reactive oxygen species in the regulation of Arabidopsis seed dormancy. Plant Cell Physiol. 2012, 53, 96–106. [Google Scholar] [CrossRef]
- Ogawa, K.; Iwabuchi, M. A mechanism for promoting the germination of Zinnia elegans seeds by hydrogen peroxide. Plant Cell Physiol. 2001, 42, 286–291. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, X.; Peng, J.; Li, F.; Ali, F.; Wang, Z. Regulation of seed germination: ROS, epigenetic, and hormonal aspects. J. Adv. Res. 2025, 71, 107–125. [Google Scholar] [CrossRef]
- Tsukagoshi, H. Control of root growth and development by reactive oxygen species. Curr. Opin. Plant Biol. 2016, 29, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Muhlemann, J.K.; Younts, T.L.B.; Muday, G.K. Flavonols control pollen tube growth and integrity by regulating ROS homeostasis during high-temperature stress. Proc. Natl. Acad. Sci. USA 2018, 115, E11188–E11197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zeng, L. Crosstalk between ubiquitination and other post-translational protein modifications in plant immunity. Plant Commun. 2020, 1, 100041. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Liu, L.; Xie, Q. In vitro protein ubiquitination assay. Methods Mol. Biol. 2012, 876, 163–172. [Google Scholar]
- Bueso, E.; Rodriguez, L.; Lorenzo-Orts, L.; Gonzalez-Guzman, M.; Sayas, E.; Muñoz-Bertomeu, J.; Ibañez, C.; Serrano, R.; Rodriguez, P.L. The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 2014, 80, 1057–1071. [Google Scholar] [CrossRef]
- Chen, Y.T.; Liu, H.; Stone, S.; Callis, J. ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. 2013, 75, 965–976. [Google Scholar] [CrossRef]
- Liu, H.; Yang, W.; Zhao, X.; Kang, G.; Li, N.; Xu, H. Genome-wide analysis and functional characterization of CHYR gene family associated with abiotic stress tolerance in bread wheat (Triticum aestivum L.). BMC Plant Biol. 2022, 22, 204. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, Y.; Ma, Z.; Wang, D.; Yan, F.; Liu, Y.; Li, J.; Yang, X.; Gao, Z.; Liu, X.; et al. Genome-wide identification of CHYR gene family in Sophora alopecuroides and functional analysis of SaCHYR4 in response to abiotic stress. Int. J. Mol. Sci. 2024, 25, 6173. [Google Scholar] [CrossRef]
- Wu, T.; Wang, Y.; Jin, J.; Zhao, B.; Wu, S.; Jia, B.; Sun, X.; Zhang, D.; Sun, M. Soybean RING-type E3 ligase GmCHYR16 ubiquitinates the GmERF71 transcription factor for degradation to negatively regulate bicarbonate stress tolerance. New Phytol. 2025, 246, 1128–1146. [Google Scholar] [CrossRef]
- Ding, S.; Zhang, B.; Qin, F. Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. Plant Cell 2015, 27, 3228–3244. [Google Scholar] [CrossRef]
- Liu, H.; Liu, B.; Lou, S.; Bi, H.; Tang, H.; Tong, S.; Song, Y.; Chen, N.; Zhang, H.; Jiang, Y.; et al. CHYR1 ubiquitinates the phosphorylated WRKY70 for degradation to balance immunity in Arabidopsis thaliana. New Phytol. 2021, 230, 1095–1109. [Google Scholar] [CrossRef] [PubMed]
- Hindt, M.N.; Akmakjian, G.Z.; Pivarski, K.L.; Punshon, T.; Baxter, I.; Salt, D.E.; Guerinot, M.L. BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode important negative regulators of the iron deficiency response in Arabidopsis thaliana. Metallomics 2017, 9, 876–890. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Celma, J.; Connorton, J.M.; Kruse, I.; Green, R.T.; Franceschetti, M.; Chen, Y.T.; Cui, Y.; Ling, H.Q.; Yeh, K.C.; Balk, J. Arabidopsis BRUTUS-LIKE E3 ligases negatively regulate iron uptake by targeting transcription factor FIT for recycling. Proc. Natl. Acad. Sci. USA 2019, 116, 17584–17591. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Seo, P.J. The Arabidopsis MIEL1 E3 ligase negatively regulates ABA signalling by promoting protein turnover of MYB96. Nat. Commun. 2016, 7, 12525. [Google Scholar] [CrossRef]
- Zhao, C.Y.; Xue, H.W. PI4Kγ2 Interacts with E3 Ligase MIEL1 to Regulate Auxin Metabolism and Root Development. Plant Physiol. 2020, 184, 933–944. [Google Scholar] [CrossRef]
- Huang, Y.; Li, C.Y.; Pattison, D.L.; Gray, W.M.; Park, S.; Gibson, S.I. SUGAR-INSENSITIVE3, a RING E3 ligase, is a new player in plant sugar response. Plant Physiol. 2010, 152, 1889–1900. [Google Scholar] [CrossRef]
- Aoyama, S.; Terada, S.; Sanagi, M.; Hasegawa, Y.; Lu, Y.; Morita, Y.; Chiba, Y.; Sato, T.; Yamaguchi, J. Membrane-localized ubiquitin ligase ATL15 functions in sugar-responsive growth regulation in Arabidopsis. Biochem. Biophys. Res. Commun. 2017, 491, 33–39. [Google Scholar] [CrossRef]
- Luo, Y.; Aoyama, S.; Fukao, Y.; Chiba, Y.; Sato, T.; Yamaguchi, J. Involvement of the membrane-localized ubiquitin ligase ATL8 in sugar starvation response in Arabidopsis. Plant Biotechnol. 2019, 36, 107–112. [Google Scholar] [CrossRef]
- Wang, S.; Chen, H.; Huang, Y.; Zhang, X.; Chen, Y.; Du, H.; Wang, H.; Qin, F.; Ding, S. Ubiquitin E3 ligase AtCHYR2 functions in glucose regulation of germination and post-germinative growth in Arabidopsis thaliana. Plant Cell Rep. 2023, 42, 989–1002. [Google Scholar] [CrossRef]
- Jamsheer, K.M.; Laxmi, A. Expression of Arabidopsis FCS-Like Zinc finger genes is differentially regulated by sugars, cellular energy level, and abiotic stress. Front. Plant Sci. 2015, 6, 746. [Google Scholar] [CrossRef]
- Wang, T.J.; Huang, S.; Zhang, A.; Guo, P.; Liu, Y.; Xu, C.; Cong, W.; Liu, B.; Xu, Z.Y. JMJ17-WRKY40 and HY5-ABI5 modules regulate the expression of ABA-responsive genes in Arabidopsis. New Phytol. 2021, 230, 567–584. [Google Scholar] [CrossRef]
- Joshi, S.; Hill, K.; Chakrabarti, M.; Perry, S.E. Regulatory mechanisms of the LBD40 transcription factor in Arabidopsis thaliana somatic embryogenesis. Plant Direct. 2023, 7, e547. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, B.J.; Schuurmans, J.A.; Smeekens, S.C. Glucose delays seed germination in Arabidopsis thaliana. Planta 2004, 218, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Singh, M.; Laxmi, A. Interaction between glucose and brassinosteroid during the regulation of lateral root development in Arabidopsis. Plant Physiol. 2015, 168, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Sewelam, N.; Kazan, K.; Schenk, P.M. Global plant stress signaling: Reactive oxygen species at the cross-road. Front. Plant Sci. 2016, 7, 187. [Google Scholar] [CrossRef]
- Park, J.J.; Yi, J.; Yoon, J.; Cho, L.H.; Ping, J.; Jeong, H.J.; Cho, S.K.; Kim, W.T.; An, G. OsPUB15, an E3 ubiquitin ligase, functions to reduce cellular oxidative stress during seedling establishment. Plant J. 2011, 65, 194–205. [Google Scholar] [CrossRef]
- Wolyniec, K.; Levav-Cohen, Y.; Jiang, Y.H.; Haupt, S.; Haupt, Y. The E6AP E3 ubiquitin ligase regulates the cellular response to oxidative stress. Oncogene 2013, 32, 3510–3519. [Google Scholar] [CrossRef]
- Mazzucotelli, E.; Belloni, S.; Marone, D.; De Leonardis, A.; Guerra, D.; Di Fonzo, N.; Cattivelli, L.; Mastrangelo, A. The e3 ubiquitin ligase gene family in plants: Regulation by degradation. Curr. Genomics 2006, 7, 509–522. [Google Scholar] [CrossRef]
- Price, J.; Laxmi, A.; St Martin, S.K.; Jang, J.C. Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 2004, 16, 2128–2150. [Google Scholar] [CrossRef]
- Nunes, C.; O’Hara, L.E.; Primavesi, L.F.; Delatte, T.L.; Schluepmann, H.; Somsen, G.W.; Silva, A.B.; Fevereiro, P.S.; Wingler, A.; Paul, M.J. The trehalose 6-phosphate/SnRK1 signaling pathway primes growth recovery following relief of sink limitation. Plant Physiol. 2013, 162, 1720–1732. [Google Scholar] [CrossRef]
- Zhai, Z.; Keereetaweep, J.; Liu, H.; Feil, R.; Lunn, J.E.; Shanklin, J. Trehalose 6-phosphate positively regulates fatty acid synthesis by stabilizing WRINKLED1. Plant Cell 2018, 30, 2616–2627. [Google Scholar] [CrossRef]
- Peixoto, B.; Moraes, T.A.; Mengin, V.; Margalha, L.; Vicente, R.; Feil, R.; Höhne, M.; Sousa, A.G.G.; Lilue, J.; Stitt, M.; et al. Impact of the SnRK1 protein kinase on sucrose homeostasis and the transcriptome during the diel cycle. Plant Physiol. 2021, 187, 1357–1373. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Cai, Z.; He, F.; Feng, X.; Liang, T.; Wang, H.; Ding, S.; Tian, X. Transcriptomic analysis reveals important roles of lignin and flavonoid biosynthetic pathways in rice thermotolerance during reproductive stage. Front. Genet. 2020, 11, 562937. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, S.; Xue, Y.; Teng, Y.; Qin, S.; Wang, H. Arabidopsis Ubiquitin E3 Ligase AtCHYR1 Promotes ROS Production in Plant Responses to Sugar Availability. Plants 2025, 14, 2617. https://doi.org/10.3390/plants14172617
Ding S, Xue Y, Teng Y, Qin S, Wang H. Arabidopsis Ubiquitin E3 Ligase AtCHYR1 Promotes ROS Production in Plant Responses to Sugar Availability. Plants. 2025; 14(17):2617. https://doi.org/10.3390/plants14172617
Chicago/Turabian StyleDing, Shuangcheng, Yuxin Xue, Yulu Teng, Simin Qin, and Hongwei Wang. 2025. "Arabidopsis Ubiquitin E3 Ligase AtCHYR1 Promotes ROS Production in Plant Responses to Sugar Availability" Plants 14, no. 17: 2617. https://doi.org/10.3390/plants14172617
APA StyleDing, S., Xue, Y., Teng, Y., Qin, S., & Wang, H. (2025). Arabidopsis Ubiquitin E3 Ligase AtCHYR1 Promotes ROS Production in Plant Responses to Sugar Availability. Plants, 14(17), 2617. https://doi.org/10.3390/plants14172617






