Abstract
Fraxinus spp. is one of the most important salt-alkali resistant tree species in the Yellow River region of China. However, the limited number of superior families and individuals, as well as the lack of a well-established parent selection system for hybrid breeding, have seriously constrained the improvement of seed orchards and the construction of advanced breeding populations. To address these issues, this study investigated 22 full-sib families of Fraxinus spp., using SSR molecular markers to calculate the genetic distance (GD) between parents. Combined with combining ability analysis, the study aimed to predict heterosis in offspring growth traits and select superior families and individuals through multi-trait comprehensive evaluation. The results showed the following: (1) Tree height (TH), diameter at breast height (DBH), and volume index (VI) exhibited extremely significant differences among families, indicating rich variation and strong selection potential. (2) The phenotypic and genotypic coefficients of variation for TH, DBH, and VI ranged from 4.34% to 16.04% and 5.10% to 17.73%, respectively. Family heritability was relatively high, ranging from 0.724 to 0.818, suggesting that growth is under strong genetic control. (3) The observed and expected heterozygosity of 15 parents were 0.557 and 0.410, respectively, indicating a moderate level of heterozygosity. Nei’s genetic diversity index and Shannon’s information index were 0.488 and 0.670, respectively, indicating relatively high genetic diversity. GD between parents ranged from 0.155 to 0.723. (4) Correlation analysis revealed significant or highly significant positive correlations between family heterosis and growth traits, combining ability, and GD, with specific combining ability (SCA) showing the strongest predictive power. Regression analysis further demonstrated significant linear correlations between GD and heterosis of TH and VI, and between SCA and heterosis of TH, DBH, and VI, establishing a GD threshold (≤0.723) and SCA-based co-selection strategy. In addition, four superior Fraxinus families and 11 elite individuals were selected. Their genetic gains for TH, DBH, and VI reached 2.28%, 3.30%, and 9.96% (family selection), and 1.98%, 2.11%, and 4.00% (individual selection), respectively. By integrating genetic distance (GD) and quantitative genetic combining ability (SCA), this study established a quantifiable prediction model and proposed the “GD–SCA dual-index parent selection method”, offering a new paradigm for genetic improvement in tree breeding.
1. Introduction
Ash trees (Fraxinus spp.) are broad-leaved deciduous trees in the genus Fraxinus (Oleaceae, Lamiales), exhibiting self- or cross-pollination and being either monoecious or dioecious [1]. They have a somatic chromosome number of 2n = 22 and belong to the “1A” cytological type [2]. Fraxinus species are widely distributed across Asia, the Americas, and Europe [3,4], demonstrating broad ecological adaptability. In China, they are important tree species for windbreaks, sand fixation, and the reclamation of saline-alkaline lands in the northern regions [5]. Among the most widely distributed species of F. pennsylvanica, F. velutina and F. americana exhibit high phenotypic plasticity, cold tolerance, salt tolerance, waterlogging tolerance, ease of vegetative propagation, and rapid growth [6,7,8]. They were introduced into northern and northeastern China in the mid-20th century as the parents of the breeding program for soil amelioration and ecological restoration [9,10]. In addition, due to their excellent wood properties and texture, they are widely used in the production of furniture, sporting goods, and tool handles [11]. However, owing to overharvesting, slow growth, and abiotic stresses such as drought and salinity, many Fraxinus species have been classified as Class II endangered plants in China [12,13]. With the increasing urgency of global sustainable forestry and the demand for high-quality tree breeding, the genetic improvement and conservation of Fraxinus germplasm resources face significant challenges [14,15].
Heterosis (H) is a widespread biological phenomenon and a fundamental basis of hybrid breeding [16,17]. Artificial hybrids derived from genetically diverse parents often exhibit traits superior to those of their parents, such as yield, growth rate, and stress resistance [18,19]. More importantly, distant hybridization among different species within the same genus can result in the integration of superior traits and the creation of novel germplasm [20]. Studies have shown that interspecific hybridization is an effective approach for developing fast-growing and high-quality Fraxinus cultivars [5]. For instance, clonal hybrids such as F. mandshurica × F. americana and F. mandshurica × F. velutina exhibit significantly higher growth rates and stress resistance than their maternal parents [21,22,23]. However, hybrid breeding practices indicate that trait improvement in plants is often goal-directed, and not all natural materials are suitable as ideal parents. Therefore, parent selection should follow high standards from the outset [24,25]. Strong combining ability is essential for effective parental selection and the development of superior hybrid combinations [26,27]. General combining ability (GCA) refers to the average performance of a parent in hybrid progeny, representing the additive genetic effects that can be stably inherited across generations [28]; specific combining ability (SCA), derived from allele interactions, reflects non-additive genetic effects that are heritable but not fixable, and is particularly important for the development of commercial hybrids [29,30,31]. Hence, parent selection based on both GCA and SCA is crucial for ensuring the stability and consistency of hybrid progeny, and for minimizing segregation and variability in subsequent generations [27,32].
In addition, genetic distance (GD) quantifies the genetic divergence between parents and serves as an important indicator in parent selection. With the advancement of modern biotechnology, predicting heterosis using GD calculated from molecular markers has become increasingly scientific and reliable [33,34]. Typically, there is a certain degree of correlation between GD and H in plant populations [35,36]. The improvement potential of hybrid progeny largely depends on the magnitude of H and the variability of H deviation. In general, the greater the genetic divergence between parental populations, the lower the variability in heterosis and the higher the overall heterotic effect [37]. In Fraxinus breeding, enhancing growth-related traits has become a key objective. However, the traditional approach—constructing numerous hybrid combinations and evaluating phenotypes—is labor-intensive, time-consuming, and costly. More notably, the lengthy process from hybridization to new variety development/quantitative maturity, taking years or decades, limits forest tree production [38]. Addressing this requires early identification of high-potential individuals for target traits, optimizing resources, reducing ineffective screening, and shortening the breeding cycle [39]. Previous studies have demonstrated that utilizing molecular markers to assess genetic distance, combined with combining ability for heterosis prediction, enhances early selection efficiency and accelerates breeding of adaptable varieties [40]. However, systematic reports on this technology for Fraxinus breeding remain limited.
To make up for this shortcoming, this study selected 15 Fraxinus parents with diverse genetic backgrounds and conducted an incomplete factorial mating design. Growth traits of 22 full-sib families were evaluated, genetic parameters were calculated, and SSR markers were used to estimate GD among the parents. The main objectives of this study were the following: (1) to evaluate the growth traits of 22 full-sib Fraxinus families, quantify the genetic variation among families, and assess their potential for genetic improvement; (2) to explore the relationships among H, GCA, SCA, and GD, in order to identify superior parents and predict progeny performance, thereby laying a theoretical foundation for accelerating Fraxinus improvement; and (3) to identify superior families and elite individuals based on multi-trait evaluation, providing materials for elite variety breeding and advanced-generation breeding population development. The results of this study will improve the efficiency and accuracy of elite parent selection in Fraxinus and offer valuable insights for the genetic improvement of other forest tree species.
2. Results
2.1. Genetic Variation in Growth Traits Among Full-Sib Fraxinus Families
The linear model effects and genetic parameters for tree height (TH), diameter at breast height (DBH), and volume index (VI) among full-sib Fraxinus families are presented in Table 1. The family variance components (σF2) for TH, DBH, and VI ranged from 0.00279% to 21.70%, while the block × family interaction variances (σBF2) ranged from 0.00022% to 3.71%. The environmental variance components (σE2) were considerably high, particularly for TH (81.36%) and DBH (320.87%), indicating that environmental effects had a substantial influence on these traits. The phenotypic coefficient of variation (PCV) and genotypic coefficient of variation (GCV) ranged from 4.34% to 16.04% and from 5.10% to 17.73%, respectively. Among the traits, VI exhibited the highest level of genetic variation and family heritability, followed by DBH and TH. All three growth traits showed highly significant differences among full-sib families (p < 0.01), suggesting abundant genetic variation within the population.

Table 1.
Variance components and genetic parameters of different traits. TH tree height, DBH diameter at breast height, VI volume index, σF2 variance components of family, σBF2 variance components of interaction of block and family, σE2 variance components of environment. ** F value < 0.01. The block degrees of freedom and the family degrees of freedom are 3 and 21, respectively. The units of each trait—TH (m), DBH (cm), VI (m3), coefficient of variation (%)—are the same for tables below. PCV phenotypic coefficient of variation, GCV genotypic coefficient of variation, FH family heritability, SH single tree heritability.
Table 2 and Figure 1 illustrate the growth performance and variation across families. The average TH across all families was 5.87 m, ranging from 5.25 m to 6.35 m. The average DBH was 7.04 cm, ranging from 5.86 cm to 8.00 cm. The average VI was 0.03293 m3, with a range of 0.02191 m3 to 0.04419 m3. Among all families, P6P8 exhibited the best growth performance, with phenotypic values of 6.35 m for TH, 7.99 cm for DBH, and 0.0412 m3 for VI, making it a highly promising candidate for selection as a superior family.

Table 2.
The average performance of 22 hybrid combinations.
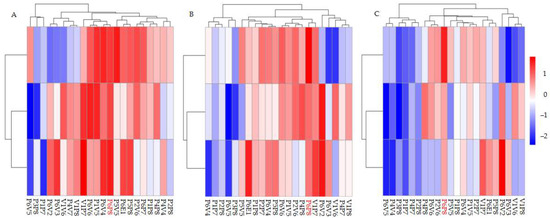
Figure 1.
Heatmap of growth trait differences among families. Heatmaps about tree height (A), diameter at breast height (B) and volume index (C).
2.2. Analysis of Genetic Correlation Among Fraxinus Parents
As shown in Table 3, the number of alleles (Na) amplified by each pair of SSR primers ranged from 1.667 to 3.333, with a total of 34 alleles amplified. On average, each primer pair amplified 2.429 alleles, and the average effective number of alleles (Ne) was 1.977. Among them, seven primer pairs had a polymorphic information content (PIC) greater than 0.5, indicating a relatively high level of polymorphism. The observed heterozygosity (Ho) ranged from 0.167 to 1.000, with an average of 0.557, while the expected heterozygosity (He) ranged from 0.178 to 0.585, with an average of 0.410, reflecting a moderate level of heterozygosity among the parental genotypes. Nei’s genetic diversity index (h) ranged from 0.242 to 0.718 (average 0.488), and Shannon’s information index (I) ranged from 0.326 to 0.981 (average 0.670), indicating a high level of genetic diversity among the tested parents.

Table 3.
Polymorphic analysis of the 14 SSR markers.
As shown in Table 4, the genetic distance (GD) between the parents of the 22 families ranged from 0.155 to 0.723, with an average GD of 0.286. Some hybrid combinations shared similar GD values. The greatest genetic distance (0.723) was observed between the parental combinations of P6E1 and P4P7. In contrast, nine families with V5 or P8 as male parents exhibited the lowest and most similar GD values (0.155), suggesting that these parental combinations were more closely related.

Table 4.
Parental genetic distance (GD).
2.3. Analysis of Heterosis and Combining Ability for Growth Traits Among Fraxinus Families
The degree of heterosis varied considerably among families for all traits (Table 5), with the greatest variation observed for the volume index (VI), ranging from –40.11% to 73.67%. Among the progeny, the combinations P2V6, P2P7, and P5P8 exhibited the highest heterosis for tree height (TH), at 20.87%, 18.62%, and 25.53%, respectively. For diameter at breast height (DBH), the highest heterosis was observed in P5V5 (21.45%), P2V6 (15.87%), and P5P8 (21.46%). Similarly, for VI, P5V5 (62.15%), P2V6 (65.21%), and P5P8 (73.67%) showed the highest values. Notably, P2V6 and P5P8 exhibited strong heterosis across all three traits, indicating their potential as superior hybrid combinations.

Table 5.
Heterosis of hybrid progeny growth traits. HH, DH, and VIH, respectively, represent the hybrid superiority degrees of tree height, diameter at breast height, and volume index.
The combining ability for TH, DBH, and VI is presented in Table 6. Among the maternal parents, combinations involving P6, P2, V1, and P5 had positive GCAP1 values for TH, DBH, and VI in seven, two, three, and two combinations, respectively. Notably, only the P2P8 combination showed a positive GCAP1 for DBH, while the GCAP1 values for its other traits were negative. Among the five combinations with P6 as the female parent (P6V4, P6V5, P6V2, P6V6, and P6P8), GCAP1 values for TH and VI were the highest, reaching 0.29 and 0.0035, respectively. For DBH, the highest GCAP1 values were observed in P5V5 and P5P8 (both 0.26), indicating that P6 and P5 are among the most promising female parents for breeding purposes.

Table 6.
Combining ability of hybrid combination progeny traits. GCAP1, GCAP2, GCA, and SCA, respectively, represent the general compatibility of the maternal parent, the general compatibility of the paternal parent, the general compatibility, and the special compatibility.
Six combinations had positive GCAP2 values for TH, DBH, and VI simultaneously, with P7 and V6 as the male parents. Only two combinations (P6E1 and P4P7) showed positive GCAP2 values for both TH and DBH. Combinations with P8 and V2 as male parents had positive GCAP2 for TH and VI, while those with V4 and V5 had positive GCAP2 values only for DBH. Among these, P6V2 showed the highest GCAP2 values for TH (0.78) and VI (0.0091), and combinations involving V6 as the male parent (P6V6, P2V6, and V1V6) had the highest GCAP2 values for DBH (0.15), suggesting that V2 and V6 are the most promising male breeding parents.
In terms of overall general combining ability (GCA), among the 22 families, P6V2 showed the highest GCA for TH (1.07) and VI (0.0127), while P6P8 had the highest GCA for DBH (0.97), indicating that these hybrids are capable of producing vigorous growth performance. Regarding specific combining ability (SCA), P6P8 exhibited the highest SCA values across all three traits—TH (0.97), DBH (0.37), and VI (0.0129)—highlighting its potential as a superior cross for the development of high-growth Fraxinus cultivars.
2.4. Correlation and Regression Analysis
Correlation analysis (Figure 2) showed that genetic distance (GD) between parents was positively correlated with the heterosis degree of all traits, and the correlations with tree height (TH) heterosis and volume index (VI) heterosis were statistically significant (p < 0.05). Both general combining ability (GCA) and specific combining ability (SCA) were positively correlated with the heterosis degree of the corresponding traits. Specifically, the correlation between SCA and heterosis was significant for TH (p < 0.05), DBH (p < 0.05), and VI (p < 0.05). Only the correlation between DBH, GCA, and DBH heterosis was extremely significant (p < 0.01). These results suggest that, compared to GCA, heterosis performance in hybrid progeny is more strongly associated with GD and SCA.
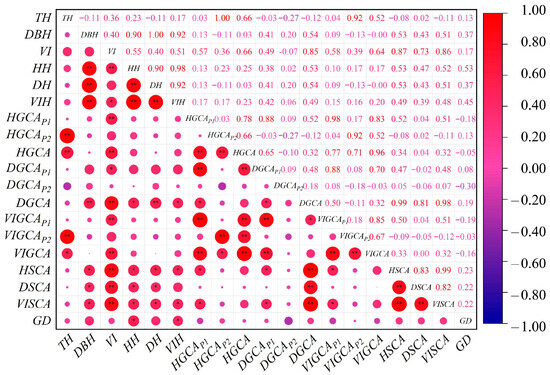
Figure 2.
Correlation between combining ability, genetic distance, and heterosis degree of hybrid progeny. HH stands for tree height heterosis degree, DH stands for diameter at breast height heterosis degree, VIH stands for the volume index heterosis degree, GCAP1 stands for parental general performance, GCAP2 stands for parental strain’s general performance coefficient, GCA stands for general combining ability, SCA stands for specific combining ability and GD stands for genetic distance. * means the probability of significance is 0.05, and ** means the probability of significance is 0.01.
Regression analysis (Figure 3) revealed linear positive relationships between heterosis degrees of growth traits and both parental GD and SCA. Significance tests of the linear regression models indicated that, except for the non-significant relationship between GD and DBH heterosis, all other models—including TH and VI heterosis vs. GD and heterosis vs. SCA for all traits—were statistically significant (p < 0.05). Among them, the regression between SCA and TH heterosis showed the best fit, with the highest coefficient of determination (R2 = 0.249). Scatter plots showed that heterosis values increased with rising GD and SCA, followed by a decline, indicating a nonlinear trend.
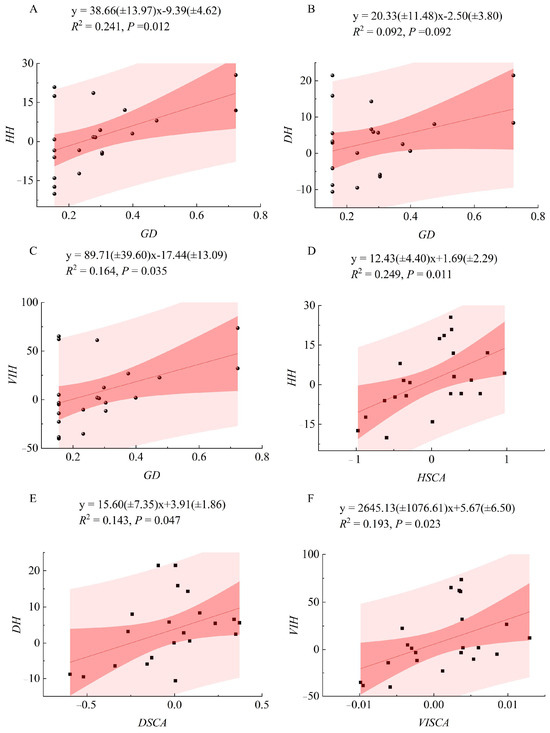
Figure 3.
Linear regression analysis between specific combining ability, genetic distance, and heterosis degree of hybrid progeny. x represents the corresponding horizontal coordinate, y represents the corresponding vertical coordinate, R2 is the coefficient of determination, and P indicates significance. (A–C) represent the regression relationships between the GD among parents and the heterosis degree for TH, DBH, and VI, respectively. (D–F) represent the regression relationships between the specific combining ability of TH, DBH, and VI and their corresponding heterosis degree, respectively.
Within the confidence interval, the highest heterosis was observed when GD (range: 0.155–0.723) approached approximately 0.7. For SCA of tree height (range: −0.98–0.97), the optimal range was 0.5–1.0; for DBH (range: −0.60–0.37), the optimal SCA range was 0.0–0.25; and for VI (range: 0.0099–0.0129), the highest heterosis occurred when SCA was between 0.005 and 0.010.
2.5. Selection of Superior Families and Elite Individuals in Fraxinus
A comprehensive evaluation of all traits was conducted across the 22 full-sib families. The Qi values for each family are shown in Table 7. The highest Qi value (1.732) was observed in family P6P8, while the lowest (1.438) was found in P6V5. Based on a 20% selection intensity threshold, four families were selected as superior: P6P8, V1P7, P5P8, and P6V6. The average tree height (TH), diameter at breast height (DBH), and volume index (VI) of the selected families were 6.25 m, 7.71 cm, and 0.04076 m3, respectively—representing increases of 6.34%, 9.63%, and 23.78% over the population means. The corresponding genetic gains were 2.28% (TH), 3.30% (DBH), and 9.96% (VI).

Table 7.
Qi values and genetic gains of different families.
Using a more stringent 1% selection intensity threshold, 11 elite individual trees were selected from the entire population (Table 8). Compared to the overall means, the selected individuals exhibited average increases of 23.36% in TH, 66.35% in DBH, and 199.61% in VI. The corresponding genetic gains for these individuals were 1.98% (TH), 2.11% (DBH), and 4.00% (VI).

Table 8.
Qi values and genetic gains of different single plants.
3. Discussion
Genetic variation is the prerequisite for genetic improvement in plants, and its effectiveness determines the potential of a species in long-term breeding programs [41]. Understanding the patterns of genetic variation is fundamental to selecting appropriate breeding strategies [42,43]. Moreover, tree age correlation indicates growth stability by measuring juvenile–mature trait consistency. When strong, it enables effective early selection for mature performance, improving genetic breeding efficiency [44,45]. Fraxinus data confirm this: genetic height (0.89) and diameter (0.92) correlations are high at age 7–8 (F. excelsior) [46], while juvenile–mature volume and progeny height adult volume correlations are significant (F. mandshurica) [47]. This demonstrates early growth stability and supports early selection feasibility. In this study, highly significant differences in growth traits were observed among 8-year-old full-sib families of Fraxinus, indicating strong genetic control over progeny traits [48]. Heritability analysis showed that family heritability values exceeded 0.7, indicating a high level of genetic control [31]. These values are much higher than the heritability of tree height (0.30) and diameter at breast height (0.27) observed in 8-year-old F. excelsior progeny [46]. Moreover, family-level heritability was higher than individual-tree heritability, indicating that greater genetic gains can be achieved through family selection in Fraxinus, consistent with the findings of Weng et al. [49]. These results suggest that hybridizing the selected Fraxinus parents can create a base population with sufficient variation for the next generation, providing opportunities for selecting superior families.
Heterosis is the phenomenon where hybrids resulting from crosses between genetically distinct parents outperform their parents in various traits [16]. It is one of the key innovations in modern plant breeding and genetic improvement, playing an important role in global wood and food security [50]. However, the superiority of hybrids depends not only on the performance of the parents but also on the combining ability between them [51]. Combining ability analysis is an effective method to evaluate and select superior parents and hybrid combinations, thereby producing large numbers of high-quality families or progeny [52,53]. In this study, maternal parent P6 and paternal parent V6 showed high general combining ability (GCA) values for growth traits. The hybrids from these parents, such as P6V6, P6P8, P6V4, and P2V6, exhibited high specific combining ability (SCA) values and significant heterosis, suggesting that strong heterotic combinations are more likely when at least one parent has high GCA or when the combination has high SCA—consistent with results in maize [54], Camellia oleifera [55], and Populus tomentosa [56]. Therefore, when breeding for superior Fraxinus genotypes, it is advisable to select parents with both high GCA and high SCA values to increase the probability of producing highly heterotic combinations.
Furthermore, this study found that in some highly heterotic combinations—such as P2P7 and P2V6 (with high heterosis in tree height), and P5P8 and P5V5 (with high heterosis in DBH)—the parental GCA values were high, while SCA values were not. Conversely, although the combination P6P8 showed the highest GCA and SCA values for all three growth traits, its heterosis performance was weak. This indicates that forming highly heterotic combinations in Fraxinus cannot rely solely on parental GCA or hybrid SCA; instead, a comprehensive evaluation of individual traits and their contributing factors is necessary. This is consistent with the findings of Josue et al. [57] in maize. Additionally, correlation analysis between heterosis and combining ability in this study revealed that only DBH heterosis was significantly positively correlated with GCA, while all three traits showed significant positive correlations with SCA, with higher correlation coefficients—indicating that SCA plays a more important predictive role for growth-related traits in Fraxinus. This is in line with findings by Umakanth et al. [58] in sweet sorghum, where SCA variance components for total biomass, juice yield, and grain yield were greater than GCA, and by Chai et al. [59] in C. oleifera, where SCA was more important than GCA for most traits. However, in practical breeding, parental selection should consider both GCA and SCA along with the genetic background of the parental material to increase the likelihood of producing superior hybrids.
In recent years, with the rapid development of molecular markers, breeders have commonly used molecular marker-based genetic distance to predict heterosis in crops. Since its first application in Zea mays L. [60], significant progress has been made in tree species such as Populus [61], Salix [36], Pinus [62], and Liriodendron chinense [63] for predicting heterosis in traits such as growth and resistance. Simple sequence repeat (SSR) markers or microsatellites have been considered markers of choice due to their co-dominant inheritance, high polymorphism, multi-allelic nature, and high reproducibility [64,65,66]. In this study, the average genetic distance (GD) among parents based on SSR markers was 0.286, which is considered moderate compared to previous studies [62,67]. Among strong heterotic combinations, P2P7 and P2V6 had relatively large parental GD values, whereas combinations like P5V5 and P5P8 had smaller GDs yet showed strong heterosis. Correlation analysis showed that GD was significantly positively correlated with heterosis in tree height and volume index, but not DBH, suggesting that greater parental GD tends to result in stronger heterosis. This finding aligns with studies in Larix decidua [68], chili pepper [69], and Pennisetum glaucum [70], but contrasts with studies in Cucumis melo [71], certain Pennisetum glaucum lines [72], and Brassica napus [73]. These discrepancies may be attributed to differences in plant materials, marker types and quantities, parental genetic relationships, phenotypic traits, and environmental factors. The observed positive relationship between GD and heterosis may be explained by either (1) increased distance resulting from the accumulation of favorable alleles, or (2) greater differences in gene frequencies at key loci between parental populations as GD increases [74,75]. Our results suggest that GD can be used to predict heterosis in tree height and volume index. However, the relationship between GD and heterosis is complex and requires further investigation. Moreover, due to the limited number of SSR markers used in this study, it is difficult to fully represent genome-wide information. Future work may incorporate higher-density markers (e.g., SNPs) or genome-wide association studies to improve the accuracy of heterosis prediction.
4. Materials and Methods
4.1. Test Materials
The experimental materials consisted of eight-year-old full-sib families of Fraxinus established in a trial plantation. In October 2015, seeds from 22 full-sib families were obtained through an incomplete factorial-controlled pollination design; details of the parental lines are shown in Table 9. It is important to emphasize that all the hybrid parents discussed in this study are superior varieties that have been selected by our team or collaborating units. This selection process followed preliminary evaluations of phenotypic traits, including tree height, diameter at breast height, and salt-alkali tolerance. In March 2016, the seeds were sown in non-woven container bags at the nursery of the Shandong Academy of Forestry. In March 2017, one-year-old seedlings (average basal diameter ~6.11 cm) were transplanted to the experimental base of Bohua Ecological Agriculture Co., Ltd., Shandong Province (Shandong, China). The site is located on the Yellow River alluvial plain, at an elevation of 9.66 m with a slope gradient of less than 1.75%. It has a temperate continental monsoon climate, with an annual average temperature of 13.6 °C, a frost-free period of 191 days, 2479.8 h of annual sunshine, average annual precipitation of 582.8 mm, and an annual evaporation of 1686.6 mm. The soil type is salinized fluvo-aquic soil with a pH of 8.48, salt content of 3.43 g·kg−1, available nitrogen 121.23 mg·kg−1, available phosphorus 32.02 mg·kg−1, and available potassium 149.67 mg·kg−1 [76]. Figure 4 shows the trial site location and general overview. A randomized complete block design was applied, with three blocks in total. Each plot contained 20 trees planted at a spacing of 3 m × 4 m.

Table 9.
The parental combinations of 22 full-sibling families.
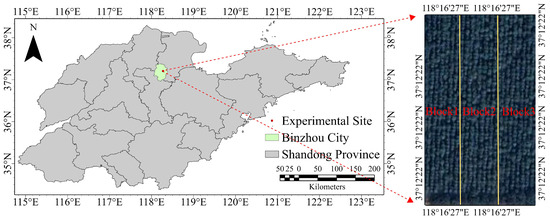
Figure 4.
Location of the experimental site and overview of the experimental forest.
4.2. Growth Trait Measurements
In November 2023 (after leaf fall in autumn), tree height (TH) and diameter at breast height (DBH) were measured for 22 eight-year-old full-sib families. Measurements were conducted using a tower ruler (accuracy: 0.01 m; Saiwei Geological Survey and Mapping Co., Ltd., Xuzhou, China) and a vernier caliper (accuracy: 0.01 cm; Mitutoyo Precision Measuring Instruments Co., Ltd., Shanghai, China). Survival rates for each family are shown in Table 10.

Table 10.
Survival rate of families.
4.3. DNA Extraction and SSR Analysis
Young leaves from the current-year shoots of 15 parent trees were collected for DNA extraction and SSR marker analysis. Genomic DNA was extracted using a modified CTAB method, in which 2% β-mercaptoethanol and 2% PVP were added to the extraction buffer [77]. DNA concentration and purity were assessed using a NanoDrop-2000 ultramicro spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and the samples were diluted to 30 ng·μL−1 and stored at −20 °C for further use.
A total of 14 SSR primer pairs were selected from 42 pairs previously developed by the research group and synthesized by Shandong VON Biotechnology Co., Ltd. (Yantai, China) (Table S1). Fluorescently labeled PCR amplification and capillary electrophoresis were conducted following the procedure described by Yan [78]. Raw electropherogram files were imported into GeneMarker (Version 2.2.0, SoftGenetics LLC, State College, PA, USA) for fragment scoring and allele peak analysis.
4.4. Data Processing
A nested PCR product size was read from the amplified DNA samples. SSR genotype data were formatted using GeneAlEx (Version 6.503, Australian National University, Canberra, Australia) to calculate the number of alleles (Na), effective number of alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He), Nei’s genetic diversity index (h), and Shannon’s information index (I). Polymorphism information content (PIC) and genetic distance (GD) were computed using PowerMarker (Version 3.25, North Carolina State University, Raleigh, NC, USA).
Statistical analyses of phenotypic traits were conducted using SPSS (Version 21.0), where ANOVA F-tests were used to assess the significance of fixed effects. Trait data were organized in Excel (Version 2016) (Table 11). Origin (Version 2021b) was used to evaluate the phenotypic values and heterosis of hybrid progenies, calculate correlation coefficients between heterosis and parental combining ability or genetic distance, construct scatterplots illustrating the relationships between heterosis and parental parameters, assess the goodness-of-fit of simple linear regression models, and generate the corresponding regression equations.

Table 11.
Phenotypic/genetic parameters and model calculation formulas. VI: volume index; TH: tree height; DBH: diameter at breast height; XijK: performance of the k-th tree in family i within block j; μ: general mean; Fi: effect of the i-th family; Bj: effect of the j-th block; FBij: interaction effect between family i and block j; eijk: random error; PCV: phenotypic coefficient of variation; σp2: phenotypic variance component; GCV: genotypic coefficient of variation; σg2: genetic variance component; X: mean of trait; H2: family heritability; h2: individual tree heritability; σF2: family variance component; σFB2: family × block interaction variance component; σe2: error variance component; B: number of blocks; N: total number of families. ΔGH: genetic gain from family selection; ΔGh: genetic gain from individual tree selection; S: selection differential; H: heterosis; Fi: mean value of family I; : mean value of all families sharing the same parents as family i; GCAP1i: general combining ability of the i-th female parent; P1: mean trait value of all families with female parent i; GCAP2j: general combining ability of the j-th male parent; P2j: mean trait value of all families with male parent j; : mean trait value of all families; GCAij: general combining ability of the fixed family of female parent i and male parent j; SCAij: specific combining ability of the hybrid combination of female parent i and male parent j; ij: mean trait value of the hybrid combination of female parent i and male parent j; Qi: comprehensive evaluation index value; xij: mean value of trait j for family i; xjmax: maximum value of trait j; n: number of traits.
5. Conclusions
In this study, we conducted a comprehensive analysis of growth traits in 22 full-sib families of Fraxinus spp., clarifying the patterns of genetic variation, combining ability effects, heterosis expression, and their relationship with parental genetic distance. The main conclusions are as follows: tree height (TH), diameter at breast height (DBH), and volume index (VI) showed highly significant differences among families, with family heritability values exceeding 0.7, indicating strong genetic control over growth traits and the potential for early selection. Heterosis was closely associated with combining ability, with specific combining ability (SCA) showing significantly positive correlations with heterosis across all three traits and providing better predictive value than general combining ability (GCA). SSR analysis revealed that genetic distance (GD) among parents was moderate, averaging 0.286. GD was significantly positively correlated with heterosis in TH and VI, demonstrating its reference value for heterosis prediction. Four elite families (P6P8, V1P7, P5P8, P6V6) and eleven superior individuals were selected, with growth traits exceeding the overall mean by 6.34%, 9.63%, and 23.78%, respectively, and genetic gains reaching 2.28%, 3.30%, and 9.96%. Outstanding parental lines included maternal parents P6, P2, P5, and paternal parents V6, V2, all exhibiting high GCA and SCA effects, making them suitable as core parents for advanced-generation breeding. The eight selected individuals exhibited improvements of 23.36%, 66.35%, and 199.61% in TH, DBH, and VI, respectively, with corresponding genetic gains of 1.98%, 2.11%, and 4.00%, demonstrating high breeding value. In conclusion, intra-specific hybridization in Fraxinus exhibits notable heterosis. Coordinated prediction of superior combinations using GD and SCA is a feasible strategy for enhancing breeding efficiency and provides technical support for elite parent selection and germplasm improvement.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants14162601/s1, Table S1: 14 pairs of primer information; Figure S1: the electrophoresis for some screened SSR primers.
Author Contributions
L.Y. and C.G. contributed equally to this study. Conceptualization, L.Y., C.G., C.L. and Y.W.; methodology, L.Y. and C.G.; software, L.Y., C.L. and N.L.; validation, L.Y., X.Z. and N.L.; formal analysis, C.G., N.L. and F.L.; investigation, Y.W., X.Z. and F.L.; resources, C.L.; data curation, L.Y.; writing—original draft preparation, L.Y. and C.G.; writing—review and editing, L.Y., C.G., C.L., Y.W. and N.L.; visualization, L.Y. and C.G.; supervision, C.L.; project administration, Y.W.; funding acquisition, L.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This project was financially supported by the Improved Variety Program of Shandong Province of China (2023LZGC012).
Data Availability Statement
The data underlying this article are available in the article and in its Supplementary Materials.
Acknowledgments
We are grateful for the scientific research platform and support provided by Shandong Provincial Academy of Forestry, the Research Institute of Forestry, Chinese Academy of Forestry and the State Key Laboratory of Tree Genetics and Breeding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Li, E.; Wang, Y.; Liu, K.; Liu, Y.; Xu, C.; Dong, W.; Zhang, Z. Historical climate change and vicariance events contributed to the intercontinental disjunct distribution pattern of ash species (Fraxinus, Oleaceae). Commun. Biol. 2024, 7, 603. [Google Scholar] [CrossRef]
- Huff, M.; Seaman, J.; Wu, D.; Zhebentyayeva, T.; Kelly, L.J.; Faridi, N.; Nelson, C.D.; Cooper, E.; Best, T.; Steiner, K.; et al. A high-quality reference genome for Fraxinus pennsylvanica for ash species restoration and research. Mol. Ecol. Resour. 2022, 22, 1284–1302. [Google Scholar] [CrossRef] [PubMed]
- Wallander, E. Systematics and floral evolution in Fraxinus (Oleaceae). Belg. Dendrologie Belge 2012, 2012, 39–58. [Google Scholar]
- Sollars, E.S.; Harper, A.L.; Kelly, L.J.; Sambles, C.M.; Ramirez-Gonzalez, R.H.; Swarbreck, D.; Kaithakottil, G.; Cooper, E.D.; Uauy, C.; Havlickova, L.; et al. Genome sequence and genetic diversity of European ash trees. Nature 2017, 541, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.N.; Yan, L.; Chai, Z.; Liang, Q.; Dong, Y.H.; Wang, C.X.; Li, X.C.; Li, C.Y.; Mu, Y.T.; Gong, A.D.; et al. Pan-genome analyses of 11 Fraxinus species provide insights into salt adaptation in ash trees. Plant Commun. 2024, 6, 101137. [Google Scholar] [CrossRef] [PubMed]
- Mao, P.; Tang, Q.; Cao, B.; Liu, J.; Shao, H.; Cao, Z.; Hao, M.; Zhu, Z. Eco-physiological adaptability in mixtures of Robinia pseudoacacia and Fraxinus velutina and coastal eco-engineering. Ecol. Eng. 2017, 106, 109–115. [Google Scholar] [CrossRef]
- Liu, H. Under siege: Ash management in the wake of the emerald ash borer. J. Integr. Pest Manag. 2018, 9, 5. [Google Scholar] [CrossRef]
- Abhainn, E.A.; Shirley, D.L.; Stanley, R.K.; Scarpato, T.; Koch, J.L.; Romero-Severson, J. Gene flow from Fraxinus cultivars into natural stands of Fraxinus pennsylvanica occurs range-wide, is regionally extensive, and is associated with a loss of allele richness. PLoS ONE 2024, 19, e0294829. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Z.; Wang, M.; Yang, Y.; Wang, Y.; Nie, Q.; Liang, F.; Qin, H.; Zhang, Z. The chromosome-level genome assembly of Fraxinus americana provides insights into the evolution of Oleaceae plants. Int. J. Biol. Macromol. 2023, 253, 127132. [Google Scholar] [CrossRef]
- Plumb, W.J.; Kelly, L.J.; Mullender, J.; Powell, R.F.; Csiba, L.; Nemesio-Gorriz, M.; Carey, D.; Mason, M.E.; Crowther, W.; Koch, J.; et al. Preliminary genetic barcodes for ash (Fraxinus) species and generation of new wide hybrids. Plants People Planet 2025, 1–14. [Google Scholar] [CrossRef]
- Zhu, Z.; Qi, F.H.; Yan, C.F.; Zhan, Y.G. Sexually different morphological, physiological and molecular responses of Fraxinus mandshurica flowers to floral development and chilling stress. Plant Physiol. Biochem. 2016, 99, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Xue, G.; Liu, F.Z.; Gong, X.L. Immunosuppressive effect of extracts from leaves of Fraxinus mandshurica Rupr. Bioengineered 2017, 8, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.K.; Jin, J.M. China Plant Red Data Book—Rare and Endangered Plants, Volume 1; Science Press: Beijing, China, 1992. [Google Scholar]
- Diao, S.; Geng, Y.D. Efficiency evaluation and nonlinear multi-objective optimization of forestry industry transformation in the Heilongjiang state-owned forest region. Sci. Rep. 2023, 13, 21216. [Google Scholar] [CrossRef]
- Yang, W.; Min, Z.; Yang, M.X.; Yan, J. Exploration of the implementation of carbon neutralization in the field of natural resources under the background of sustainable development—An overview. Int. J. Environ. Res. Public Health 2022, 19, 14109. [Google Scholar] [CrossRef] [PubMed]
- Moll, R.H.; Lonnquist, J.H.; Fortuno, J.V.; Johnson, E.C. The Relationship of heterosis and genetic divergence in Maize. Genetics 1965, 52, 139–144. [Google Scholar] [CrossRef]
- Wang, B.B.; Hou, M.; Shi, J.P.; Ku, L.X.; Song, W.; Li, C.H.; Ning, Q.; Li, X.; Li, C.Y.; Zhao, B.B.; et al. De novo genome assembly and analyses of 12 founder inbred lines provide insights into maize heterosis. Nat. Genet. 2023, 55, 312–323. [Google Scholar] [CrossRef]
- Lu, H.; Chen, M.J.; Fu, M.; Yan, J.L.; Su, W.L.; Zhan, Y.G.; Zeng, F.S. Brassinosteroids affect wood development and properties of Fraxinus mandshurica. Front. Plant Sci. 2023, 14, 1167548. [Google Scholar] [CrossRef]
- Al-Ahmad, H. Biotechnology for bioenergy dedicated trees: Meeting future energy demands. Z. Naturforsch. C. J. Biosci. 2018, 73, 15–32. [Google Scholar] [CrossRef]
- Alix, K.; Gérard, P.R.; Schwarzacher, T.; Heslop-Harrison, J.S.P. Polyploidy and interspecific hybridization: Partners for adaptation, speciation and evolution in plants. Ann. Bot. 2017, 120, 183–194. [Google Scholar] [CrossRef]
- Zeng, F.S.; Li, L.L.; Liang, N.S.; Wang, X.; Li, X.; Zhan, Y.G. Salt tolerance and alterations in cytosine methylation in the interspecific hybrids of Fraxinus velutina and Fraxinus mandshurica. Euphytica 2015, 205, 721–737. [Google Scholar] [CrossRef]
- Zeng, F.S.; Zhou, S.; Zhan, Y.G.; Dong, J. Drought resistance and DNA methylation of interspecific hybrids between Fraxinus mandshurica and Fraxinus americana. Trees 2014, 28, 1679–1692. [Google Scholar] [CrossRef]
- He, Z.; Zhan, Y.; Zeng, F.; Zhao, X.T.; Wang, X. Drought physiology and gene expression characteristics of fraxinus interspecific hybrids. Plant Growth Regul. 2016, 78, 179–193. [Google Scholar] [CrossRef]
- Gu, Z.; Han, B. Unlocking the mystery of heterosis opens the era of intelligent rice breeding. Plant Physiol. 2024, 196, 735–744. [Google Scholar] [CrossRef]
- Guo, T.; Guo, M.; Li, C.; Wang, Z.; Zheng, W.; Xu, J.; Zhao, X.; Wang, X. Combination scale, variety selection probability and parental selection of soybean crossbreeding. Heilongjiang Agric. Sci. 2024, 4, 1–10. (In Chinese) [Google Scholar]
- Reddy, B.V.S.; Ramesh, S.; Reddy, P.S.; Ramaiah, B. Combining ability and heterosis as influenced by male-sterility inducing cytoplasms in sorghum [Sorghum bicolor(L.) Moench]. Euphytica 2007, 154, 153–164. [Google Scholar] [CrossRef]
- Rodríguez-Llanes, Y.; Pérez-Brito, D.; Guzmán-Antonio, A.; Mijangos-Cortés, J.O.; Iglesias-Andreu, L.G.; Canto- Flick, A.; Avilés-Viñas, S.A.; Pijeira-Fernández, G.; Santana-Buzzy, N. Combining ability, heterosis, and heterobeltiosis to select highly productive F1 hybrids of habanero pepper (Capsicum chinense Jacq.). Plant Genet. Res. 2023, 21, 1–11. [Google Scholar] [CrossRef]
- Chen, J.X.; Zhou, H.; Xie, W.B.; Xia, D.; Gao, G.J.; Zhang, Q.L.; Wang, G.W.; Lian, X.M.; Xiao, J.H.; He, Y.Q. Genome-wide association analyses reveal the genetic basis of combining ability in rice. Plant Biotechnol. J. 2019, 17, 2211–2222. [Google Scholar] [CrossRef]
- Muneera Parveen, A.B.; Muthupandi, M.; Kumar, N.; Chauhan, S.S.; Vellaichamy, P.; Senthamilselvam, S.; Rajasugunasekar, D.; Nagarajan, B.; Mayavel, A.; Bachpai, V.K.W.; et al. Quantitative genetic analysis of wood property traits in bipa rental population of Eucalyptus camaldulensis × E. tereticornis. J. Genet. 2021, 100, 46. [Google Scholar] [CrossRef]
- Vázquez-González, C.; López-Goldar, X.; Alía, R.; Bustingorri, G.; José, L.F.; Lema, M.; Mata, R.; Sampedro, L.; Touza, R.; Zas, R. Genetic variation in resin yield and covariation with tree growth in maritime pine. For. Ecol. Manag. 2021, 482, 118843. [Google Scholar] [CrossRef]
- Yousaf, M.F.; Tomar, V.; Romé, H.; Bagge, M.; Timmermann, M.; Chu, T.T.; Jensen, J. Rate of double reduction and genetic variability in yield, quality, and senescence related traits in tetraploid potato (Solanum tuberosum L.). Front. Plant Sci. 2025, 16, 1560123. [Google Scholar] [CrossRef]
- Gedil, M.; Menkir, A. An integrated molecular and conventional breeding scheme for enhancing genetic gain in maize in Africa. Front. Plant Sci. 2019, 10, 1430. [Google Scholar] [CrossRef]
- Charcosset, A.; Lefort-Buson, M.; Gallais, A. Relationship between heterosis and heterozygosity at marker loci: A theoretical computation. Theor. Appl. Genet. 1991, 81, 571–575. [Google Scholar] [CrossRef]
- Sun, X.P.; Jiao, C.; Schwaninger, H.D.; Chao, C.T.; Ma, Y.M.; Duan, N.B.; Khan, A.; Ban, S.; Xu, K.N.; Cheng, L.L.; et al. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 2020, 52, 1423–1432. [Google Scholar] [CrossRef]
- Li, M.; Shi, J.S.; Gan, S.M.; He, Z.X.; Li, L.; Yi, N.J. Correlation between RAPD-based parental genetic distance and filial performance of Chinese fir. For. Res. 2001, 14, 35–40. (In Chinese) [Google Scholar]
- Kopp, R.F.; Smart, L.B.; Maynard, C.A.; Tuskan, G.A.; Abrahamson, L.P. Predicting within family variability in juvenile height growth of Salix based upon similarity among parental AFLP fingerprints. Theor. Appl. Genet. 2002, 105, 106–112. [Google Scholar] [CrossRef]
- Legarra, A.; Gonzalez-Dieguez, D.O.; Charcosset, A.; Vitezica, Z.G. Impact of interpopulation distance on dominance variance and average heterosis in hybrid populations within species. Genetics 2023, 224, iyad059. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Liu, H.; Wang, H.; Lu, Z.; Wang, Y.; Mullan, D.; Hamblin, J.; Liu, C. Accelerated generation of selfed pure line plants for gene identification and crop breeding. Front. Plant Sci. 2017, 8, 1786. [Google Scholar] [CrossRef]
- Keller, B.; Soto, J.; Steier, A.; Portilla-Benavides, A.E.; Raatz, B.; Studer, B.; Walter, A.; Muller, O.; Urban, M.O. Linking photosynthesis and yield reveals a strategy to improve light use efficiency in a climbing bean breeding population. J. Exp. Bot. 2024, 75, 901–916. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.D.; Asyraf Md Hatta, M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhao, Y.; Zhang, Y.; Wang, R.; Li, X.; Liu, Z.; Wang, W.; Zhu, S.; Fan, Y.; Xu, L.; et al. Insights into the genomic divergence of maize heterotic groups in China. J. Integr. Plant Biol. 2025, 67, 1467–1486. [Google Scholar] [CrossRef]
- Zhu, Z.D. Forest Tree Genetics Foundation; Forestry Publishing House: Beijing, China, 1989; pp. 190–191. [Google Scholar]
- Kang, X.Y. On conventional and unconventional tree breeding and their relationships. J. Beijing For. Univ. 2023, 48, 1–7. (In Chinese) [Google Scholar]
- Delpierre, N.; Guillemot, J.; Dufrêne, E.; Cecchini, S.; Nicolas, M. Tree phenological ranks repeat from year to year and correlate with growth in temperate deciduous forests. Agric. For. Meteorol. 2017, 234, 1–10. [Google Scholar] [CrossRef]
- Rizvi, R.H.; Gupta, V.K.; Gurunathan, N.; Datta, A. Age-age correlation models for early selection of Azadirachta indica provenances planted in semi-arid region. Indian J. Agrofor. 2012, 14, 1–5. [Google Scholar]
- Mwase, W.F.; Savill, P.S.; Hemery, G. Genetic parameter estimates for growth and form traits in common ash (Fraxinus excelsior L.) in a breeding seedling orchard at Little Wittenham in England. New For. 2008, 36, 225–238. [Google Scholar] [CrossRef]
- Fu, M.; Liang, R.; Wang, P.; Feng, Q.; Li, C.; Xin, Y.; Zhan, Y.; Zeng, F. Evaluation and selection of superior Fraxinus mandshurica families for genetic improvement using growth traits, morphological characteristics, and wood properties. Euphytica 2025, 221, 86. [Google Scholar] [CrossRef]
- Wei, X.; Chen, M.J.; Zhang, Q.; Gong, J.Y.; Liu, J.; Yong, K.C.; Wang, Q.; Fan, J.J.; Chen, S.H.; Hua, H.; et al. Genomic investigation of 18,421 lines reveals the genetic architecture of rice. Science 2024, 385, eadm8762. [Google Scholar] [CrossRef]
- Weng, Y.; Ford, R.; Tong, Z.; Krasowski, M. Genetic parameters for bole straightness and branch angle in jack pine estimated using linear and generalized linear mixed models. For. Sci. 2017, 63, 111–117. [Google Scholar] [CrossRef]
- Hochholdinger, F.; Yu, P. Molecular concepts to explain heterosis in crops. Trends Plant Sci. 2025, 30, 95–104. [Google Scholar] [CrossRef]
- Ene, C.O.; Ogbonna, P.E.; Agbo, C.U.; Chuakwudi, U.P. Heterosis and combining ability in cucumber (Cucumis sativus L.). Inf. Process. Agric. 2019, 6, 150–157. [Google Scholar] [CrossRef]
- Rahimi, M.; Rabiei, B.; Samizadeh, H.; Ghasemi, A.K. Combining ability and heterosis in rice (Oryza sativa L.) cultivars. J. Agric. Sci. Technol. 2010, 12, 223–231. [Google Scholar]
- Han, Y.Y.; Wang, K.Y.; Liu, Z.Q.; Pan, S.H.; Zhao, X.Y.; Zhang, Q.; Wang, S.F. Research on hybrid crop breeding information management system based on combining ability analysis. Sustainability 2020, 12, 4938. [Google Scholar] [CrossRef]
- Yu, K.C.; Wang, H.; Liu, X.G.; Xu, C.; Li, Z.W.; Xu, X.J.; Liu, J.C.; Wang, Z.H.; Xu, Y.B. Large-scale analysis of combining ability and heterosis for development of hybrid maize breeding strategies using diverse germplasm resources. Front. Plant Sci. 2020, 11, 660. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.Y.; Wang, K.L.; Xie, Y.H.; Wang, A.N.; Zhong, H.Q.; Yao, X.H.; Lin, P. Combining ability from complete diallel design of Camellia oleifera: Implications for the utility of GCA and SCA in oil-related traits breeding. Ind. Crops Prod. 2023, 205, 117434. [Google Scholar] [CrossRef]
- Bai, F.Y.; Kang, N.; Zhang, P.D.; Kang, X.Y. Selection of female parents with high fertility and high combining abilities for cross-breeding Populus tomentosa. J. For. Res. 2019, 30, 445–450. [Google Scholar] [CrossRef]
- Josue, A.D.L.; Brewbaker, J.L. Diallel analysis of grain filling rate and grain filling period in tropical maize (Zea mays L.). Euphytica 2018, 214, 39. [Google Scholar] [CrossRef]
- Ulmakanth, A.V.; Datil, I.V.; Rani, C.; Gadalch, C.R.; Kumar, C.C.; Ran, C.C.; Kotarthana, T.V. Comhininn ahility and hataro-cir ouar anuironmante for ctalk and cunar ralatad traite in cua et sorghum (Sorghum bicolor (L) Moench). Sugar Technol. 2012, 14, 237–246. [Google Scholar]
- Chai, J.Y.; Wang, K.L.; Yao, X.H.; Teng, J.H.; Lin, P. Genetic analysis of the fruit and oil related traits on hybrid offspring of nested mating of camellia oleifera. For. Res. 2023, 36, 1–10. (In Chinese) [Google Scholar]
- Zhang, Q.; Gao, Y.J.; Yang, S.H.; Ragab, R.A.; Maroof, M.A.; Li, Z.B. A diallel analysis of heterosis in elite hybrid rice based on RFLP and microsatellite. Theor. Appl. Genet. 1994, 89, 185–192. [Google Scholar] [CrossRef]
- Li, S.W.; Zhang, Z.Y.; Yu, Z.S.; He, C.Z.; An, X.M.; Li, B.L. Correlation between molecular genetic distances among parents and growth traits of progenies in Populus. For. Res. 2008, 44, 150–154. (In Chinese) [Google Scholar]
- Zhang, Y.; Yang, Q.; Zhou, Z.C.; Jin, G.Q. Divergence among masson pine parents revealed by geographical origins and SSR markers and their relationships with progeny performance. New For. 2013, 44, 341–355. [Google Scholar] [CrossRef]
- Li, Y.L.; Zhao, F.C.; Wu, H.S.; Zhang, Y.Z.; Li, F.M.; Zhong, S.Y.; Li, X.Z.; Cai, J. Relationship between growth traits heterosis and genetic distance among parents of Pinus elliottii × P. caribaea based on SSR molecular markers. For. Res. 2012, 25, 138–143. (In Chinese) [Google Scholar]
- Akaogu, I.C.; Badu-Apraku, B.; Adetimirin, V.O.; Vroh-Bi, I.; Oyekunle, M.; Akinwale, R.O. Genetic diversity assessment of extra-early maturing yellow maize inbreds and hybrid performance in Striga-infested and Striga-free environments. J. Agric. Sci. 2013, 151, 519–537. [Google Scholar] [CrossRef]
- Nyaligwa, L.; Hussein, S.; Amelework, B.; Ghebrehiwot, H. Genetic diversity analysis of elite maize inbred lines of diverse sources using SSR markers. Maydica 2015, 60, M29. [Google Scholar]
- Sserumaga, J.P.; Ji, H.; Njoroge, K.; Muthomi, J.; Chemining’wa, G.; Si-myung, L.; Kim, H.; Asea, G.; Makumbi, D. Molecular characterization of tropical maize inbred lines using microsatellite DNA markers. Maydica 2014, 59, 267–274. [Google Scholar]
- Moriguchi, Y.; Yamazaki, Y.; Taira, H.; Tsumura, Y. Mating patterns in an indoor miniature Cryptomeria japonica seed orchard as revealed by microsatellite markers. New For. 2010, 39, 261–273. [Google Scholar] [CrossRef]
- Arcade, A.; Faivre-rampant, P.; Le Guerroue, B.; Pâques, L.E.; Prat, D. Heterozygosity and hybrid performance in larch. Theor. Appl. Genet. 1996, 93, 1274–1281. [Google Scholar] [CrossRef] [PubMed]
- He, J.W.; Jiang, H.; Yang, H.; Su, D.; Liu, Y.X. Relationship between SSR genetic distance and heterosis in hot pepper. Agric. Sci. Technol. 2013, 14, 1224–1227. [Google Scholar]
- Singh, S.; Gupta, S.K. Formation of heterotic pools and understanding relationship between molecular divergence and heterosis in pearl millet [Pennisetum glaucum (L.) R. Br.]. PLoS ONE 2019, 14, e0207463. [Google Scholar] [CrossRef]
- Napolitano, M.; Terzaroli, N.; Kashyap, S.; Russi, L.; Jones-Evans, E.; Albertini, E. Exploring Heterosis in Melon (Cucumis melo L.). Plants 2020, 9, 282. [Google Scholar] [CrossRef]
- Gupta, S.K.; Nepolean, T.; Shaikh, C.G.; Rai, K.; Hash, C.T.; Das, R.R.; Rathore, A. Phenotypic and molecular diversity-based prediction of heterosis in pearl millet (Pennisetum glaucum L. (R.) Br.). The Crop J. 2018, 6, 271–281. [Google Scholar] [CrossRef]
- Tian, H.Y.; Channa, S.A.; Hu, S.W. Relationships between genetic distance, combining ability and heterosis in rapeseed (Brassica napus L.). Euphytica 2016, 213, 1. [Google Scholar] [CrossRef]
- Snehi, S.; Prakash, N.R.; Pant, U.; Singh, P.K.; Kumar, S.; Jeena, A.S.; Bhajan, R. Prediction of heterotic combinations using correlation between genetic distance, heterosis and combining ability in yellow sarson (Brassica rapa var. yellow sarson Prain). Vegetos 2024, 37, 1552–1564. [Google Scholar] [CrossRef]
- Ali, M.; Copeland, L.O.; Elias, S.G.; Kelly, J.D. Relationship between genetic distance and heterosis for yield and morphological traits in winter canola (Brassica napus L.). Theor. Appl. Genet. 1995, 91, 118–121. [Google Scholar] [CrossRef]
- Wang, S.H. Effects of Different Amendments on Salt Removal and Organic Matter Enhancement of Saline-Alkali Soil in the Yellow River Delta. Master’s Thesis, Nanjing Agricultural University, Nanjing, China, 2021. (In Chinese). [Google Scholar]
- Ding, H.; Zheng, T.; Liang, D.; Qu, G. A simple and efficient method of DNA extraction from different plant samples. Bull. Bot. Res. 2015, 35, 457–461. (In Chinese) [Google Scholar]
- Yan, L.P.; Wu, D.J.; Mao, X.H.; Yao, J.X.; Ren, F.; Li, S.W.; Wang, K.F.; Wang, Y.H.; Liu, C.L. Construction of core collection of Fraxinus based on SSR molecular markers. J. Cent. South Univ. For. Technol. 2019, 39, 1–9. (In Chinese) [Google Scholar]
- Dong, H.Y.; Liu, Q.H.; Zhou, Z.C.; Jin, G.Q.; Shen, D.Y.; Song, X.H. Correlation between heterosis in the growth of progeny and combining ability and genetic distance of the parents for Pinus massoniana. For. Res. 2017, 53, 65–75. (In Chinese) [Google Scholar]
- Xu, J.R. Trees Quantitative Genetics; Higher Education Press: Beijing, China, 2006; pp. 42–43. [Google Scholar]
- Mohamed, L.M.; Mimoun, M.; Peter, J.M.; Eric, N.J.; Ouafae, B. Morphological variability, heritability and correlation studies within an argan tree population (Argania spinosa (L.) Skeels) preserved in situ. Int. J. Agric. For. 2017, 7, 42–51. [Google Scholar]
- Liu, Z.B.; Jiang, J.B.; Ren, A.; Xu, X.Y.; Zhang, H.; Zhao, T.T.; Jiang, X.M.; Sun, Y.G.; Li, J.F.; Yang, H.H. Heterosis and combining ability analysis of fruit yield, early maturity, and quality in tomato. Agronomy 2021, 11, 807. [Google Scholar] [CrossRef]
- Wang, M.X. Forest Tree Genetic Breeding; China Forestry Publishing House: Beijing, China, 2001. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).