The Scarlet Alchemy of Survival: Integrated Transcriptomic and Metabolomic Analysis of Leaf Coloration in Endangered Parrotia subaequalis
Abstract
1. Introduction
2. Results
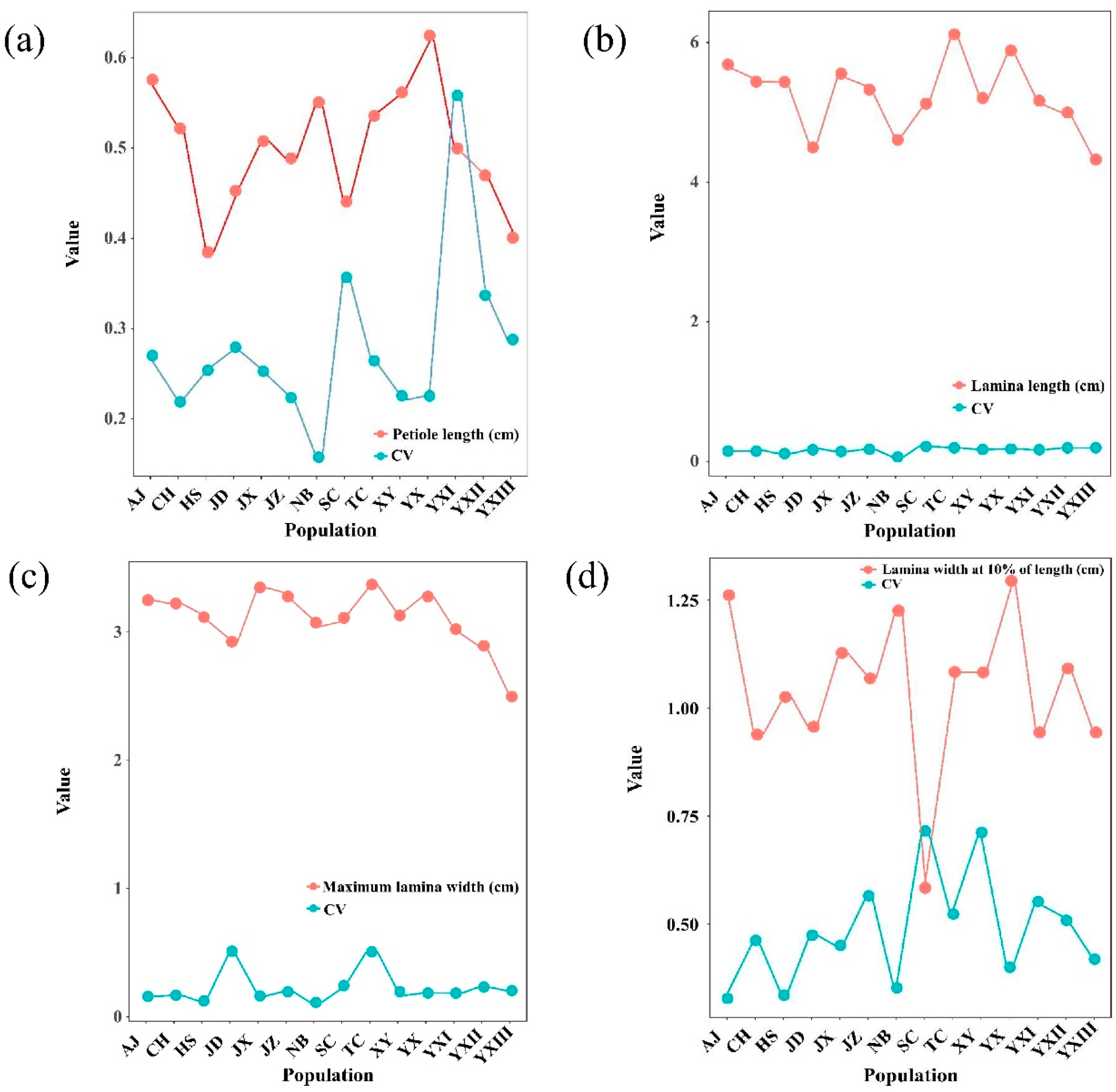
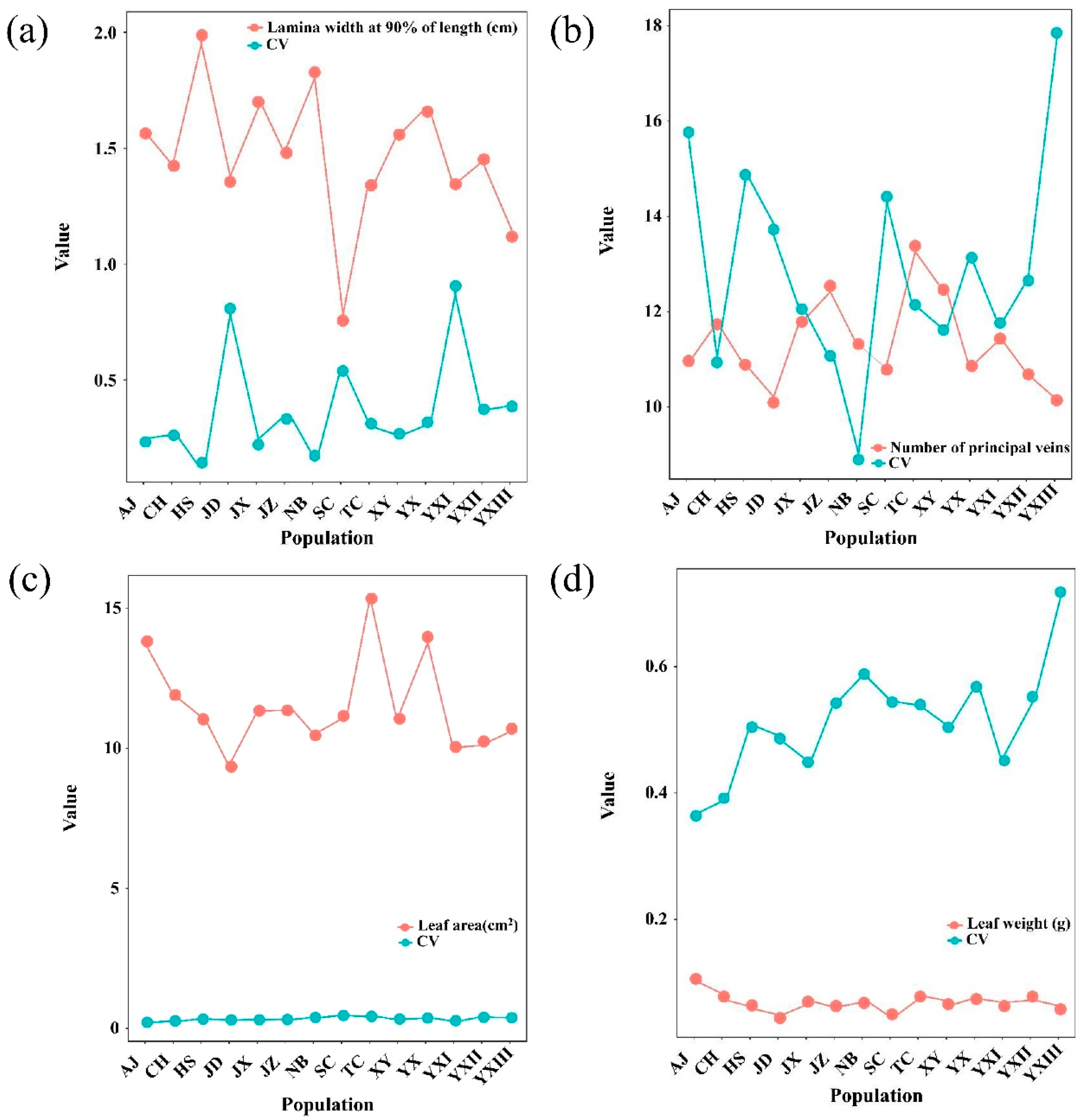
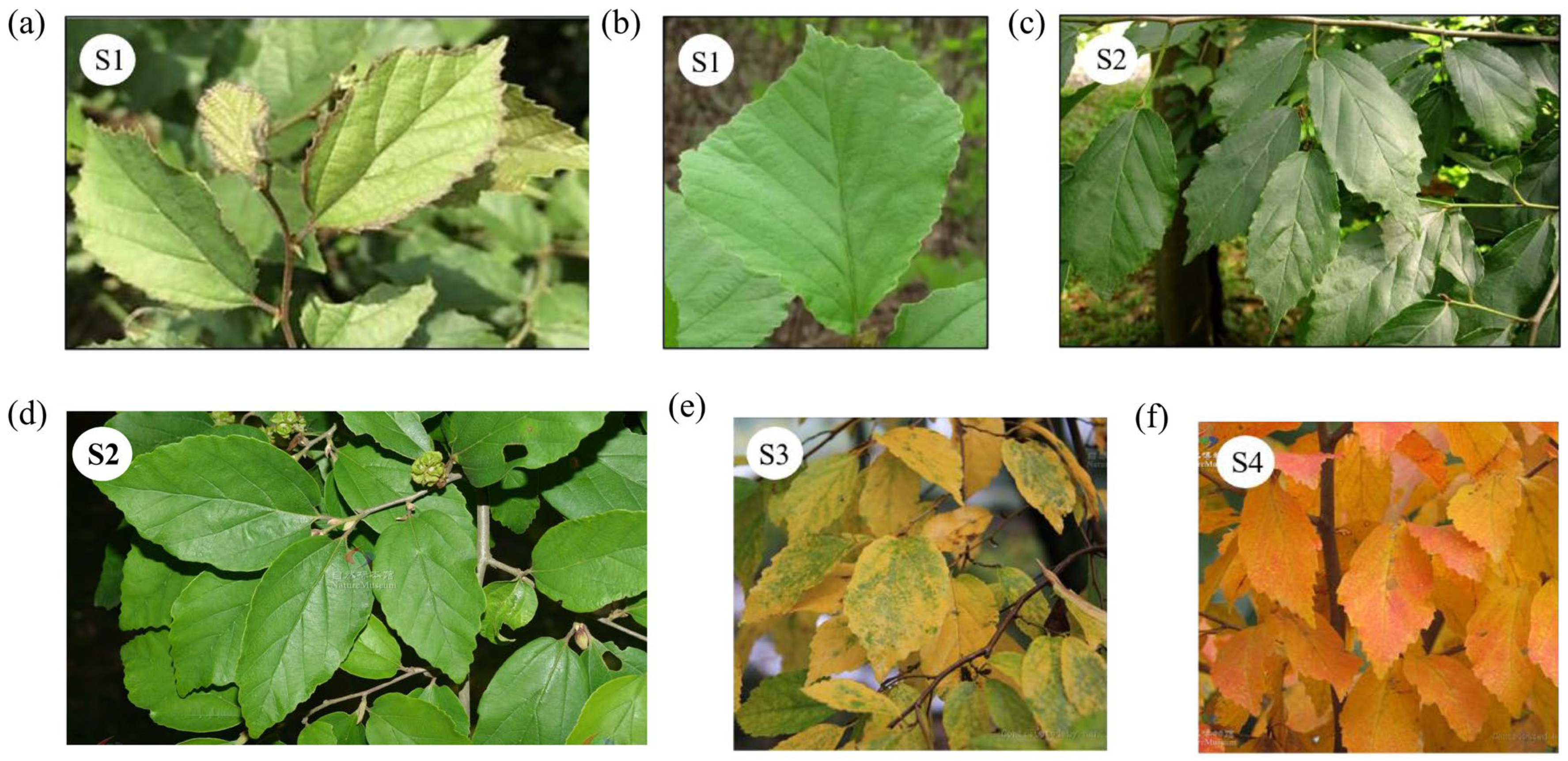
2.1. Phenotypic Variation in Leaves Across Different Populations
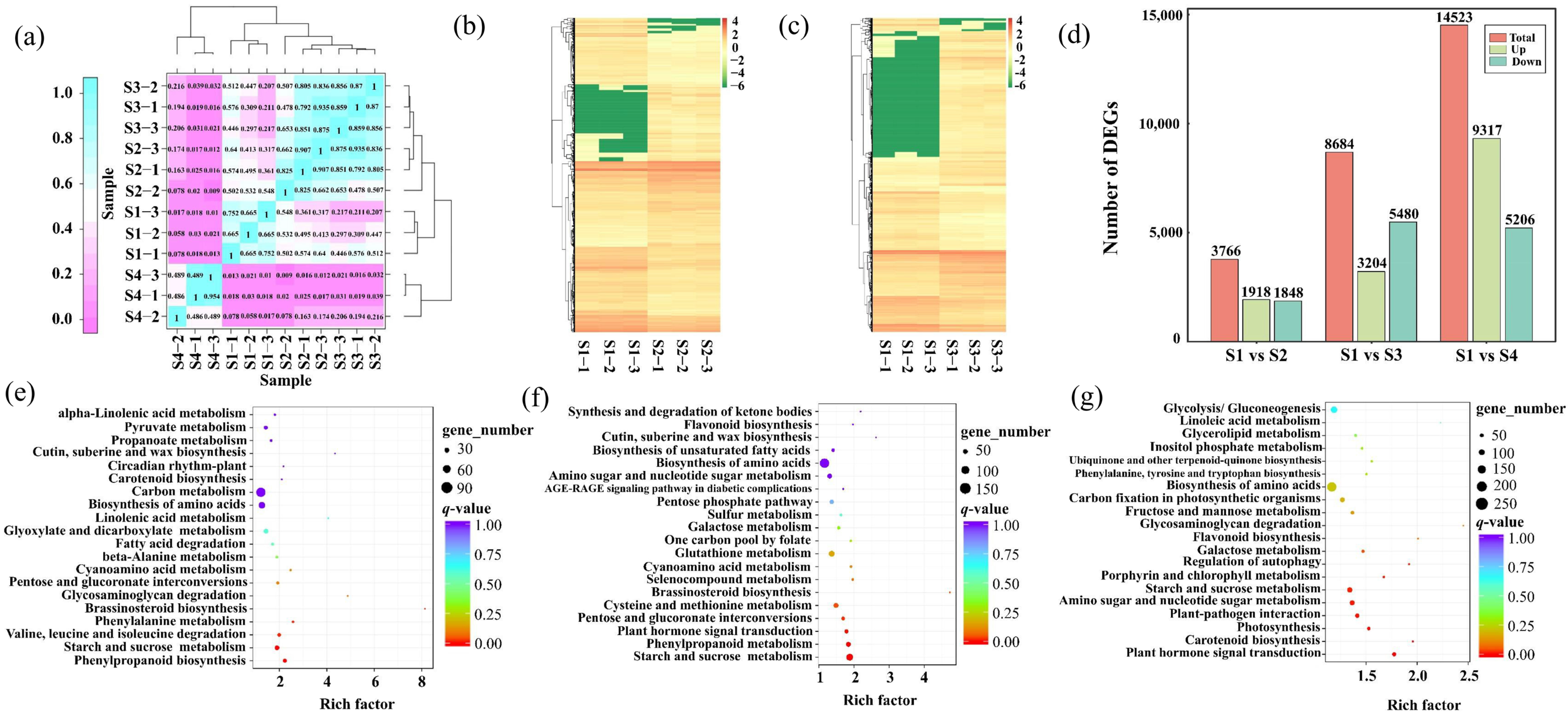
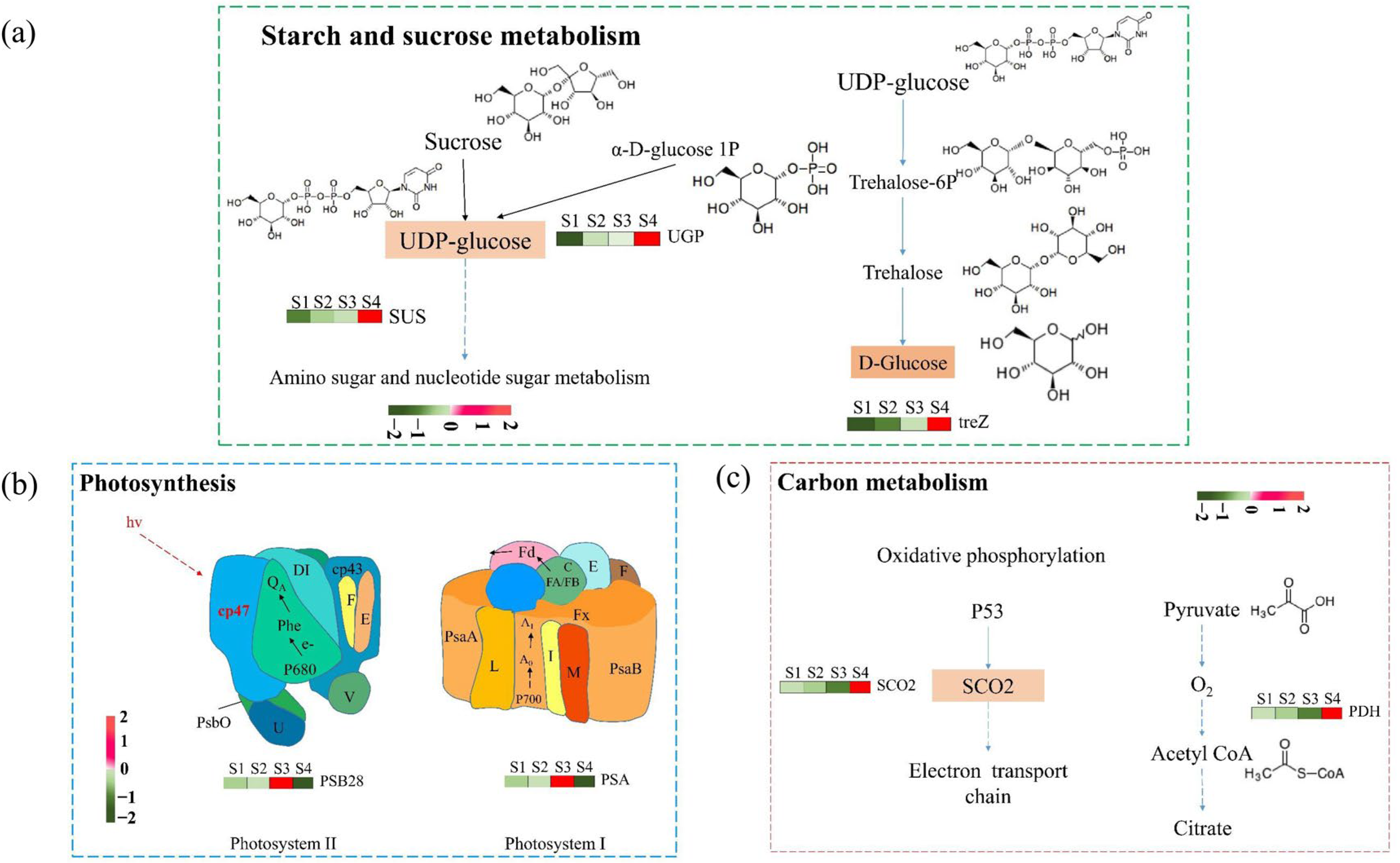
2.2. Differentiation of Gene Expression of Leaf Pigment Change
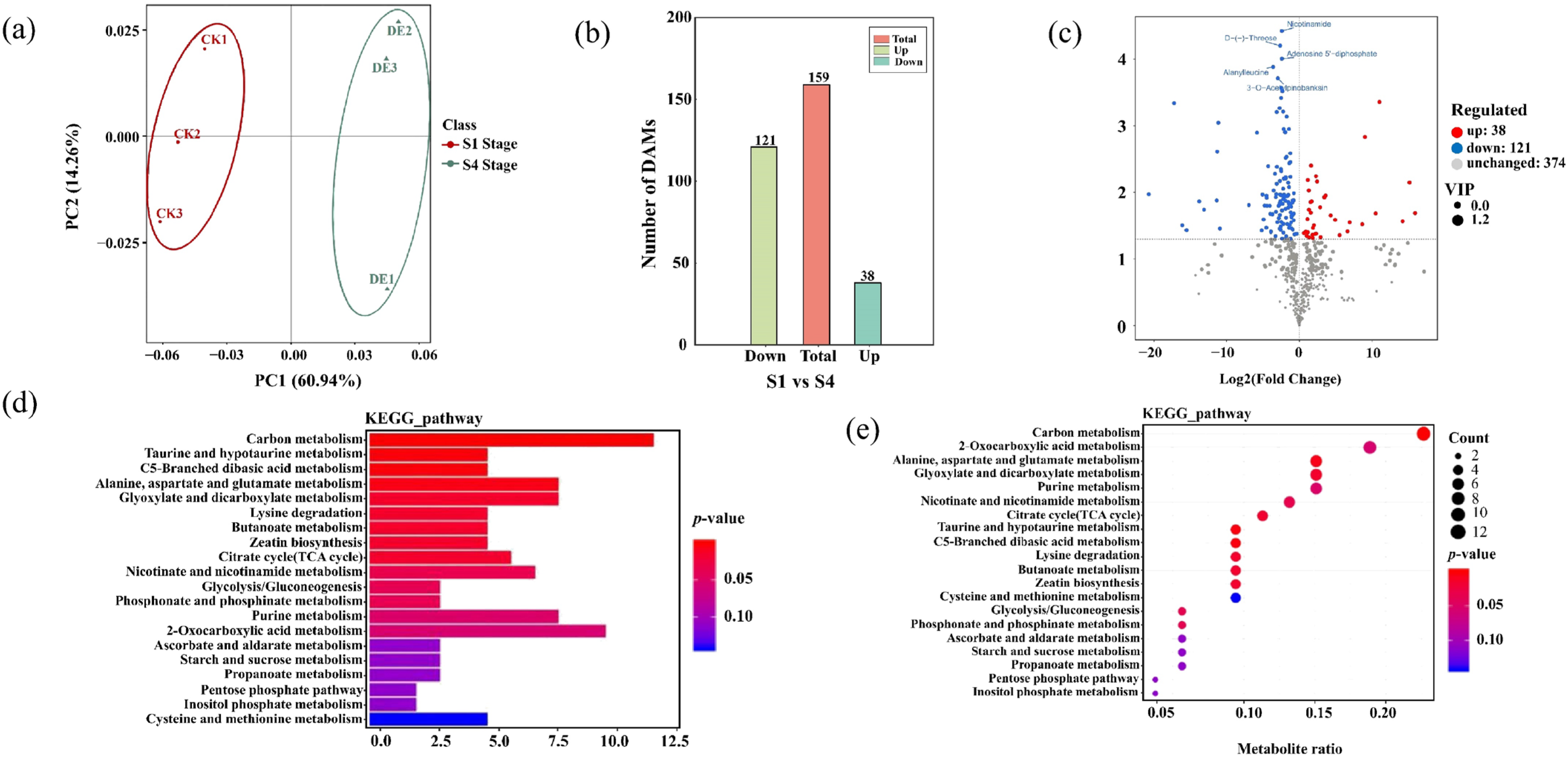
2.3. Metabolome Profiling of P. subaequalis
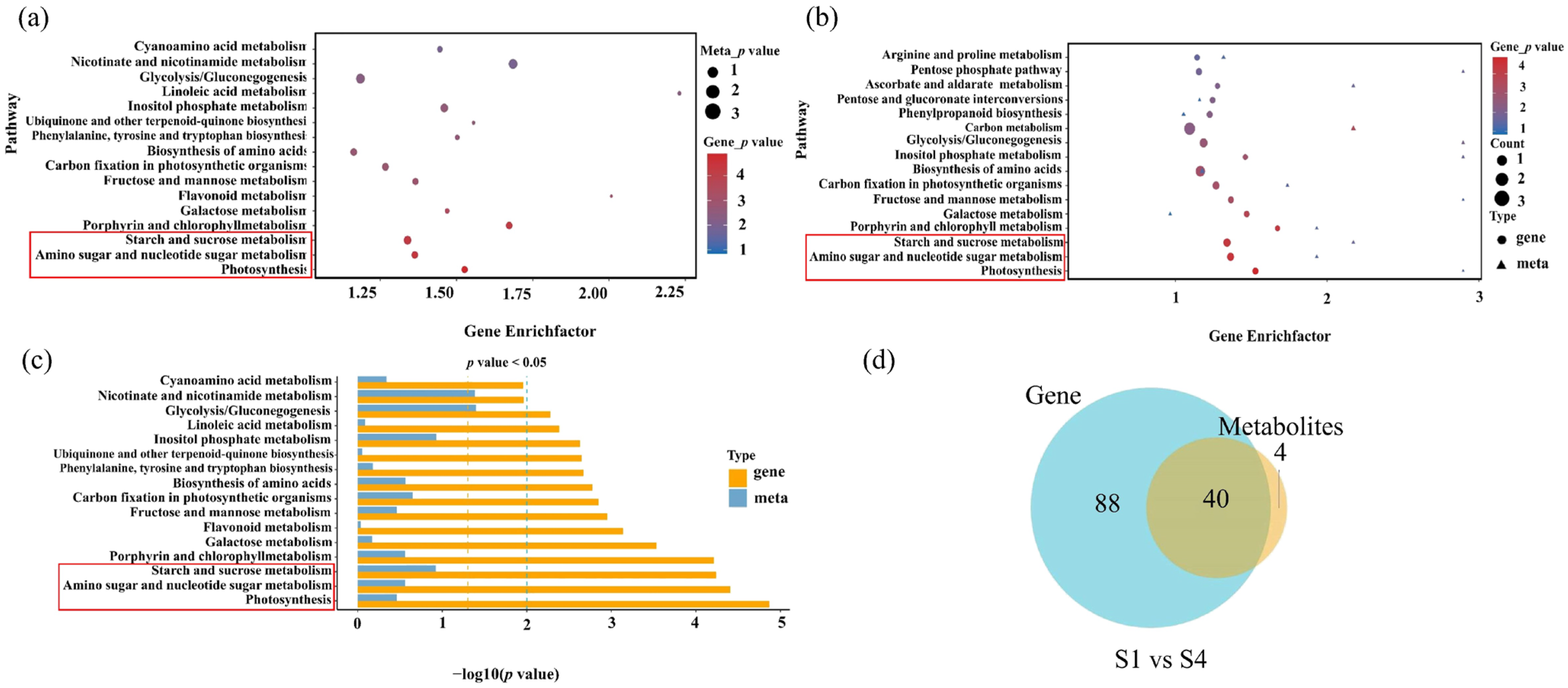
2.4. Correlated Transcriptome and Metabolome Analyses of Leaf Pigment Change
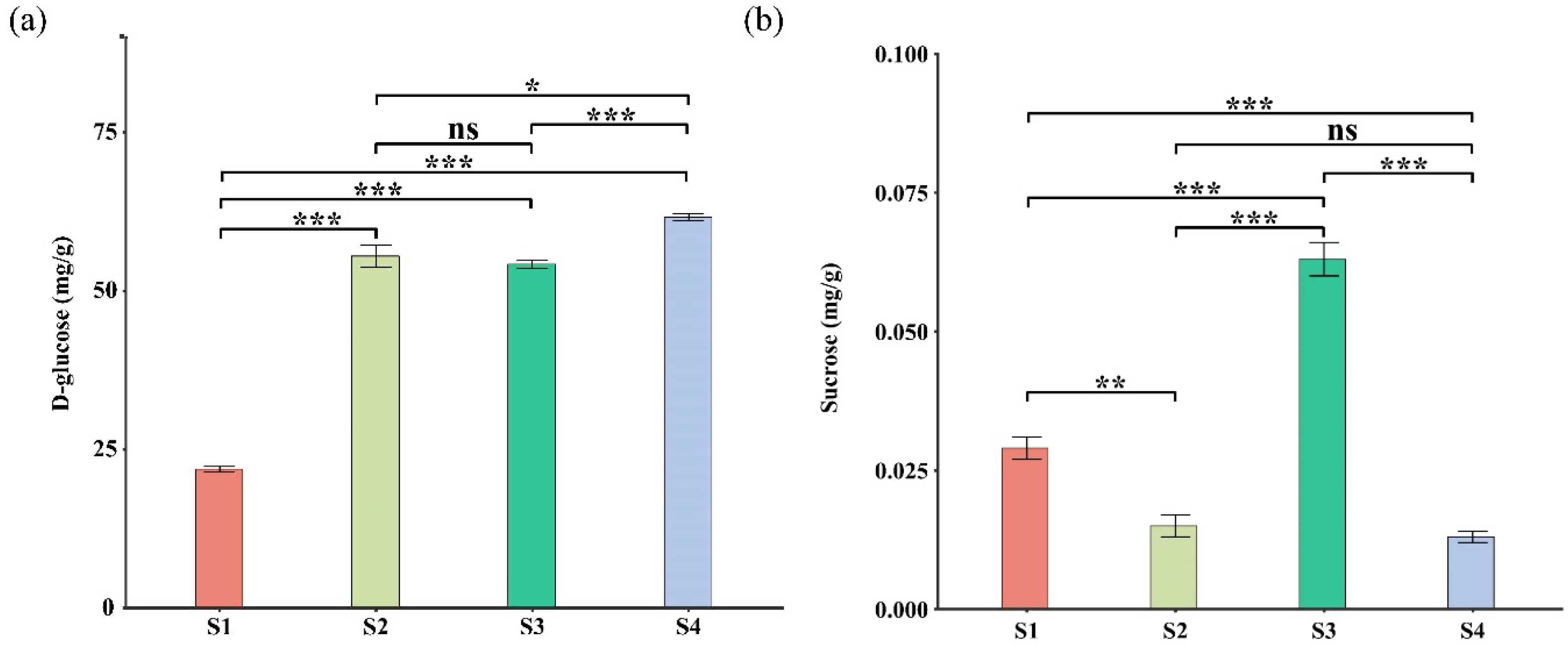
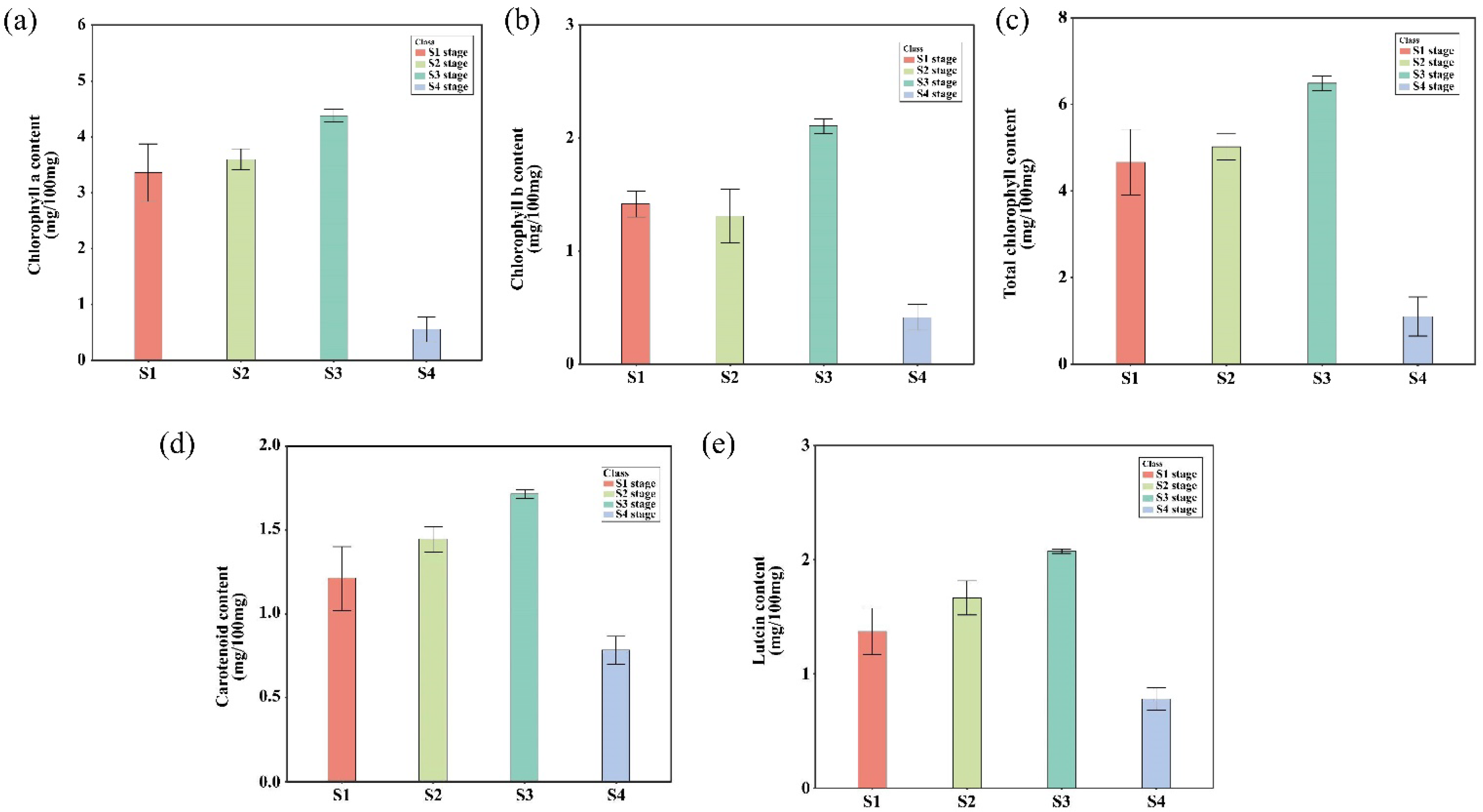
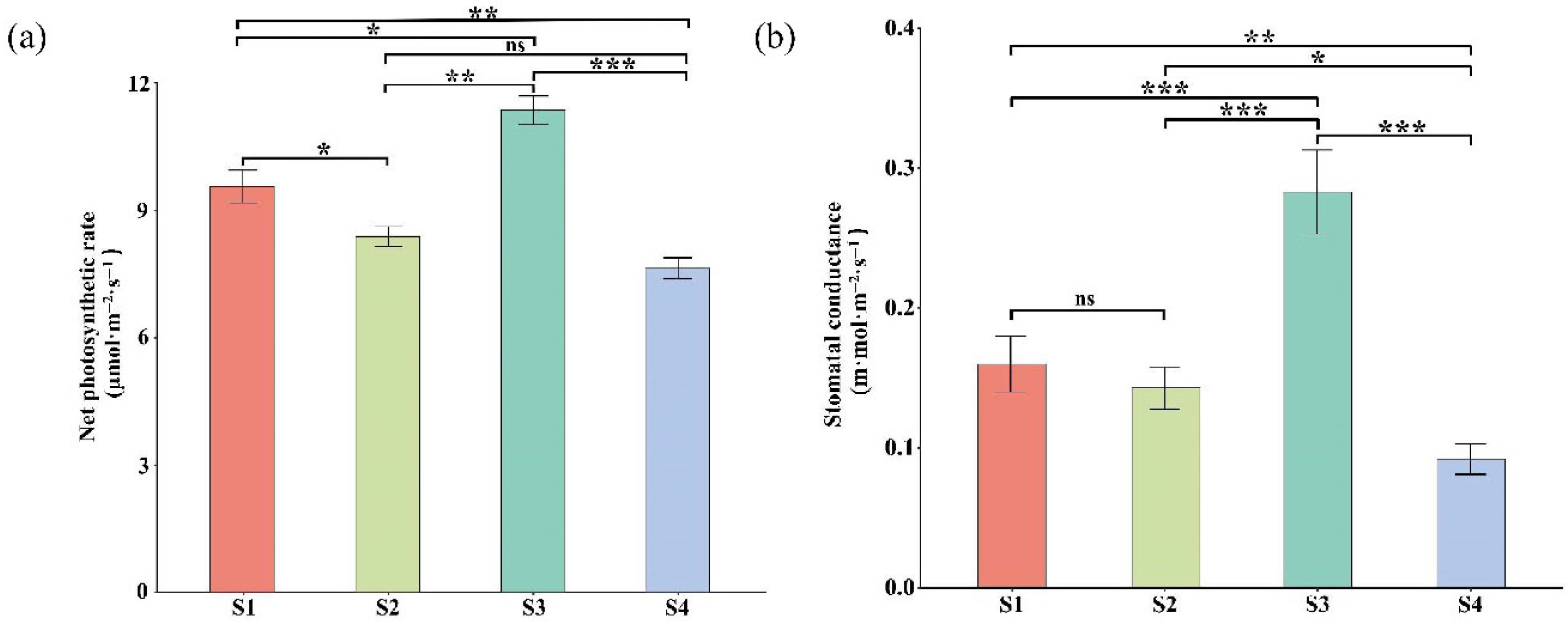
2.5. Leaf Pigment Change Enhanced Photosynthetic Pigments Content and Organic Compound
3. Discussion
3.1. Phenotypic Variation in Leaves Across P. subaequalis Populations
3.2. The Genes and Metabolites Involved in Leaf Pigmentation of P. subaequalis
4. Materials and Methods
4.1. Plant Material
4.2. Phenotypic Measurements
4.3. Physiological Measurements
4.4. Transcriptome Sequencing and Analysis
4.5. Metabolomic Analysis
4.6. Correlation Analysis of Transcriptome and Metabolome
4.7. qRT-PCR
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Y.; He, N.P.; Hou, J.H.; Li, X.; Liu, C.C.; Zhang, J.H.; Wang, Q.; Zhang, X.; Wu, X. Factors influencing leaf chlorophyll content in natural forests at the biome scale. Front. Ecol. Evol. 2018, 6, 64. [Google Scholar] [CrossRef]
- Yang, W.X.; Yang, S.B.; He, Z.C.; Jin, B. Research advances in the regulatory mechanisms of leaf coloration. Acta Hort. Sin. 2017, 44, 14. [Google Scholar]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Korban, S.S. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in f lowers, leading to loss of anthocyanin. J. Exp. Bot. 2012, 63, 2437–2447. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ge, Y.; Xie, X.; Atefi, A.; Wijewardane, N.K.; Thapa, S. High throughput analysis of leaf chlorophyll content in sorghum using RGB, hyperspectral, and fluorescence imaging and sensor fusion. Plant Methods 2022, 18, 60. [Google Scholar] [CrossRef]
- Umakanta, S.; Shinya, O. Augmentation of leaf color parameters, pigments, vitamins, phenolic acids, flavonoids and antioxidant activity in selectedamaranthus tricolorundersalinity stress. Sci. Rep. 2018, 8, 12349. [Google Scholar]
- Huang, D.; Wang, X.; Tang, Z.Z.; Yuan, Y.; Xu, Y.T. Subfunctionalization of the ruby2-ruby1 gene cluster during the domestication of citrus. Nat. Plants 2018, 4, 930. [Google Scholar] [CrossRef]
- Lee, D.W. Anthocyanins in autumn leaf senescence. Adv. Bot. Res. 2002, 37, 147–165. [Google Scholar]
- Kornyushenko, G.A.; Solov’eva, L.V. The ecological analysis of pigment contents in the leaves of mountain-tundra herb. Bot. Zhurnal 1994, 79, 80–101. [Google Scholar]
- Biancalani, T.; Scalia, G.; Buffoni, L.; Avasthi, R.; Lu, Z.; Sanger, A.; Tokcan, N.; Vanderburg, C.R.; Segerstolpe, A.; Zhang, M.; et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with tangram. Nat. Methods 2021, 18, 1352. [Google Scholar] [CrossRef]
- Migliavacca, M.; Musavi, T.; Mahecha, M.D.; Nelson, J.A.; Knauer, J.; Baldocchi, D.D.; Perez-Priego, O.; Christiansen, R.; Jonas Peters, J.; Anderson, K.; et al. The three major axes of terrestrial ecosystem function. Nature 2021, 598, 468–472. [Google Scholar] [CrossRef]
- Chen, H.; Chiu, T.Y.; Sahu, S.K.; Sun, H.; Wen, J.; Sun, J.; Li, Q.; Tang, Y.; Jin, H.; Liu, H. Transcriptomic analyses provide new insights into green and purple color pigmentation in Rheum tanguticum medicinal plants. PeerJ 2022, 10, e14265. [Google Scholar] [CrossRef]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technology. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef] [PubMed]
- Reid, N.M.; Proestou, D.A.; Clark, B.W.; Warren, W.C.; Colbourne, J.K.; Shaw, J.R.; Karchner, S.I.; Hahn, M.E.; Nacci, D.; Oleksiak, M.F.; et al. The genomic landscape of rapid repeated evolutionary adaptation to toxic pollution in wild fish. Science 2016, 354, 1305–1308. [Google Scholar] [CrossRef] [PubMed]
- Burda, J.E.; O’Shea, T.M.; Ao, Y.; Suresh, K.B.; Wang, S.; Bernstein, A.M.; Chandra, A.; Deverasetty, S.; Kawaguchi, R.; Kim, J.H. Divergent transcriptional regulation of astrocyte reactivity across disorders. Nature 2022, 606, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, L.; Wang, J.; Zhang, M.; Song, Z.; Ni, B.; You, Y. Correction to: Identification of key biomarkers and immune infiltration in systemic lupus erythematosus by integrated bioinformatics analysis. J. Transl. Med. 2021, 19, 64. [Google Scholar] [CrossRef]
- Fu, M.; Yang, X.; Zheng, J.; Wang, L.; Xu, F. Unraveling the regulatory mechanism of color diversity in camellia japonica petals by integrative transcriptome and metabolome analysis. Front. Plant Sci. 2021, 12, 685136. [Google Scholar] [CrossRef]
- Islam, E.S.; Dong, L.; Kenong, X. Transcriptome analysis of an apple (malus × domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. J. Exp. Bot. 2015, 22, 7359–7376. [Google Scholar]
- Huang, H.; Huang, W.; Zhang, Z.; An, B.; Zhang, B.; Zhao, L. Integrated analysis of the metabolome and transcriptome provides insights into anthocyanin biosynthesis of cashew apple. Food Res. Int. 2024, 175, 113711. [Google Scholar] [CrossRef]
- Chen, Y.; Li, E.M.; Xu, L.Y. Guide to metabolomics analysis: A bioinformatics workflow. Metabolites 2022, 12, 357. [Google Scholar] [CrossRef]
- Utpott, M.; Rodrigues, E.; Rios, A.D.O.; Mercali, G.D.; Flres, S.H. Metabolomics: An analytical technique for food processing evaluation. Food Chem. 2021, 5, 130685. [Google Scholar] [CrossRef]
- Li, S.; Tian, Y.; Jiang, P.; Lin, Y.; Yang, H. Recent advances in the application of metabolomics for food safety control and food quality analyses. Crit. Rev. Food Sci. Nutr. 2020, 61, 1448–1469. [Google Scholar] [CrossRef]
- Liu, H.W.; Brettell, L.E.; Qiu, Z.G.; Singh, B.K. Microbiome-mediated stress resistance in plants. Trends Plant Sci. 2020, 25, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Saito, K. Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Biol. 2015, 24, 10–16. [Google Scholar] [CrossRef]
- He, G.R.; Zhang, R.; Jiang, S.H.; Wang, H.H.; Ming, F. The MYB transcription factor RcMYB1 plays a central role in rose anthocyanin biosynthesis. Hortic. Res. 2023, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.H.; Kim, C.K. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant. 2021, 172, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB-bHLH-WD Repeat (MBW) pigment regulatory model: Addition of a WRKY Factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Xie, X.B.; Li, S.; Zhang, R.F.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Yao, Y.X.; You, C.X.; Zhang, X.S.; Hao, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit coloration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, W.; Li, C.; Wang, Z.; Lu, C.; Cheng, J.; Wei, S.; Yang, J.; Yang, Q. Integrated transcriptomic and metabolomic analyses elucidate the mechanism of flavonoid biosynthesis in the regulation of mulberry seed germination under salt stress. BMC Plant Biol. 2024, 5, 24. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, S.H.; Fu, Q.Q.; Wang, Z.D.; Liu, X.J.; Sun, L.L.; Zhao, Z.Y. Transcriptomic and metabolomic analysis reveals a protein module involved in pre-harvest apple peel browning. Plant Physiol. 2023, 192, 2102–2122. [Google Scholar] [CrossRef]
- Yu, X.; Xiao, J.; Chen, S.; Yu, Y.; Ma, J.; Lin, Y.; Li, R.; Lin, J.; Fu, Z.; Zhou, Q.; et al. Metabolite signatures of diverse camellia sinensis tea populations. Nat. Commun. 2020, 11, 5586. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.D. Hamamelidaceae. In Flora Republicae Popularis Sinica; Chang, H.D., Ed.; Science Press: Alexandria, NSW, Australia, 1997; Volume 35, pp. 73–74. [Google Scholar]
- Wang, S.; Xie, Y. China Species Red List; Higher Education Press: Beijing, China, 2004; Volume 1. [Google Scholar]
- Ellie, M.W.; Jayanthi, N.; Yang, X.Y. Plant species with extremely small populations (PSESP) in China: 499 a seed and sporebiology perspective. Plant Divers. 2016, 38, 209–220. [Google Scholar]
- Gong, B.; Xia, Y.J.; Zhang, G.F.; Lu, Y.; Sun, G. Population structure and spatial pattern of Parrotia subaequalis, a rare endangered species endemic to China. J. Ecol. Rural Environ. 2012, 28, 638–646. [Google Scholar]
- Zhang, G.F.; Yao, R.; Jiang, Y.Q.; Chen, F.C.; Zhang, W.Y. Intraspecific and interspecific competition intensity of Parrotia subaequalis in different habitats from Wanfoshan Nature Reserve, Anhui Province. Chin. J. Ecol. 2016, 35, 1744–1750. [Google Scholar]
- Deng, M.B.; Wei, H.T.; Wang, X.Q. Shaniodendron, a new genus of Hamamelidoideae from China. Acta Phytotax. Sin. 1992, 30, 57–61. [Google Scholar]
- Si, G.C.; Zhang, Y.L.; Zhao, B.; Xu, H. Phenotypic variation of natural population in Rhododendron purdomii in Qinling Mountains. Acta Bot. Boreali-Occident. Sin. 2012, 32, 1560–1566. [Google Scholar]
- Li, Z.Y.; Zhang, H.Y. Morphological variation and diversity in populations of Paeonia lutea. J. Northwest For. Univ. 2011, 26, 117–122. [Google Scholar]
- Gallusci, P.; Dai, Z.W.; Genard, M.; Gauffretau, A.; Leblanc-Fournier, N.; Richard-Molard, C.; Vile, D.; Brunel-Muguet, S. Epigenetics for plant improvement: Current knowledge and modeling avenues. Trends Plant Sci. 2017, 22, 610–623. [Google Scholar] [CrossRef]
- Arnold, P.A.; Kruuk, L.E.B.; Nicotra, A.B. How to analyse plant phenotypic plasticity in response to a changing climate. New Phytol. 2019, 222, 1235–1241. [Google Scholar] [CrossRef]
- Meril, J.; Hendry, A. Climate change, adaptation, and phenotypic plasticity: The problem and the evidence. Evol. Appl. 2014, 7, 1–14. [Google Scholar] [CrossRef]
- Chevin, L.M.; Hoffmann, A.A. Evolution of phenotypic plasticity in extreme environments. Philos. Trans. R. Soc. B 2017, 372, 138. [Google Scholar] [CrossRef]
- Oostra, V.; Saastamoinen, M.; Zwaan, B.J.; Wheat, C.W. Strong phenotypic plasticity limits potential for evolutionary responses to climate change. Nat. Commun. 2018, 9, 1005. [Google Scholar] [CrossRef]
- Ji, X.; Tang, J.; Zhang, J. Effects of salt stress on the morphology, growth and physiological parameters of Juglans microcarpa L. Seedlings. Plants 2022, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Jahan, M.S.; Shu, S.; Wang, Y.; Hasan, M.M.; El-Yazied, A.A.; Alabdallah, N.M.; Hajjar, D.; Altaf, M.A.; Sun, J.; Guo, S. Melatonin pretreatment confers heat tolerance and repression of heat-induced senescence in tomato through the modulation of ABA and GA-mediated pathways. Front. Plant Sci. 2021, 12, 650955. [Google Scholar] [CrossRef] [PubMed]
- Dooner, H.K.; Robbins, T.P. Genetic and development control of anthocyanin biosynthesis. Annu. Rev. Genet. 1991, 25, 179–199. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Tao, R.; Yin, L.; Ni, J.; Yang, Q.; Yan, X.; Yang, F.; Guo, X.; Li, H.; Teng, Y. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 2019, 100, 1208–1223. [Google Scholar] [CrossRef]
- Song, X.; Duan, X.; Chang, X.; Xian, L.; Liu, Y. Molecular and metabolic insights into anthocyanin biosynthesis during leaf coloration in autumn. Environ. Exp. Bot. 2021, 190, 104584. [Google Scholar] [CrossRef]
- Pei, Z.; Huang, Y.; Ni, J.; Liu, Y.; Yang, Q. For a colorful life: Recent advances in anthocyanin biosynthesis during leaf senescence. Biology 2024, 13, 329. [Google Scholar] [CrossRef]
- Dizaji, S.Z.; Fariman, G.A.; Zahedi, M.M. Pigment content analysis in two hab forming dinoflagellate species during the growth period. J. Appl. Phycol. 2021, 33, 807–817. [Google Scholar] [CrossRef]
- Kumar, J.V.; Chowdhury, R.S.; Kantamraju, P.; Dutta, S.; Pal, K.; Ghosh, S.; Das, S.; Mandal, R.; Datta, S.; Choudhury, A.; et al. Anthocyanin profiling of genetically diverse pigmented potato (Solanum tuberosum L.) clonal accessions from north-eastern sub-Himalayan plateau of India. Heliyon 2024, 10, e36730. [Google Scholar]
- Li, M.; Wang, Z.; Chen, L.Q.; Wang, J.J.; Li, H.Y.; Han, Y.H.; Zhang, B. The relationship between the photosynthetic pigments, carotenoids and yield of broomcorn millet (panicum miliaceum; Poaceae). Appl. Ecol. Environ. Res. 2021, 19, 191–203. [Google Scholar] [CrossRef]
- Kreslavski, V.D.; Strokina, V.V.; Khudyakova, A.Y.; Shirshikova, G.N.; Kosobryukhov, A.A.; Pashkovskiy, P.P.; Alwasel, S.; Allakhverdiev, S.I. Effect of high-intensity light and UV-B on photosynthetic activity and the expression of certain light-responsive genes in A. thaliana phyA and phyB mutants. Biochim. Biophys. Acta Bioenerg. 2021, 1862, 148445. [Google Scholar] [CrossRef] [PubMed]
- Ashikhmin, A.; Pashkovskiy, P.; Kosobryukhov, A.; Khudyakova, A.; Abramova, A.; Vereshchagin, M.; Bolshakov, M.; Kreslavski, V. The role of pigments and cryptochrome 1 in the adaptation of solanum lycopersicum photosynthetic apparatus to high-intensity blue light. Antioxidants 2024, 13, 605. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Z.; Yuan, L.; Zhou, H.; Hou, X.; Liu, T. Cold acclimation can specifically inhibit chlorophyll biosynthesis in young leaves of Pakchoi. BMC Plant Biol. 2021, 21, 172. [Google Scholar] [CrossRef]
- Wang, F.F.; Chen, N.Z.; Shen, S.H. iTRAQ-based quantitative proteomics analysis reveals the mechanism of golden-yellow leaf mutant in hybrid paper mulberry. Int. J. Mol. Sci. 2021, 23, 127. [Google Scholar] [CrossRef] [PubMed]
- Adeleke, I.A.; Kavalappara, S.R.; Codod, C.B.; Kharel, P.; Luckew, A.; McGregor, C.; Simmons, A.M.; Srinivasan, R.; Bag, S. Evaluation of plant introduction lines of yellow squash (Cucurbita pepo) for resistance against single infection of cucurbit chlorotic yellows virus and cucurbit leaf crumple virus. Hortscience 2024, 59, 949–956. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, P.; Fan, X.; Yang, H. Characterization and rna-seq analysis of yellow-green leaf mutants in tomato. Agronomy 2024, 14, 828. [Google Scholar] [CrossRef]
- Wang, P.; Karg, C.A.; Frey, N.; Frdrich, J.; Vollmar, A.M.; Moser, S. Phyllobilins as a challenging diverse natural product class: Exploration of pharmacological activities. Arch. Pharm. 2021, 354, 1–10. [Google Scholar] [CrossRef]
- Durrett, T.P.; Welti, R. The tail of chlorophyll: Fates for phytol. J. Biol. Chem. 2021, 296, 100802. [Google Scholar] [CrossRef]
- Lee, S.; Masclauxdaubresse, C. Current understanding of leaf senescence in rice. Int. J. Mol. Sci. 2021, 22, 4515. [Google Scholar] [CrossRef]
- Zou, S.C.; Zhuo, M.G.; Abbas, F.; Hu, G.B.; Wang, H.C.; Huang, X.M. Transcription factor LcNAC002 coregulates chlorophyll degradation and anthocyanin biosynthesis in litchi. Plant Physiol. 2023, 192, 1913–1927. [Google Scholar] [CrossRef]
- Esteban, R.; Fernández-Marín, B.; Becerril, J.M.; García-Plazaola, J.I. Pigment composition and photoprotection in autumn leaves. Plant Physiol. 2015, 169, 1655–1668. [Google Scholar]
- Lee, D.W.; Gould, K.S. Why leaves turn red. Am. Sci. 2002, 90, 524–531. [Google Scholar] [CrossRef]
- Renner, S.S.; Zohner, C.M. The occurrence of red and yellow autumn leaves explained by regional differences in insolation and temperature. New Phytol. 2019, 223, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Yousefzadeh, M.; Miyamoto, S.; Ohtsubo, N.; Kawamura, Y. Effects of light intensity and temperature on the growth and physiological characteristics of Arundo donax L. for biomass production. J. Photochem. Photobiol. B Biol. 2015, 149, 168–175. [Google Scholar]
- Liu, Z.C.; Feng, A.Y.; Hu, H.L. Study on chlorophyll extraction methods of leaf in persimmon. J. Henan Inst. Sci. Technol. 2014, 42, 12–15. [Google Scholar]
- Chen, T.; Sheng, Y.; Hao, Z.; Long, X.; Fu, F.; Liu, Y.; Tang, Z.; Asif, A.; Peng, Y.; Liu, Y. Transcriptome and proteome analysis suggest enhanced photosynthesis in tetraploid Liriodendron sino-americanum. Tree Physiol. 2021, 41, 1953–1971. [Google Scholar] [CrossRef]
- Gilmore, A.M.; Yamamoto, H.Y. Resolution of lutein and zeaxanthin using a non-endcapped, lightly carbon-loaded C18 high-performance liquid chromatographic column. J. Chromatogr. 1991, 543, 137–145. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Shen, S.; Zhan, C.; Yang, C.; Fernie, A.R.; Luo, J. Metabolomics-centered mining of plant metabolic diversity and function: Past decade and future perspectives. Mol. Plant 2023, 16, 21. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.B. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Chen, T.T.; Wang, Z.Q.; Wang, J.J.; Liu, J.Q.; Zhang, R.; Jia, X.Y.; Yu, C.G.; Yin, Y.L.; Creech, D. Transcriptomic and metabolomic analyses unveil the growth advantage mechanism conferred by heterosis of Michelia ‘Zhongshanhanxiao’. Tree Physiol. 2023, 43, 1454–1466. [Google Scholar] [CrossRef]
- Ge, S.; Wang, M.M.; Chen, Y.W. An analysis of population genetic structure of Masson pine by isozyme technique. Sci. Silvae Sin. 1988, 4, 17–27. [Google Scholar]
- R Core Team. The R Project for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
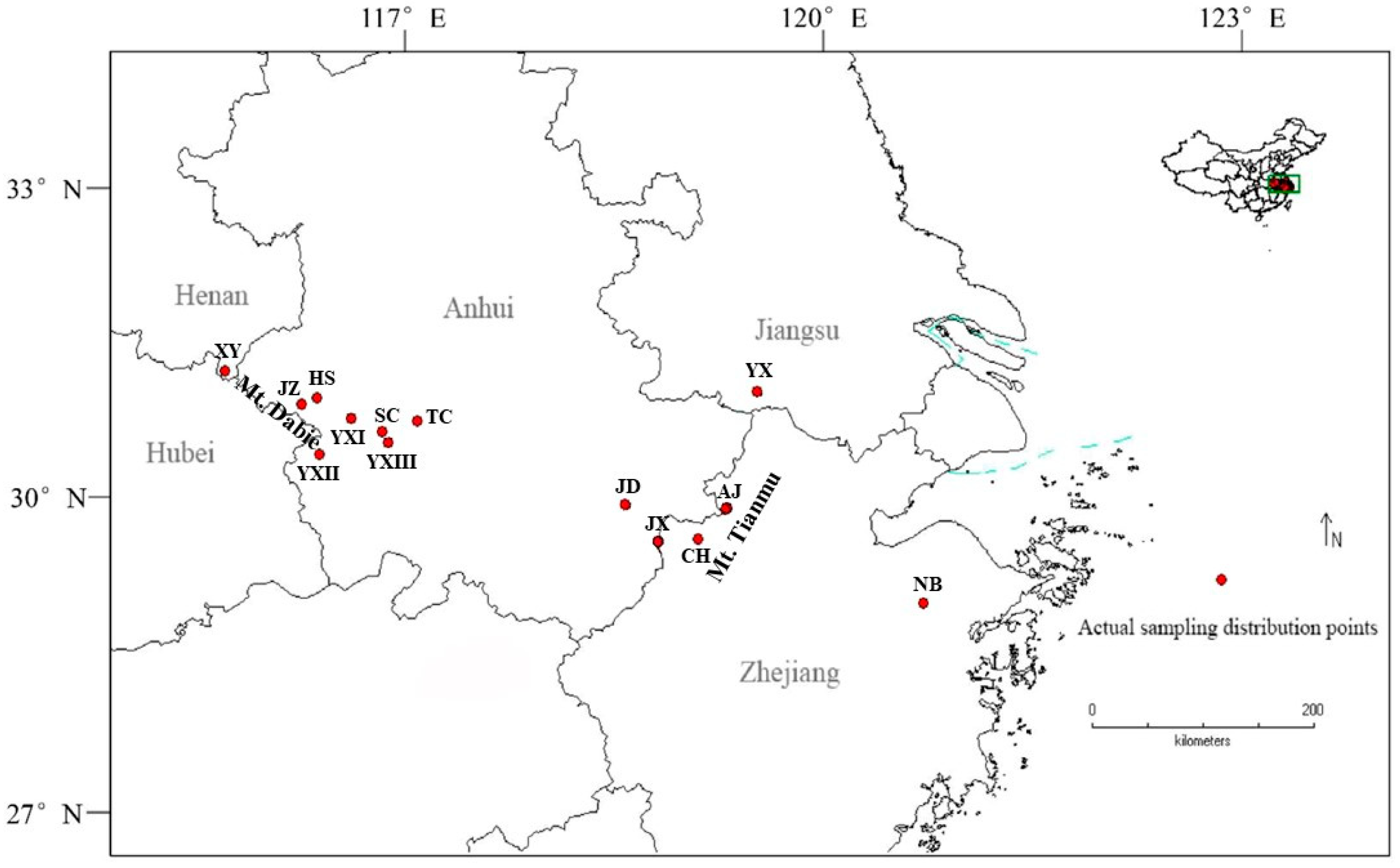
| Province | Sample Location | Population Code | Latitude (°N) | Longitude (°E) | Altitude (m) | Sample Size | Mountain | Autumn Color |
|---|---|---|---|---|---|---|---|---|
| Zhejiang | Ningbo | NB | 29.68 | 121.03 | 988 | 23 | Tianmu | Red |
| Changhua (Hangzhou) | CH | 30.16 | 119.18 | 864 | 42 | Red | ||
| Anji (Huzhou) | AJ | 30.38 | 119.4 | 820 | 30 | Purple | ||
| Jiangsu | Yixing | YX | 31.23 | 119.73 | 252 | 39 | Yellow | |
| Anhui | Jixi | JX | 30.2 | 118.88 | 684 | 23 | Dabie | Purple |
| Jingde | JD | 30.41 | 118.58 | 653 | 31 | Red | ||
| Yuexi (Huangwei) | YXI | 31.1 | 116.31 | 449 | 25 | Pink | ||
| Yuexi (Hetu) | YXII | 31.81 | 116.03 | 313 | 29 | Pink | ||
| Yuexi (Zhubo) | YXIII | 31.03 | 116.48 | 583 | 31 | Pink | ||
| Shucheng (Lu’an) | SC | 31.06 | 116.55 | 584 | 29 | Red | ||
| Tongcheng (Anqing) | TC | 31.08 | 116.85 | 270 | 24 | Red | ||
| Jinzhai (Lu’an) | JZ | 31.2 | 115.9 | 450 | 24 | Purple | ||
| Huoshan (Lu’an) | HS | 31.25 | 116.01 | 530 | 27 | Pink | ||
| Henan | Xinyang | XY | 31.45 | 115.26 | 192 | 28 | Red |
| Sample Location | Latitude (N) | Longitude (E) | Altitude (m) | Mean Soil Depth (cm) | Average Annual Precipitation (mm) | Average Annual Air Temperature (°C) |
|---|---|---|---|---|---|---|
| CH | 30°10′ | 119°11′ | 864 | 25.5 | 1123.6 | 12.2 |
| AJ | 30°23′ | 119°24′ | 820 | 11 | 1220.1 | 15 |
| NB | 29°41′ | 121°02′ | 988 | 20 | 1110.4 | 11.1 |
| YX | 31°14′ | 119°44′ | 252 | 27 | 1294.6 | 14.1 |
| JX | 30°12′ | 118°53′ | 684 | 39 | 1357.2 | 12.1 |
| JD | 30°25′ | 118°35′ | 653 | 21 | 1286.1 | 14.1 |
| TC | 31°05′ | 116°51′ | 270 | 34 | 1290.5 | 14 |
| HS | 31°15′ | 116°01′ | 530 | 12 | 1351.3 | 14.6 |
| SC | 31°4′ | 116°33′ | 584 | 28 | 1171.8 | 12.8 |
| JZ | 31°12′ | 115°54′ | 450 | 42 | 1419.9 | 15.9 |
| YXIII | 31°02′ | 116°29′ | 583 | 26 | 1296.4 | 14.6 |
| YXII | 30°49′ | 116°02′ | 313 | 24 | 1290.4 | 14.6 |
| YXI | 31°06′ | 116°19′ | 449 | 25 | 1290.4 | 14.6 |
| XY | 31°27′ | 115°16′ | 192 | 26 | 1380.5 | 15.8 |
| Traits | Mean Square | F-Value | |||
|---|---|---|---|---|---|
| Among Populations | Within Populations | Random Error | Among Populations | Within Populations | |
| Petiole length | 0.397 | 0.117 | 0.017 | 23.152 ** | 7.718 ** |
| Lamina length | 37.264 | 5.133 | 0.888 | 41.955 ** | 8.529 ** |
| Maximum lamina width | 4.063 | 1.369 | 0.842 | 4.828 ** | 2.631 ** |
| Lamina width at 10% of length | 3.087 | 0.412 | 0.208 | 14.804 ** | 2.703 ** |
| Lamina width at 90% of length | 2.827 | 0.526 | 0.748 | 3.779 ** | 2.469 ** |
| Number of principal veins | 66.422 | 15.673 | 2.138 | 31.060 ** | 9.153 ** |
| Traits | Populations | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CH | AJ | NB | YX | JX | JD | TC | HS | SC | JZ | YXIII | YXⅠI | YXI | XY | Mean | |
| Petiole length/% | 22.11 | 27.24 | 15.98 | 22.76 | 25.49 | 28.16 | 26.67 | 25.63 | 35.91 | 22.57 | 29.02 | 33.93 | 56.06 | 22.78 | 28.58 |
| Lamina length/% | 17.61 | 17.75 | 9.46 | 20.91 | 17.10 | 19.61 | 22.56 | 14.07 | 24.45 | 20.40 | 22.61 | 22.45 | 19.66 | 20.06 | 19.13 |
| Maximum lamina width/% | 18.35 | 17.43 | 12.74 | 20.12 | 17.86 | 52.80 | 25.24 | 13.92 | 25.86 | 21.08 | 21.95 | 24.75 | 19.98 | 21.17 | 22.47 |
| Lamina width at 10% of length/% | 46.64 | 33.17 | 35.63 | 40.40 | 45.49 | 47.84 | 52.72 | 33.93 | 71.97 | 56.95 | 42.28 | 51.27 | 55.62 | 71.63 | 47.22 |
| Lamina width at 90% of length/% | 26.97 | 24.10 | 18.14 | 32.60 | 22.92 | 81.70 | 32.03 | 15.11 | 54.76 | 34.07 | 39.46 | 38.19 | 91.46 | 27.60 | 39.35 |
| Number of principal veins/% | 10.98 | 15.81 | 8.94 | 13.18 | 12.10 | 13.77 | 12.19 | 14.92 | 14.46 | 11.12 | 17.90 | 12.70 | 11.81 | 11.66 | 13.07 |
| Leaf area | 33.98 | 29.47 | 46.57 | 44.69 | 38.58 | 37.92 | 50.23 | 40.56 | 53.55 | 39.40 | 45.76 | 47.46 | 35.13 | 41.06 | 41.74 |
| Leaf weight | 39.50 | 36.69 | 59.15 | 57.14 | 45.20 | 48.93 | 54.32 | 50.74 | 54.71 | 54.54 | 72.13 | 55.55 | 45.45 | 50.72 | 51.74 |
| Mean/% | 27.01 | 25.21 | 25.83 | 31.47 | 28.09 | 41.34 | 34.49 | 26.11 | 41.96 | 32.51 | 36.38 | 35.78 | 41.89 | 33.33 | 32.91 |
| Population | Mean ± SD | |||||||
|---|---|---|---|---|---|---|---|---|
| LP | LL | MWL | WL (0.1) | WL (0.9) | NPV | LA | LW | |
| CH | 0.524 ± 0.116 | 5.470 ± 0.963 | 3.239 ± 0.594 | 0.943 ± 0.440 | 1.433 ± 0.387 | 11.780 ± 1.293 | 11.984 ± 4.073 | 0.081 ± 0.032 |
| AJ | 0.578 ± 0.157 | 5.716 ± 1.014 | 3.264 ± 0.569 | 1.266 ± 0.420 | 1.573 ± 0.379 | 11.010 ± 1.714 | 13.901 ± 4.097 | 0.109 ± 0.040 |
| NB | 0.553 ± 0.088 | 4.637 ± 0.439 | 3.090 ± 0.394 | 1.230 ± 0.438 | 1.837 ± 0.333 | 11.367 ± 1.016 | 10.546 ± 4.912 | 0.071 ± 0.042 |
| YX | 0.627 ± 0.143 | 5.916 ± 1.237 | 3.293 ± 0.663 | 1.299 ± 0.525 | 1.667 ± 0.543 | 10.905 ± 1.437 | 14.059 ± 6.284 | 0.077 ± 0.044 |
| JX | 0.510 ± 0.130 | 5.586 ± 0.955 | 3.364 ± 0.601 | 1.132 ± 0.515 | 1.709 ± 0.392 | 11.833 ± 1.432 | 11.425 ± 4.408 | 0.073 ± 0.033 |
| JD | 0.455 ± 0.128 | 4.528 ± 0.888 | 2.941 ± 1.552 | 0.961 ± 0.460 | 1.364 ± 1.114 | 10.140 ± 1.397 | 9.425 ± 3.574 | 0.047 ± 0.023 |
| TC | 0.538 ± 0.143 | 6.148 ± 1.387 | 3.387 ± 0.885 | 1.088 ± 0.574 | 1.349 ± 0.432 | 13.428 ± 1.636 | 15.430 ± 7.751 | 0.081 ± 0.044 |
| HS | 0.387 ± 0.099 | 5.467 ± 0.769 | 3.133 ± 0.436 | 1.030 ± 0.349 | 1.997 ± 0.302 | 10.933 ± 1.623 | 11.121 ± 4.511 | 0.067 ± 0.034 |
| SC | 0.443 ± 0.159 | 5.156 ± 1.261 | 3.127 ± 0.808 | 0.588 ± 0.423 | 0.765 ± 0.419 | 10.830 ± 1.566 | 11.237 ± 6.018 | 0.053 ± 0.029 |
| JZ | 0.491 ± 0.111 | 5.356 ± 1.093 | 3.294 ± 0.694 | 1.073 ± 0.611 | 1.489 ± 0.507 | 12.586 ± 1.399 | 11.434 ± 4.506 | 0.066 ± 0.036 |
| YXIII | 0.403 ± 0.117 | 4.356 ± 0.985 | 2.513 ± 0.552 | 0.948 ± 0.401 | 1.127 ± 0.445 | 10.186 ± 1.823 | 10.782 ± 4.934 | 0.061 ± 0.044 |
| YXII | 0.472 ± 0.160 | 5.027 ± 1.129 | 2.909 ± 0.720 | 1.096 ± 0.562 | 1.461 ± 0.558 | 10.731 ± 1.363 | 10.327 ± 4.902 | 0.081 ± 0.045 |
| YXI | 0.502 ± 0.281 | 5.197 ± 1.022 | 3.039 ± 0.607 | 0.948 ± 0.527 | 1.354 ± 1.238 | 11.480 ± 1.356 | 10.128 ± 3.558 | 0.066 ± 0.030 |
| XY | 0.564 ± 0.129 | 5.235 ± 1.050 | 3.146 ± 0.666 | 1.087 ± 0.778 | 1.568 ± 0.433 | 12.509 ± 1.458 | 11.139 ± 4.574 | 0.069 ± 0.035 |
| p-value | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Mean | 0.5 | 5.27 | 3.12 | 1.05 | 1.48 | 11.41 | 11.63 | 0.07 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Hafsi, A.; Wang, X.; Zhang, C.; Lang, Z.; Yu, M.; Fang, Y. The Scarlet Alchemy of Survival: Integrated Transcriptomic and Metabolomic Analysis of Leaf Coloration in Endangered Parrotia subaequalis. Plants 2025, 14, 2345. https://doi.org/10.3390/plants14152345
Zhang L, Hafsi A, Wang X, Zhang C, Lang Z, Yu M, Fang Y. The Scarlet Alchemy of Survival: Integrated Transcriptomic and Metabolomic Analysis of Leaf Coloration in Endangered Parrotia subaequalis. Plants. 2025; 14(15):2345. https://doi.org/10.3390/plants14152345
Chicago/Turabian StyleZhang, Lifang, Aya Hafsi, Xianting Wang, Chenyun Zhang, Zedong Lang, Mingjian Yu, and Yanming Fang. 2025. "The Scarlet Alchemy of Survival: Integrated Transcriptomic and Metabolomic Analysis of Leaf Coloration in Endangered Parrotia subaequalis" Plants 14, no. 15: 2345. https://doi.org/10.3390/plants14152345
APA StyleZhang, L., Hafsi, A., Wang, X., Zhang, C., Lang, Z., Yu, M., & Fang, Y. (2025). The Scarlet Alchemy of Survival: Integrated Transcriptomic and Metabolomic Analysis of Leaf Coloration in Endangered Parrotia subaequalis. Plants, 14(15), 2345. https://doi.org/10.3390/plants14152345








