Pyramiding Recessive Resistance Genes Enhances Bacterial Leaf Spot Resistance in Peppers by Suppressing In Planta Bacterial Growth
Abstract
1. Introduction
2. Materials and Methodology
2.1. Plant Genetic Materials
2.2. Bacterial Strains and Inoculation
2.3. Experimental Design and Data Collection
2.4. Statistical Analysis
3. Results
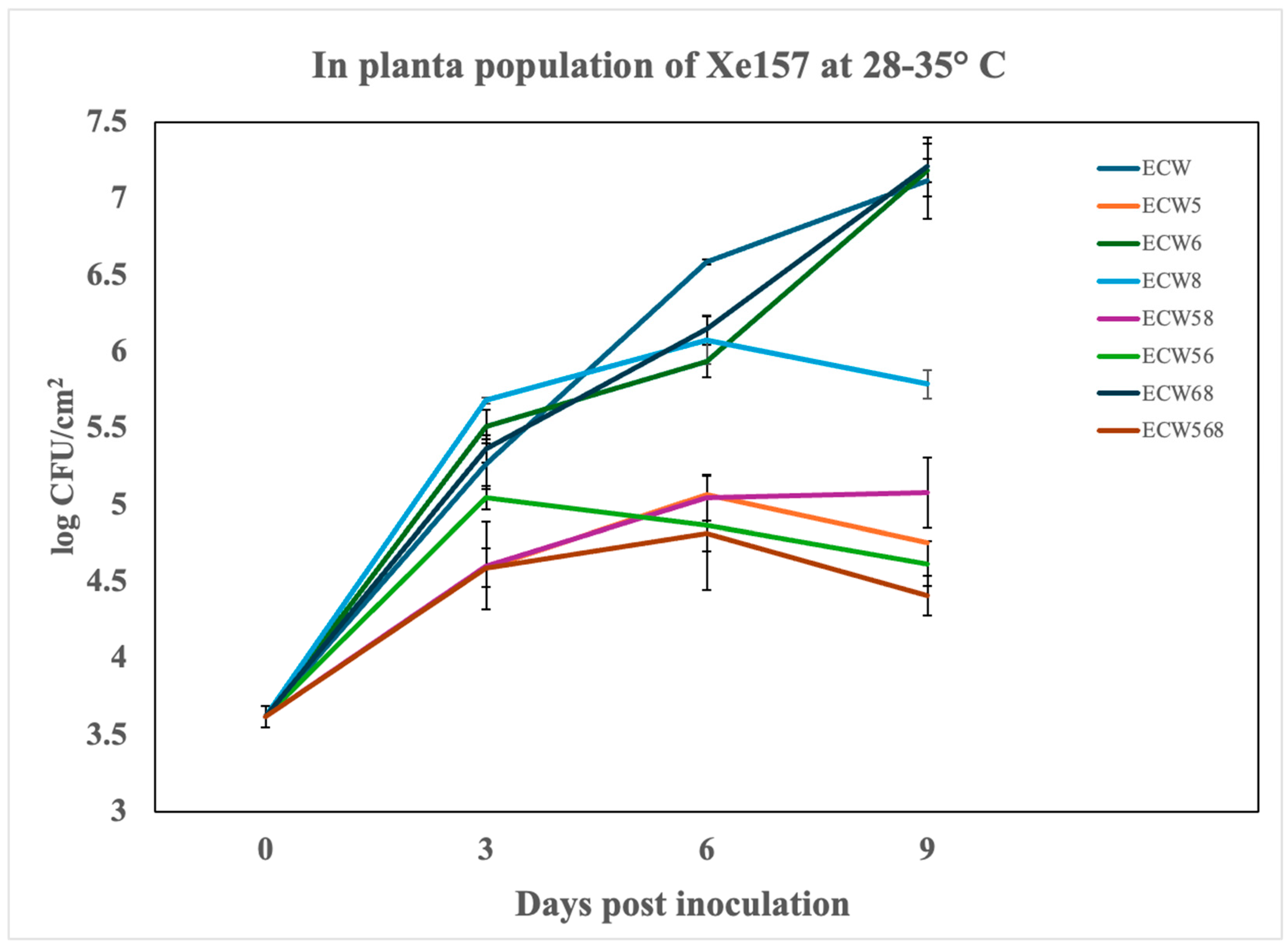
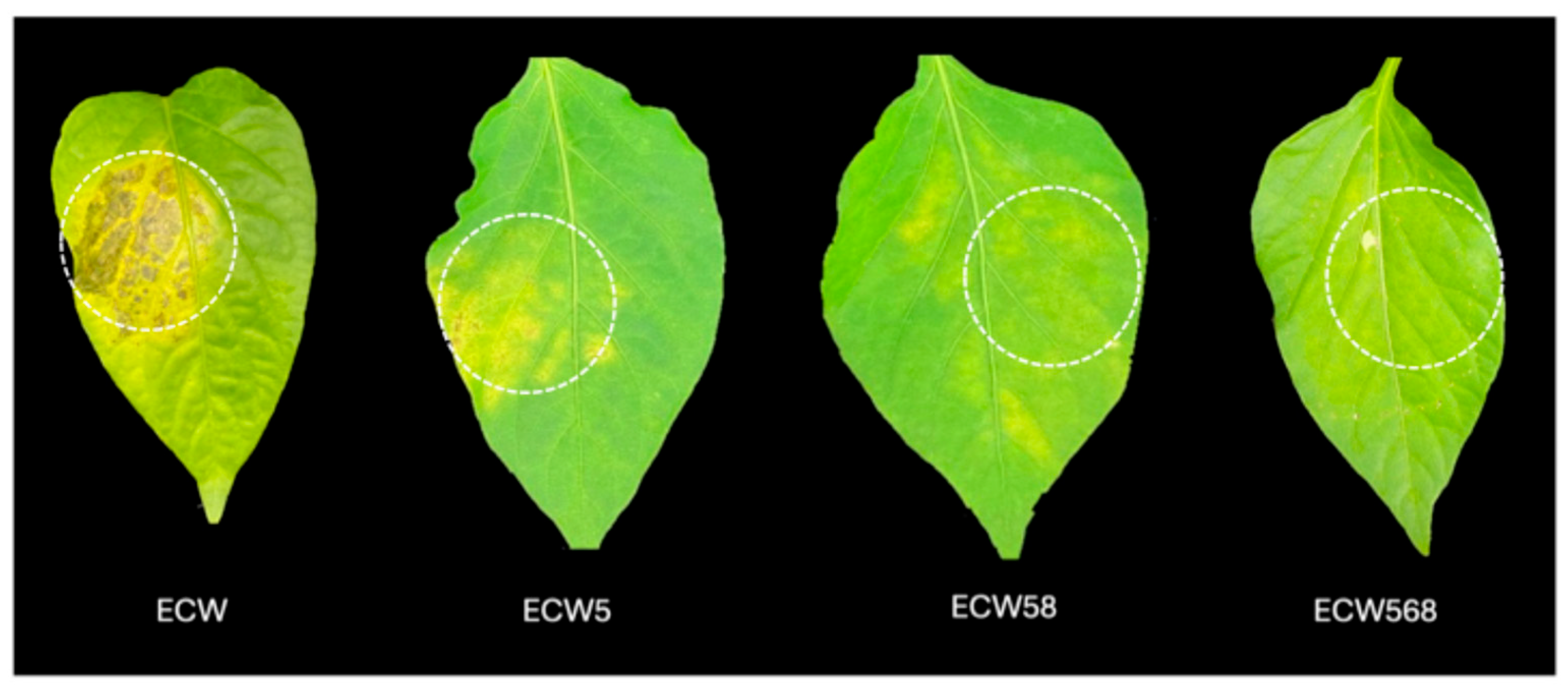
3.1. Effect of Recessive Resistance Gene Combinations on Bacterial Multiplication and Disease Development Caused by X. euvesicatoria
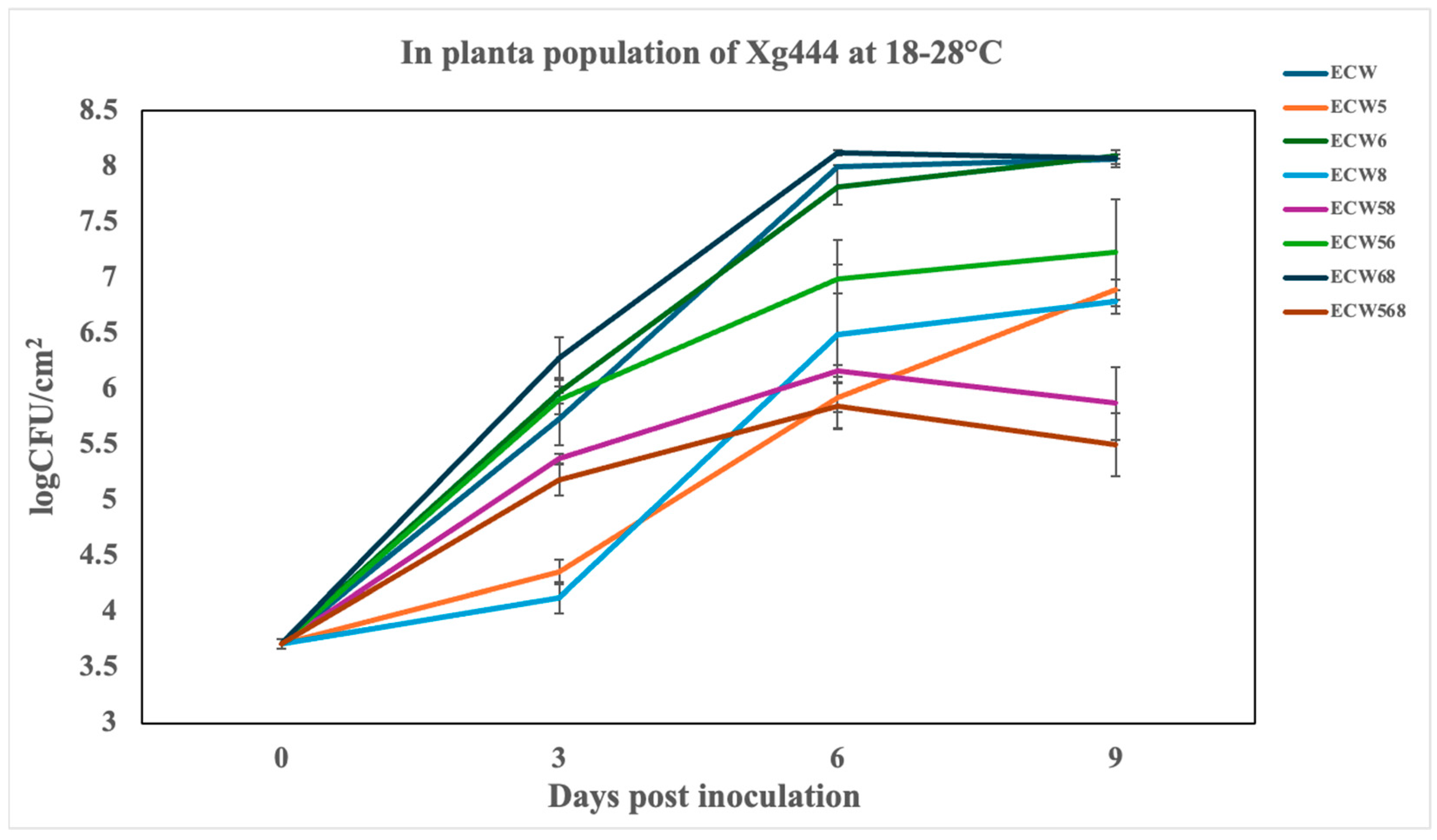
3.2. Effect of Recessive Resistance Gene Combinations on Multiplication and Disease Development Caused by X. hortorum pv. gardneri
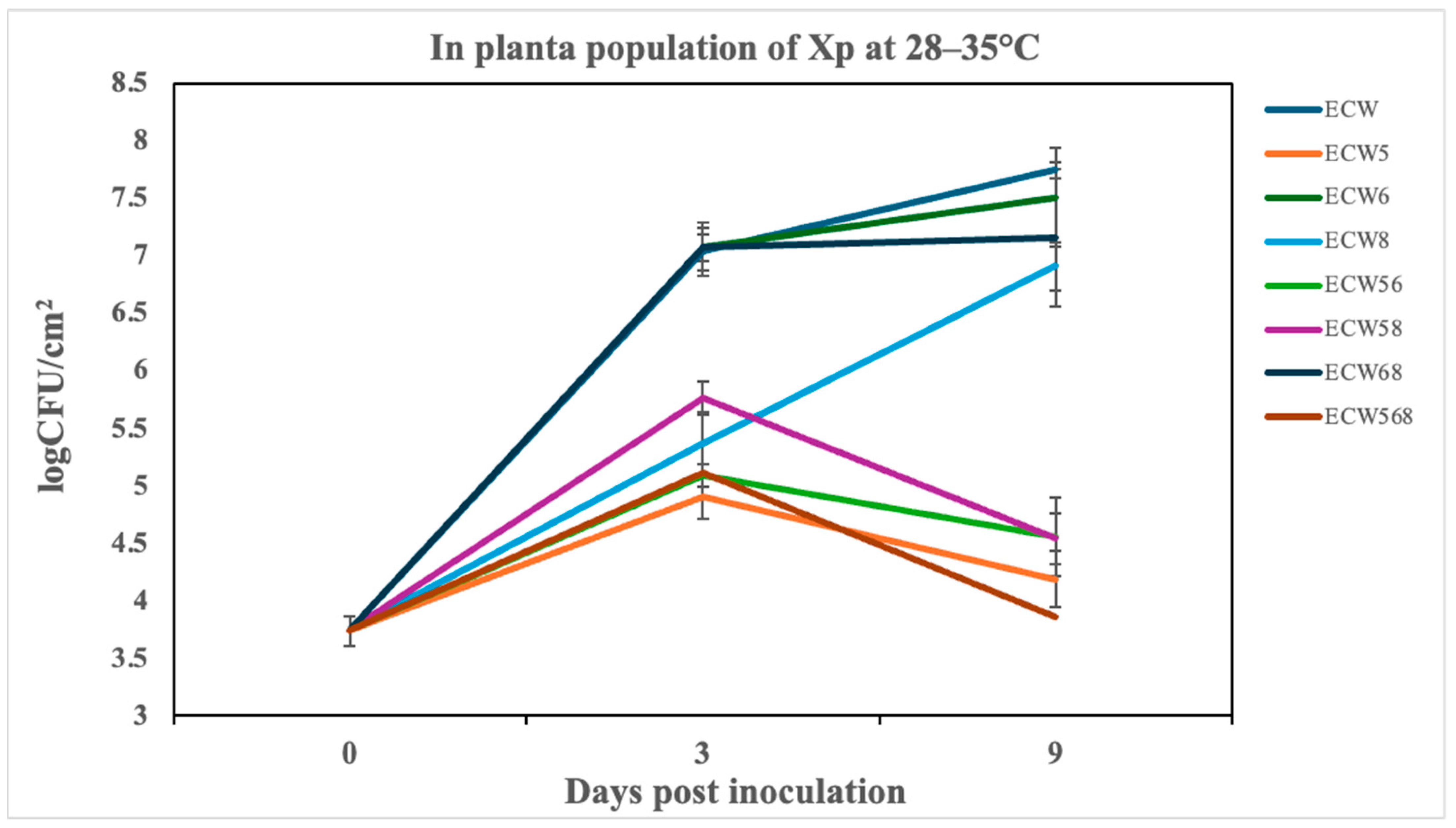
3.3. Effect of Recessive Resistance Gene Combinations on Multiplication and Disease Development Caused by X. perforans
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, J.B.; Zitter, T.A.; Momol, T.M.; Miller, S.A. (Eds.) Compendium of Tomato Diseases and Pests, 2nd ed.; The American Phytopathological Society: St. Paul, MO, USA, 2016; ISBN 978-0-89054-434-1. [Google Scholar]
- Osdaghi, E.; Jones, J.B.; Sharma, A.; Goss, E.M.; Abrahamian, P.; Newberry, E.A.; Potnis, N.; Carvalho, R.; Choudhary, M.; Paret, M.L.; et al. A centenary for bacterial spot of tomato and pepper. Mol. Plant Pathol. 2021, 22, 1500–1519. [Google Scholar] [CrossRef]
- Bashan, Y.; Azaizeh, M.; Diab, S.; Yunis, H.; Okon, Y. Crop loss of pepper plants artificially infected with Xanthomonas campestris pv. vesicatoria in relation to symptom expression. Crop Prot. 1985, 4, 77–84. [Google Scholar] [CrossRef]
- Pohronezny, K.; Volin, R.B. The effect of bacterial spot on yield and quality of fresh market tomatoes. HortScience 1983, 18, 69–70. [Google Scholar] [CrossRef]
- Jones, J.B.; Lacy, G.H.; Bouzar, H.; Stall, R.E.; Schaad, N.W. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst. Appl. Microbiol. 2004, 27, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Potnis, N.; Timilsina, S.; Strayer, A.; Shantharaj, D.; Barak, J.D.; Paret, M.L.; Vallad, G.E.; Jones, J.B. Bacterial spot of tomato and pepper: Diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol. Plant Pathol. 2015, 16, 907–920. [Google Scholar] [CrossRef] [PubMed]
- Subedi, A.; Minsavage, G.V.; Jones, J.B.; Goss, E.M.; Roberts, P.D. Exploring diversity of bacterial spot associated Xanthomonas population of pepper in southwest florida. Plant Dis. 2023, 107, 2978–2985. [Google Scholar] [CrossRef] [PubMed]
- Horvath, D.M.; Stall, R.E.; Jones, J.B.; Pauly, M.H.; Vallad, G.E.; Dahlbeck, D.; Staskawicz, B.J.; Scott, J.W. Transgenic resistance confers effective field level control of bacterial spot disease in tomato. PLoS ONE 2012, 7, e42036. [Google Scholar] [CrossRef]
- Ritchie, D. Bacterial Spot of Pepper and Tomato. Plant Health Instr. 2000, 10. [Google Scholar] [CrossRef]
- Rowell, B.; Jones, R.T.; Nesmith, W.; Satanek, A.; Snyder, J.C. Bacterial spot resistance, yield, and quality of bell and specialty peppers. HortTechnology 2001, 11, 648–657. [Google Scholar] [CrossRef]
- Araújo, E.R.; Pereira, R.C.; Ferreira, M.A.S.V.; Café-Filho, A.C.; Moita, A.W.; Quezado-Duval, A.M. Effect of temperature on pathogenicity components of tomato bacterial spot and competition between Xanthomonas perforans and X. gardneri. Acta Hortic. 2011, 914, 39–42. [Google Scholar] [CrossRef]
- Jones, J.B. Survival of Xanthomonas campestris pv. vesicatoria in Florida on tomato crop residue, weeds, seeds, and volunteer tomato plants. Phytopathology 1986, 76, 430. [Google Scholar] [CrossRef]
- Stall, R.E.; Jones, J.B.; Minsavage, G.V. Durability of resistance in tomato and pepper to xanthomonads causing bacterial spot. Annu. Rev. Phytopathol. 2009, 47, 265–284. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Minsavage, G.V.; Gill, U.S.; Hutton, S.F.; Jones, J.B. identification and mapping of bs8, a novel locus conferring resistance to bacterial spot caused by Xanthomonas gardneri. Phytopathology 2022, 112, 1640–1650. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, W.; Dahlbeck, D.; Chesnokova, O.; Minsavage, G.V.; Jones, J.B.; Staskawicz, B.J. Molecular evolution of virulence in natural field strains of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 2000, 182, 7053–7059. [Google Scholar] [CrossRef]
- Kousik, C.S.; Ritchie, D.F. Response of bell pepper cultivars to bacterial spot pathogen races that individually overcome major resistance genes. Plant Dis. 1998, 82, 181–186. [Google Scholar] [CrossRef]
- Jones, J.B.; Minsavage, G.V.; Roberts, P.D.; Johnson, R.R.; Kousik, C.S.; Subramanian, S.; Stall, R.E. A non-hypersensitive resistance in pepper to the bacterial spot pathogen is associated with two recessive genes. Phytopathology 2002, 92, 273–277. [Google Scholar] [CrossRef]
- Vallejos, C.E.; Jones, V.; Stall, R.E.; Jones, J.B.; Minsavage, G.V.; Schultz, D.C.; Rodrigues, R.; Olsen, L.E.; Mazourek, M. Characterization of Two Recessive Genes Controlling Resistance to All Races of Bacterial Spot in Peppers. Theor. Appl. Genet. 2010, 121, 37–46. [Google Scholar] [CrossRef]
- Sharma, A.; Li, J.; Wente, R.; Minsavage, G.V.; Gill, U.S.; Ortega, A.; Vallejos, C.E.; Hart, J.P.; Staskawicz, B.J.; Mazourek, M.R.; et al. Mapping of the bs5 and bs6 Non-Race-Specific Recessive Resistances against Bacterial Spot of Pepper. Front. Plant Sci. 2023, 14, 1061803. [Google Scholar] [CrossRef]
- Schwartz, A.R.; Potnis, N.; Timilsina, S.; Wilson, M.; Patané, J.; Martins, J.; Minsavage, G.V.; Dahlbeck, D.; Akhunova, A.; Almeida, N.; et al. Phylogenomics of Xanthomonas field strains infecting pepper and tomato reveals diversity in effector repertoires and identifies determinants of host specificity. Front. Microbiol. 2015, 6, 535. [Google Scholar] [CrossRef]
- Madden, L.V.; Hughes, G.; Van Den Bosch, F. The Study of Plant Disease Epidemics; APS Press: St. Paul, MO, USA, 2007; ISBN 0-89054-505-7. [Google Scholar]
- Subedi, A.; Minsavage, G.V.; Roberts, P.D.; Goss, E.M.; Sharma, A.; Jones, J.B. Insights into bs5 resistance mechanisms in pepper against Xanthomonas euvesicatoria through transcriptome profiling. BMC Genom. 2024, 25, 711. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The Plant Immune System. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]


| Name | Sequence | Position (CM334) | Position (UCD10x) | Polymorphism | Reference | |
|---|---|---|---|---|---|---|
| bs5 | JW1_F1 | ACCGAGTCTCATGCTGTTTC | ~760,000 | ~390,000 | CCAAGAG>C delCAAGAG | This study |
| JW1_R1 | CAACACTTTGCGTACAGATCATT | ~760,000 | ~390,000 | CCAAGAG>C delCAAGAG | ||
| bs6 | 190.99_F1/6g_H178.55 | TTCTCATTATCCGTATCATTACCC | 190,990,000 | 178,550,000 | AA>GG | Sharma et al. [19] |
| 190.99_R1/6g_H178.55 | CGTTCCACAAACGACATCT | 190,990,000 | 178,550,000 | AA>GG | ||
| bs8 | bsh_260.54_F1 | GGGGTACTGAGAGTGACCCTA | 260,544,417 | 232,640,000 | A>G | This study |
| bsh_260.54_R1 | CAAAGCCACGTGGTTCCG | 260,544,417 | 232,640,000 | A>G |
| Genotype | AUPPC | SNK Group |
|---|---|---|
| ECW568 | 40.26 | c |
| ECW5 | 41.53 | c |
| ECW56 | 42.00 | c |
| ECW58 | 42.10 | c |
| ECW8 | 49.11 | b |
| ECW6 | 50.55 | ab |
| ECW68 | 50.78 | ab |
| ECW | 51.66 | a |
| Genotype | AUPPC | SNK Group |
|---|---|---|
| ECW568 | 46.95 | c |
| ECW5 | 47.46 | c |
| ECW56 | 49.00 | c |
| ECW8 | 50.80 | c |
| ECW58 | 55.09 | b |
| ECW | 56.08 | b |
| ECW6 | 59.10 | ab |
| ECW68 | 60.9 | a |
| Genotype | AUPPC | SNK Group |
|---|---|---|
| ECW568 | 40.13 | d |
| ECW5 | 40.22 | d |
| ECW56 | 42.14 | d |
| ECW58 | 45.13 | c |
| ECW8 | 50.45 | b |
| ECW68 | 58.87 | a |
| ECW6 | 59.95 | a |
| ECW | 60.47 | a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poudel, M.; McDuffee, S.; Minsavage, G.V.; Hutton, S.F.; Sharma, A.; Jones, J.B. Pyramiding Recessive Resistance Genes Enhances Bacterial Leaf Spot Resistance in Peppers by Suppressing In Planta Bacterial Growth. Plants 2025, 14, 2559. https://doi.org/10.3390/plants14162559
Poudel M, McDuffee S, Minsavage GV, Hutton SF, Sharma A, Jones JB. Pyramiding Recessive Resistance Genes Enhances Bacterial Leaf Spot Resistance in Peppers by Suppressing In Planta Bacterial Growth. Plants. 2025; 14(16):2559. https://doi.org/10.3390/plants14162559
Chicago/Turabian StylePoudel, Mousami, Sophia McDuffee, Gerald V. Minsavage, Samuel F. Hutton, Anuj Sharma, and Jeffrey B. Jones. 2025. "Pyramiding Recessive Resistance Genes Enhances Bacterial Leaf Spot Resistance in Peppers by Suppressing In Planta Bacterial Growth" Plants 14, no. 16: 2559. https://doi.org/10.3390/plants14162559
APA StylePoudel, M., McDuffee, S., Minsavage, G. V., Hutton, S. F., Sharma, A., & Jones, J. B. (2025). Pyramiding Recessive Resistance Genes Enhances Bacterial Leaf Spot Resistance in Peppers by Suppressing In Planta Bacterial Growth. Plants, 14(16), 2559. https://doi.org/10.3390/plants14162559








