Pan-Genome-Based Characterization of the PYL Transcription Factor Family in Populus
Abstract
1. Introduction
2. Results
2.1. Pan-Genome Distribution of the PopPYL Gene Family
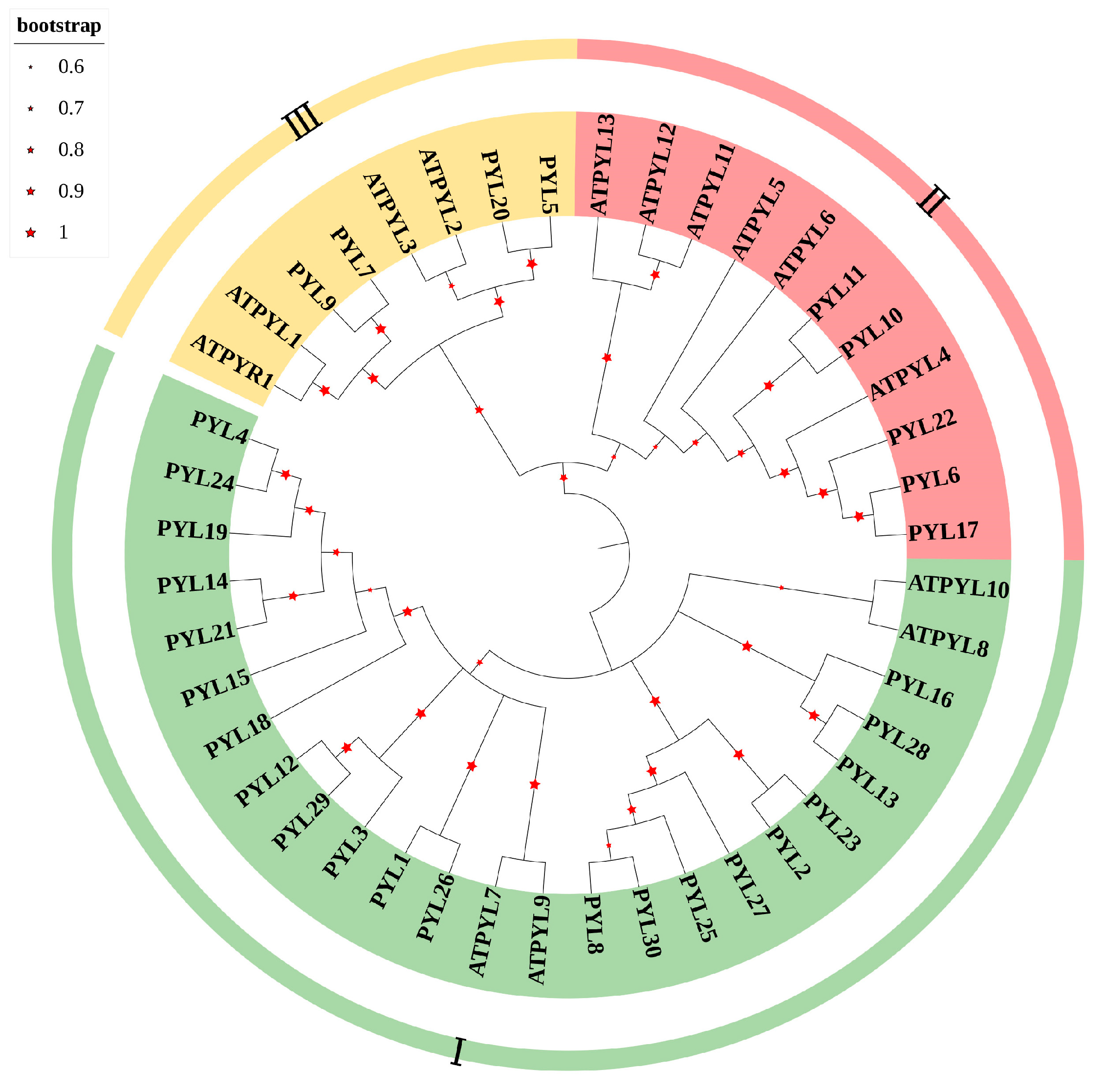
2.2. Construction of the Phylogenetic Tree of PopPYL Gene Family Members
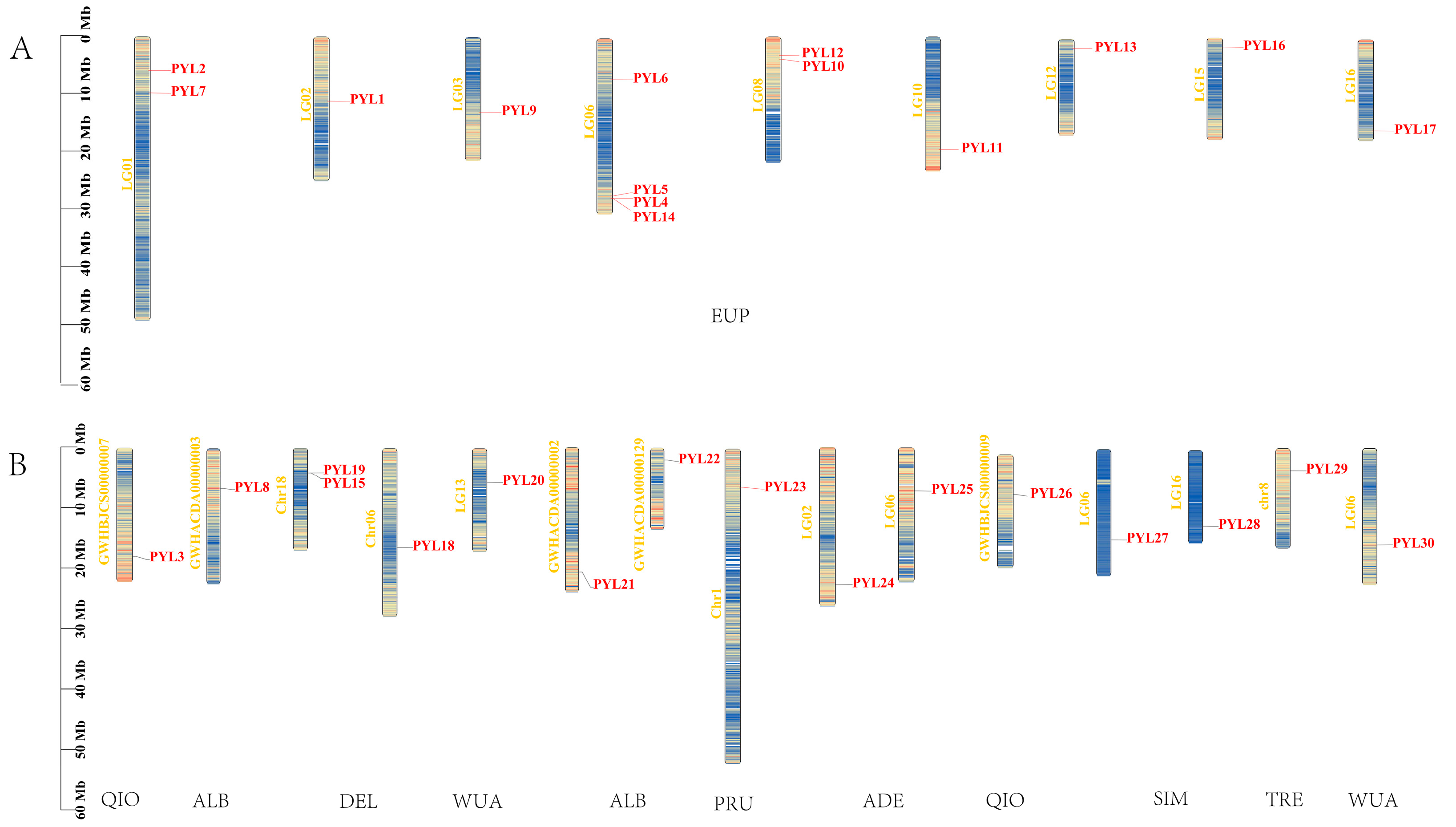
2.3. Chromosomal Location of PopPYL Genes in There Populus Species
2.4. Gene Structure and Conserved Motif Analysis
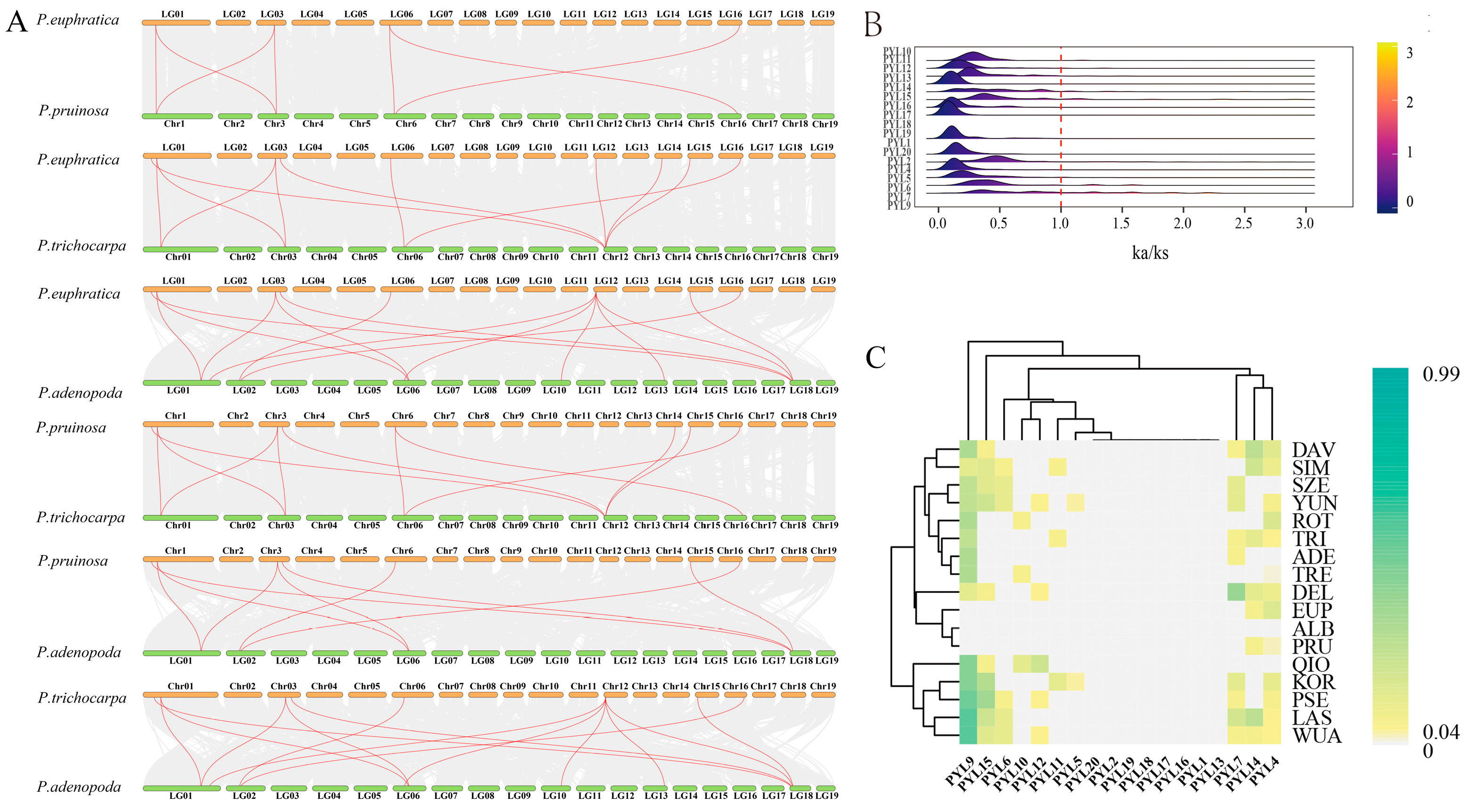
2.5. Synteny Analysis of PopPYL Genes
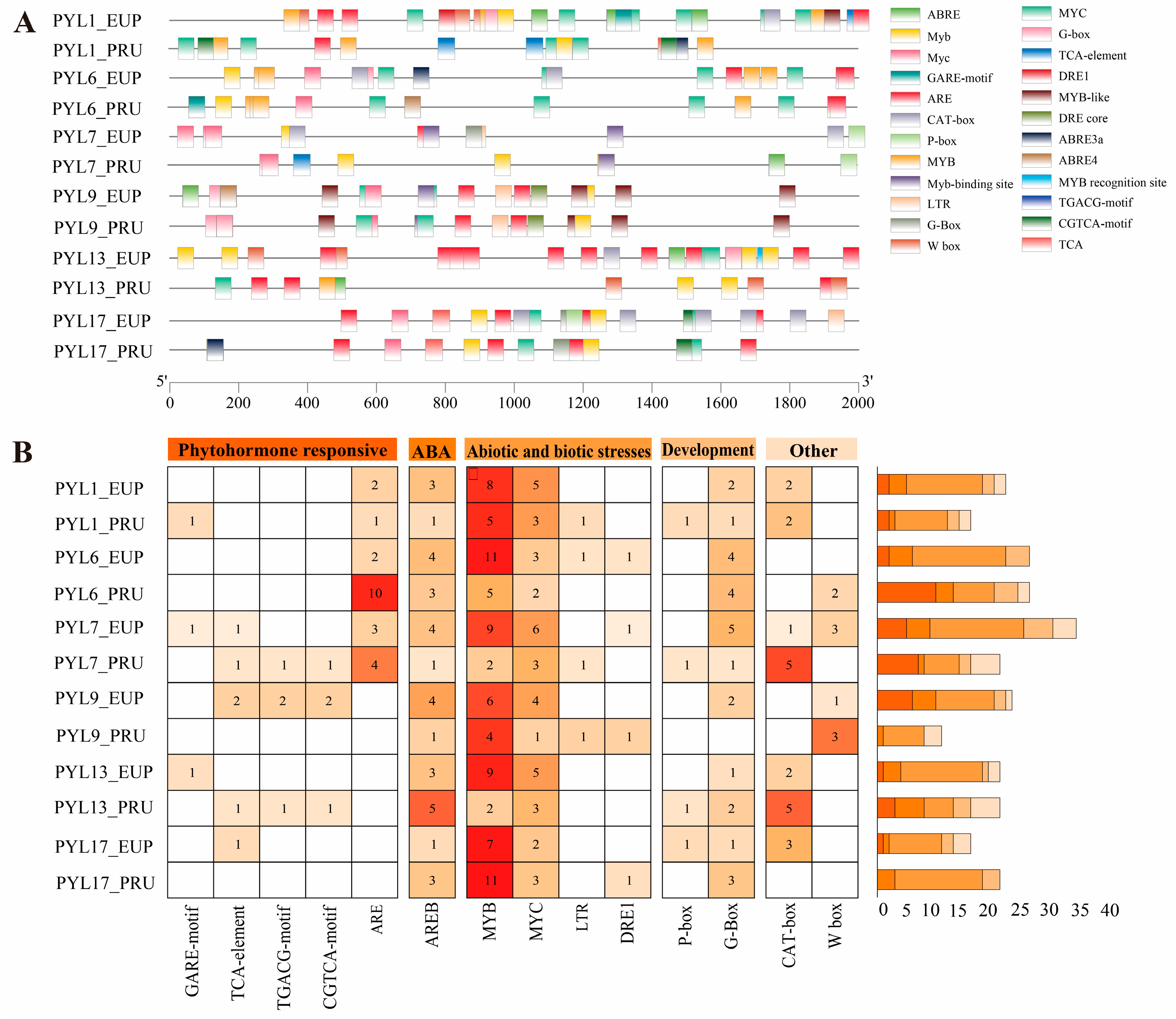
2.6. Promoter Analysis of PopPYL Genes
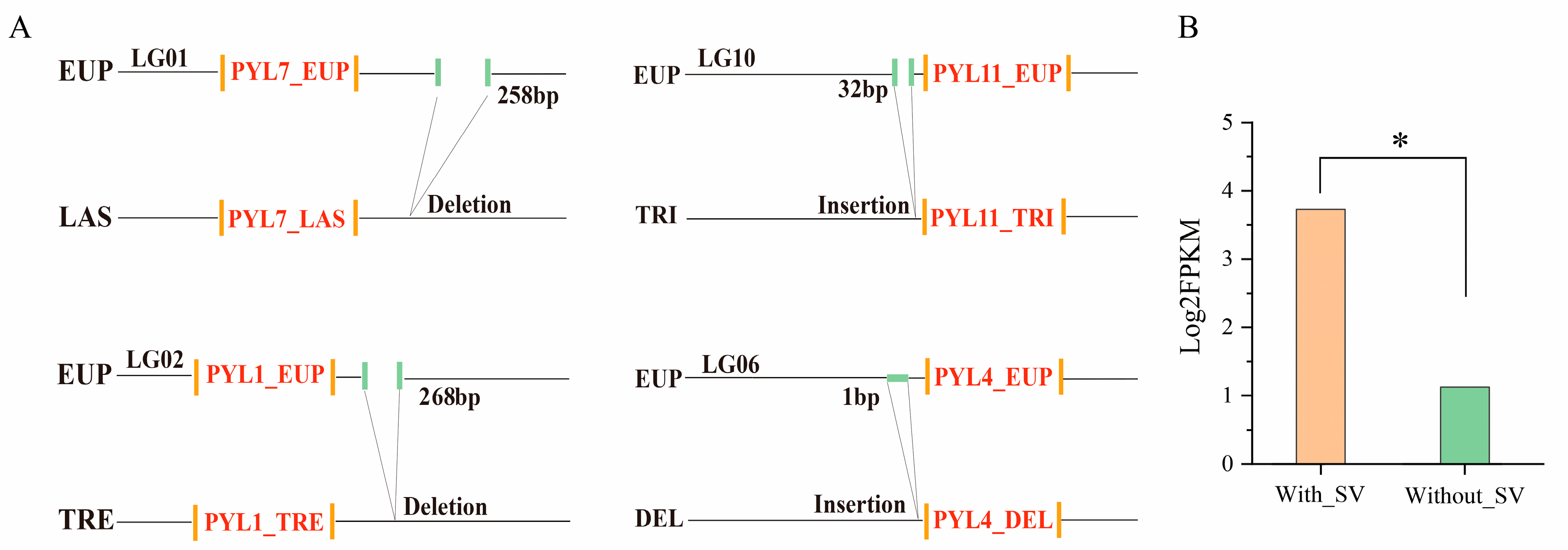
2.7. Structural Variation Analysis of PYL_EUP Genes
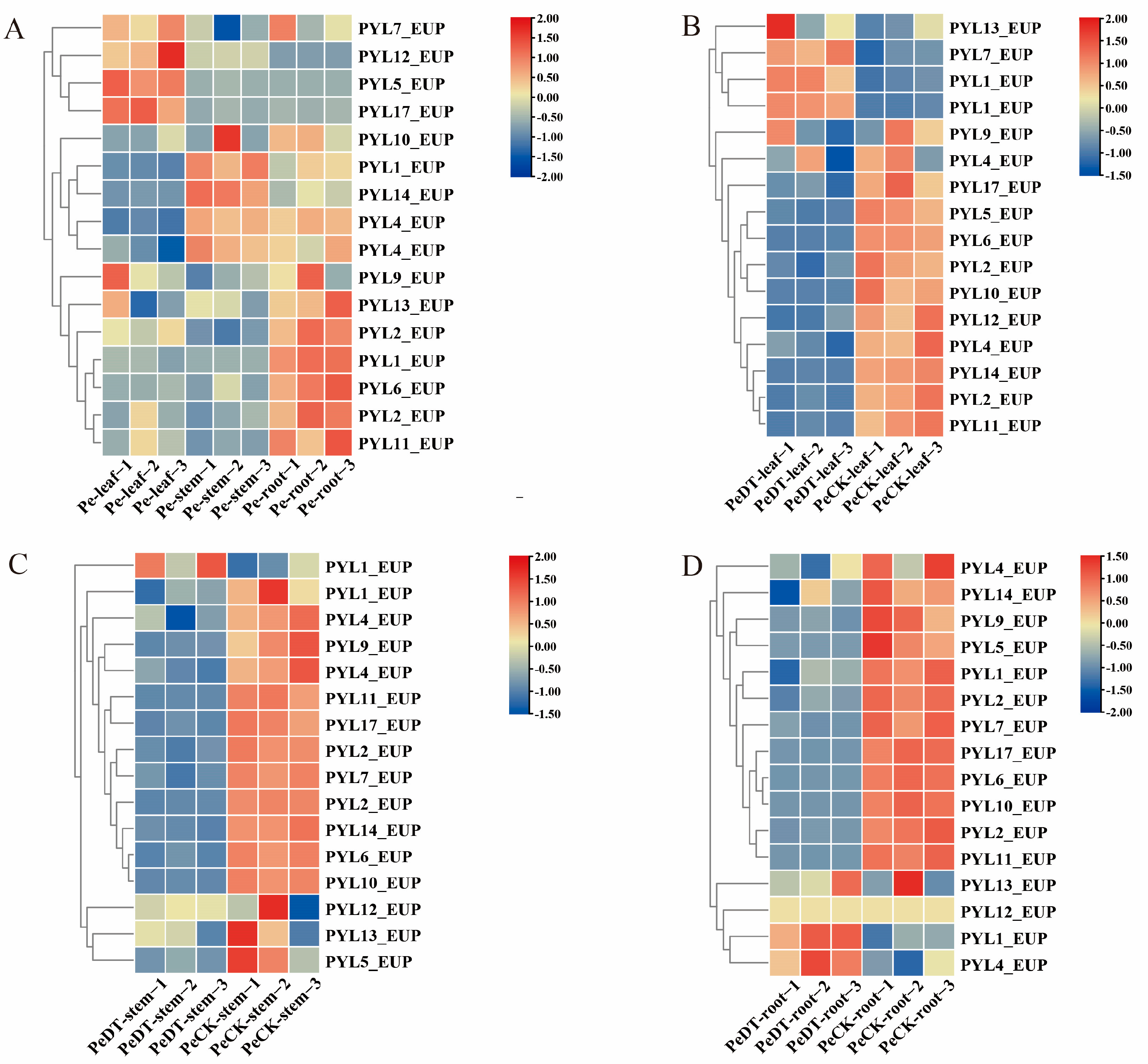
2.8. Expression Analysis of the PYL_EUP Gene Family
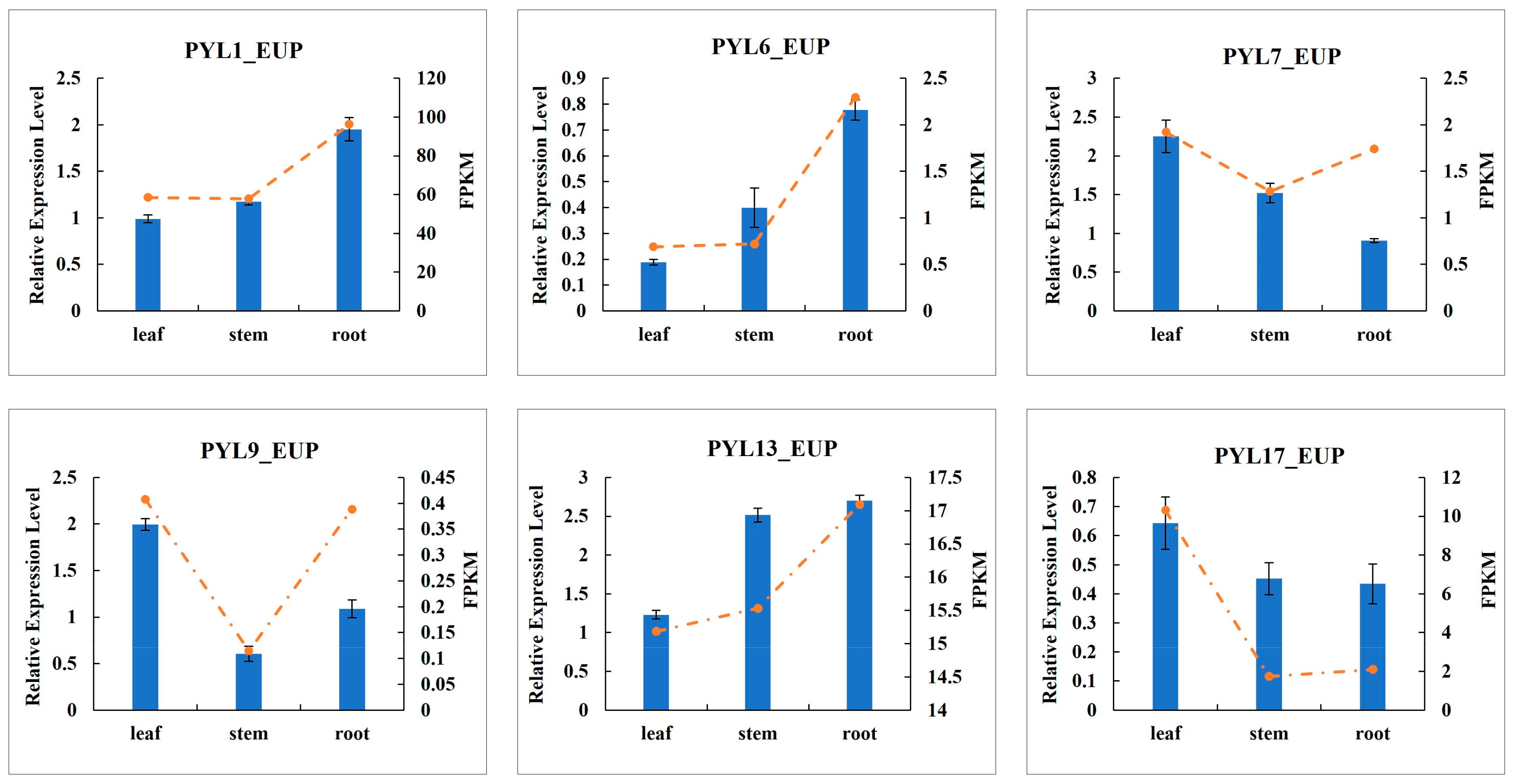
2.9. Verification of PYL Gene Expression Patterns in Different Tissues by RT-qPCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Data Acquisition
4.3. Identification of PopPYL Genes in Populus Species
4.4. Phylogenetic Analysis of PopPYL Family Protein Sequences
4.5. Motif Analysis and Gene Structure
4.6. Gene Duplication Analysis and Ka/Ks Calculation
4.7. SVs and Gene Expression Analysis of PYL_EUP
4.8. Analysis of Promoter Regions of PopPYL Genes
4.9. Analysis of Gene Expression Profiles
4.10. qRT-PCR Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef]
- Santos, A.P.; Serra, T.; Figueiredo, D.D.; Barros, P.; Lourenço, T.; Chander, S.; Oliveira, M.M.; Saibo, N.J. Transcription regulation of abiotic stress responses in rice: A combined action of transcription factors and epigenetic mechanisms. Omics J. Integr. Biol. 2011, 15, 839–857. [Google Scholar] [CrossRef]
- Friedel, S.; Usadel, B.; von Wirén, N.; Sreenivasulu, N. Reverse engineering: A key component of systems biology to unravel global abiotic stress cross-talk. Front. Plant Sci. 2012, 3, 294. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Zhu, J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernández, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef]
- Munemasa, S.; Hauser, F.; Park, J.; Waadt, R.; Brandt, B.; Schroeder, J.I. Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr. Opin. Plant Biol. 2015, 28, 154–162. [Google Scholar] [CrossRef]
- Belda-Palazon, B.; Gonzalez-Garcia, M.P.; Lozano-Juste, J.; Coego, A.; Antoni, R.; Julian, J.; Peirats-Llobet, M.; Rodriguez, L.; Berbel, A.; Dietrich, D.; et al. PYL8 mediates ABA perception in the root through non-cell-autonomous and ligand-stabilization-based mechanisms. Proc. Natl. Acad. Sci. USA 2018, 115, E11857–E11863. [Google Scholar] [CrossRef]
- Schweighofer, A.; Hirt, H.; Meskiene, I. Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 2004, 9, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, X.; Modrego, A.; Rodríguez, D.; González-García, M.P.; Sanz, L.; Nicolás, G.; Lorenzo, O. The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signaling in seeds and stress. Plant Physiol. 2010, 152, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; An, F.; Sun, Y.; Guo, R.; Pan, L.; Wan, T.; Hao, Y.; Cai, Y. Genome-wide identification of the ABA receptor PYL gene family and expression analysis in Prunus avium L. Sci. Hortic. 2023, 313, 111919. [Google Scholar] [CrossRef]
- Yuan, X.; Yin, P.; Hao, Q.; Yan, C.; Wang, J.; Yan, N. Single Amino Acid Alteration between Valine and Isoleucine Determines the Distinct Pyrabactin Selectivity by PYL1 and PYL2. J. Biol. Chem. 2010, 285, 28953–28958. [Google Scholar] [CrossRef]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132. [Google Scholar] [CrossRef]
- Choi, H.-i.; Hong, J.-h.; Ha, J.-o.; Kang, J.-y.; Kim, S.Y. ABFs, a Family of ABA-responsive Element Binding Factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, W.; Wang, X. Post-translational control of ABA signalling: The roles of protein phosphorylation and ubiquitination. Plant Biotechnology Journal. 2017, 15, 4–14. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.-C.; Liu, X.; Fu, L.; Hou, Y.-J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response. Mol. Cell 2018, 69, 100–112.e106. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Koonin, E.V.; Aravind, L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins 2001, 43, 134–144. [Google Scholar] [CrossRef]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef]
- Santiago, J.; Rodrigues, A.; Saez, A.; Rubio, S.; Antoni, R.; Dupeux, F.; Park, S.Y.; Márquez, J.A.; Cutler, S.R.; Rodriguez, P.L. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. Cell Mol. Biol. 2009, 60, 575–588. [Google Scholar] [CrossRef]
- Quan, W.; Hu, Y.; Mu, Z.; Shi, H.; Chan, Z. Overexpression of AtPYL5 under the control of guard cell specific promoter improves drought stress tolerance in Arabidopsis. Plant Physiol. Biochem. PPB 2018, 129, 150–157. [Google Scholar] [CrossRef]
- Dittrich, M.; Mueller, H.M.; Bauer, H.; Peirats-Llobet, M.; Rodriguez, P.L.; Geilfus, C.M.; Carpentier, S.C.; Al Rasheid, K.A.S.; Kollist, H.; Merilo, E.; et al. The role of Arabidopsis ABA receptors from the PYR/PYL/RCAR family in stomatal acclimation and closure signal integration. Nat. Plants 2019, 5, 1002–1011. [Google Scholar] [CrossRef]
- Zhao, Y.; Xing, L.; Wang, X.; Hou, Y.J.; Gao, J.; Wang, P.; Duan, C.G.; Zhu, X.; Zhu, J.K. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 2014, 7, ra53. [Google Scholar] [CrossRef]
- Li, H.; Gao, Z.; Chen, Q.; Li, Q.; Luo, M.; Wang, J.; Hu, L.; Zahid, M.S.; Wang, L.; Zhao, L.; et al. Grapevine ABA receptor VvPYL1 regulates root hair development in Transgenic Arabidopsis. Plant Physiol. Biochem. PPB 2020, 149, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Santosh Kumar, V.V.; Verma, R.K.; Yadav, P.; Saroha, A.; Wankhede, D.P.; Chaudhary, B.; Chinnusamy, V. Genome-wide identification and characterization of ABA receptor PYL gene family in rice. BMC Genom. 2020, 21, 676. [Google Scholar] [CrossRef]
- Bai, G.; Yang, D.H.; Zhao, Y.; Ha, S.; Yang, F.; Ma, J.; Gao, X.S.; Wang, Z.M.; Zhu, J.K. Interactions between soybean ABA receptors and type 2C protein phosphatases. Plant. Mol. Biol. 2013, 83, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.S.; Rajagopalan, N.; Risseeuw, E.P.; Surpin, M.; Ball, F.J.; Barber, C.J.; Buhrow, L.M.; Clark, S.M.; Page, J.E.; Todd, C.D.; et al. Characterization of Triticum aestivum Abscisic Acid Receptors and a Possible Role for These in Mediating Fusairum Head Blight Susceptibility in Wheat. PLoS ONE 2016, 11, e0164996. [Google Scholar] [CrossRef]
- Fan, W.; Zhao, M.; Li, S.; Bai, X.; Li, J.; Meng, H.; Mu, Z. Contrasting transcriptional responses of PYR1/PYL/RCAR ABA receptors to ABA or dehydration stress between maize seedling leaves and roots. BMC Plant Biol. 2016, 16, 99. [Google Scholar] [CrossRef]
- Yu, J.; Yang, L.; Liu, X.; Tang, R.; Wang, Y.; Ge, H.; Wu, M.; Zhang, J.; Zhao, F.; Luan, S.; et al. Overexpression of Poplar Pyrabactin Resistance-Like Abscisic Acid Receptors Promotes Abscisic Acid Sensitivity and Drought Resistance in Transgenic Arabidopsis. PLoS ONE 2016, 11, e0168040. [Google Scholar] [CrossRef]
- Guo, D.; Zhou, Y.; Li, H.-L.; Zhu, J.-H.; Wang, Y.; Chen, X.-T.; Peng, S.-Q. Identification and characterization of the abscisic acid (ABA) receptor gene family and its expression in response to hormones in the rubber tree. Sci. Rep. 2017, 7, 45157. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.-m.; Jia, H.-f.; Li, C.-l.; Dong, Q.-h.; Shen, Y.-y. FaPYR1 is involved in strawberry fruit ripening. J. Exp. Bot. 2011, 62, 5079–5089. [Google Scholar] [CrossRef]
- Zhang, G.; Lu, T.; Miao, W.; Sun, L.; Tian, M.; Wang, J.; Hao, F. Genome-wide identification of ABA receptor PYL family and expression analysis of PYLs in response to ABA and osmotic stress in Gossypium. PeerJ 2017, 5, e4126. [Google Scholar] [CrossRef]
- Hou, H.; Lv, L.; Huo, H.; Dai, H.; Zhang, Y. Genome-Wide Identification of the ABA Receptors Genes and Their Response to Abiotic Stress in Apple. Plants 2020, 9, 1028. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, X.; Yan, Y.; Shen, M.; Feng, R.; Wei, Q.; Zhang, L.; Zhang, M. Genome-wide analysis of the PYL gene family and identification of PYL genes that respond to cold stress in Triticum monococcum L. Subsp. Aegilopoides. Sci. Rep. 2024, 14, 26627. [Google Scholar] [CrossRef]
- Sun, Y.; Xiao, W.; Wang, Q.-n.; Wang, J.; Kong, X.-d.; Ma, W.-h.; Liu, S.-x.; Ren, P.; Xu, L.-n.; Zhang, Y.-J. Multiple variation patterns of terpene synthases in 26 maize genomes. BMC Genom. 2023, 24, 46. [Google Scholar] [CrossRef]
- Hufford, M.B.; Seetharam, A.S.; Woodhouse, M.R.; Chougule, K.M.; Ou, S.; Liu, J.; Ricci, W.A.; Guo, T.; Olson, A.; Qiu, Y.; et al. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science 2021, 373, 655–662. [Google Scholar] [CrossRef]
- Shi, T.; Zhang, X.; Hou, Y.; Jia, C.; Dan, X.; Zhang, Y.; Jiang, Y.; Lai, Q.; Feng, J.; Feng, J.; et al. The super-pangenome of Populus unveils genomic facets for its adaptation and diversification in widespread forest trees. Mol. Plant 2024, 17, 725–746. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.; Wu, Z.; Wang, X.; Jiang, Z.; Wang, Y.; Liu, H.; Qin, R.; Li, Z. Short-term transcriptomic responses of Populus euphratica roots and leaves to drought stress. J. For. Res. 2021, 32, 841–853. [Google Scholar] [CrossRef]
- Ottow, E.A.; Polle, A.; Brosché, M.; Kangasjärvi, J.; Dibrov, P.; Zörb, C.; Teichmann, T. Molecular characterization of PeNhaD1: The first member of the NhaD Na+/H+ antiporter family of plant origin. Plant Mol. Biol. 2005, 58, 75–88. [Google Scholar] [CrossRef]
- Ma, T.; Wang, J.; Zhou, G.; Yue, Z.; Hu, Q.; Chen, Y.; Liu, B.; Qiu, Q.; Wang, Z.; Zhang, J.; et al. Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 2013, 4, 2797. [Google Scholar] [CrossRef]
- Sun, J.; Xu, J.; Qiu, C.; Zhai, J.; Zhang, S.; Zhang, X.; Wu, Z.; Li, Z. The chromosome-scale genome and population genomics reveal the adaptative evolution of Populus pruinosa to desertification environment. Hortic. Res. 2024, 11, uhae034. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Long, Z.; Dan, X.; Feng, J.; Shi, T.; Jia, C.; Zhang, X.; Lai, Q.; Yang, G.; Zhang, H.; et al. Genomic insights into local adaptation and future climate-induced vulnerability of a keystone forest tree in East Asia. Nat. Commun. 2022, 13, 6541. [Google Scholar] [CrossRef] [PubMed]
- Long, Z.; Sang, Y.; Feng, J.; Zhang, X.; Shi, T.; Zhang, L.; Mao, K.; Rieseberg, L.H.; Liu, J.; Wang, J. Evolutionary genomics predicts adaptive genetic and plastic gene expression responses to climate change in a key alpine forest tree species. bioRxiv 2024. [Google Scholar] [CrossRef]
- Chen, L.; Jiang, Y.; Shi, T.; Jia, C.; Long, Z.; Zhang, X.; Sang, Y.; Liu, J.; Wang, J. Chromosomal-level genome assembly of Populus davidiana. bioRxiv 2023. [Google Scholar] [CrossRef]
- Schiffthaler, B.; Delhomme, N.; Bernhardsson, C.; Jenkins, J.; Jansson, S.; Ingvarsson, P.; Schmutz, J.; Street, N. An Improved Genome Assembly of the European Aspen Populus tremula. bioRxiv 2019. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, J.; Bi, H.; Yang, X.; Zhang, Z.; Su, Y.; Li, Z.; Zhang, L.; Sanderson, B.J.; Liu, J.; et al. Bioinformatic analysis of chromatin organization and biased expression of duplicated genes between two poplars with a common whole-genome duplication. Hortic. Res. 2021, 8, 62. [Google Scholar] [CrossRef]
- Li, Y.; Wang, D.; Wang, W.; Yang, W.; Gao, J.; Zhang, W.; Shan, L.; Kang, M.; Chen, Y.; Ma, T. A chromosome-level Populus qiongdaoensis genome assembly provides insights into tropical adaptation and a cryptic turnover of sex determination. Mol. Ecol. 2023, 32, 1366–1380. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Z.; Shi, T.; Dan, X.; Zhang, Y.; Liu, J.; Wang, J. Chromosomal-level genome assembly of Populus adenopoda. bioRxiv 2023. [Google Scholar] [CrossRef]
- Li, J.; Shi, C.; Sun, D.; He, Y.; Lai, C.; Lv, P.; Xiong, Y.; Zhang, L.; Wu, F.; Tian, C. The HAB1 PP2C is inhibited by ABA-dependent PYL10 interaction. Sci. Rep. 2015, 5, 10890. [Google Scholar] [CrossRef]
- Hao, Q.; Yin, P.; Li, W.; Wang, L.; Yan, C.; Lin, Z.; Wu, J.Z.; Wang, J.; Yan, S.F.; Yan, N. The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins. Mol. Cell 2011, 42, 662–672. [Google Scholar] [CrossRef]
- Sun, L.; Wang, Y.-P.; Chen, P.; Ren, J.; Ji, K.; Li, Q.; Li, P.; Dai, S.-J.; Leng, P. Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. J. Exp. Bot. 2011, 62, 5659–5669. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Lu, Y.; Zhu, J.K.; Botella, J.R. Genome editing for plant research and crop improvement. J. Integr. Plant Biol. 2021, 63, 3–33. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef]
- Tian, X.; Wang, Z.; Li, X.; Lv, T.; Liu, H.; Wang, L.; Niu, H.; Bu, Q. Characterization and Functional Analysis of Pyrabactin Resistance-Like Abscisic Acid Receptor Family in Rice. Rice 2015, 8, 28. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Z.; He, Y.; Liu, D.; Liu, Y.; Liang, C.; Xie, M.; Jia, Y.; Ke, Q.; Zhou, Y.; et al. Structural variation reshapes population gene expression and trait variation in 2,105 Brassica napus accessions. Nat. Genet. 2024, 56, 2538–2550. [Google Scholar] [CrossRef]
- Zhao, K.; Xue, H.; Li, G.; Chitikineni, A.; Fan, Y.; Cao, Z.; Dong, X.; Lu, H.; Zhao, K.; Zhang, L.; et al. Pangenome analysis reveals structural variation associated with seed size and weight traits in peanut. Nat. Genet. 2025, 57, 1250–1261. [Google Scholar] [CrossRef]
- Li, Z.; Fu, D.; Wang, X.; Zeng, R.; Zhang, X.; Tian, J.; Zhang, S.; Yang, X.; Tian, F.; Lai, J.; et al. The transcription factor bZIP68 negatively regulates cold tolerance in maize. Plant Cell 2022, 34, 2833–2851. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, R.R.; Chebotarov, D.; Duitama, J.; Smith, S.; De la Hoz, J.F.; Mohiyuddin, M.; Wing, R.A.; McNally, K.L.; Tatarinova, T.; Grigoriev, A.; et al. Structural variants in 3000 rice genomes. Genome Res. 2019, 29, 870–880. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, X.; Jia, Y.; Minkenberg, B.; Wheatley, M.; Fan, J.; Jia, M.H.; Famoso, A.; Edwards, J.D.; Wamishe, Y.; et al. The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Commun. 2018, 9, 2039. [Google Scholar] [CrossRef]
- Hao: Structural Biological Studies of the Abscisic Acid Receptor PYL Protein Family. Tsinghua University. Issue 9. 2015. Available online: https://kns.cnki.net/ (accessed on 8 April 2025).
- Fernández-Cancelo, P.; Muñoz, P.; Echeverría, G.; Larrigaudière, C.; Teixidó, N.; Munné-Bosch, S.; Giné-Bordonaba, J. Ethylene and abscisic acid play a key role in modulating apple ripening after harvest and after cold-storage. Postharvest Biol. Technol. 2022, 188, 111902. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, K.; Zhou, H.; Yan, X.; Zhan, Q.; Zheng, Y.; Song, C.-P. ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytologist. 2020, 228, 596–608. [Google Scholar] [CrossRef]
- Ramya, M.; Kwon, O.K.; An, H.R.; Park, P.M.; Baek, Y.S.; Park, P.H. Floral scent: Regulation and role of MYB transcription factors. Phytochem. Lett. 2017, 19, 114–120. [Google Scholar] [CrossRef]
- Lau, S.E.; Schwarzacher, T.; Othman, R.Y.; Harikrishna, J.A. dsRNA silencing of an R2R3-MYB transcription factor affects flower cell shape in a Dendrobium hybrid. BMC Plant Biol. 2015, 15, 194. [Google Scholar] [CrossRef]
- Wang, H.X.; Liu, T.Y.; Zhuang, W.B.; Wang, Z.Z.L.; Qu, S.C.; Zhai, H.H. Research advances in the function of anthocyanin in plant stress response. J. Agric. Biotechnol. 2020, 28, 174–183. [Google Scholar]
- Di, F.; Jian, H.; Wang, T.; Chen, X.; Ding, Y.; Du, H.; Lu, K.; Li, J.; Liu, L. Genome-Wide Analysis of the PYL Gene Family and Identification of PYL Genes That Respond to Abiotic Stress in Brassica napus. Genes 2018, 9, 156. [Google Scholar] [CrossRef]
- Li, Y.; Wang, D.; Sun, C.; Hu, X.; Mu, X.; Hu, J.; Yang, Y.; Zhang, Y.; Xie, C.G.; Zhou, X. Molecular characterization of an AtPYL1-like protein, BrPYL1, as a putative ABA receptor in Brassica rapa. Biochem. Biophys. Res. Commun. 2017, 487, 684–689. [Google Scholar] [CrossRef] [PubMed]
- Asad, M.A.U.; Zakari, S.A.; Zhao, Q.; Zhou, L.; Ye, Y.; Cheng, F. Abiotic Stresses Intervene with ABA Signaling to Induce Destructive Metabolic Pathways Leading to Death: Premature Leaf Senescence in Plants. Int. J. Mol. Sci. 2019, 20, 256. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 2018, 289660. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]

| Species | Abbreviation | Assembly Size (Mb) | Genome Data | Reference |
|---|---|---|---|---|
| Populus euphratica | EUP | 518.6 | Unpublished | |
| Populus pruinosa | PRU | 521.1 | https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA863418Pprgenome_fa/20705107/2 | [43] |
| Populus pseudoglauca | PSE | 448.7 | Database: https://bigd.big.ac.cn/gwh; Accession number: GWHBJUF00000000 | [39] |
| Populus wuana | WUA | 417.4 | Database: https://bigd.big.ac.cn/gwh; Accession number: GWHBJUE00000000 | [39] |
| Populus szechuanica | SZE | 429.1 | Database: https://bigd.big.ac.cn/gwh; Accession number: GWHBJUK00000000 | [39] |
| Populus yunnanensis | YUN | 433.7 | Database: https://bigd.big.ac.cn/gwh; Accession number: GWHBJUD00000000 | [39] |
| Populus koreana | KOR | 401.4 | Database: https://bigd.big.ac.cn/gwh; Accession number: GWHBHRS00000000 | [44] |
| Populus trichocarpa v4.1 | TRI | 392.2 | Database: https://phytozome-next.jgi.doe.gov/ | Phytozome |
| Populus deltoids v2.1 | DEL | 446.8 | Database: https://phytozome-next.jgi.doe.gov/ | Phytozome |
| Populus simonii | SIM | 408.0 | Database: https://bigd.big.ac.cn/gwh; Accession number: GWHBJTX00000000 | [39] |
| Populus lasiocarpa | LAS | 419.5 | Database: https://bigd.big.ac.cn/gwh; Accession number: GWHBJUB00000000 | [45] |
| Populus davidiana | DAV | 441.1 | Database: https://bigd.big.ac.cn/gwh; Accession number: GWHBJTW00000000 | [46] |
| Populus rotundifolia | ROT | 414.3 | Database: https://bigd.big.ac.cn/gwh; Accession number: GWHBJUA00000000 | [39] |
| Populus tremula | TRE | 408.8 | Database: http://popgenie.org | [47] |
| Populus alba var. pyramidalis | ALB | 408.1 | Database: http://bigd.big.ac.cn/bioproject; Accession number: PRJCA002423 | [48] |
| Populus qiongdaoensis | QIO | 391.3 | Database: http://bigd.big.ac.cn/bioproject; Accession number: PRJCA007862 | [49] |
| Populus adenopoda | ADE | 383.4 | Database: https://bigd.big.ac.cn/gwh; Accession number: GWHBJTV00000000 | [50] |
| Sequence ID | Number of Amino Acid | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | |
|---|---|---|---|---|---|---|
| I | PYL1 | 54~189 | 9673.47~21,213.94 | 5.89~10.17 | 38.12~43.1 | 81.9~110 |
| PYL2 | 190~205 | 21,377.46~2,3149.56 | 5.6~7.73 | 34.04~39.96 | 88.53~91.63 | |
| PYL3 | 706 | 77,272.84 | 5.22 | 30.48 | 85.44 | |
| PYL4 | 169~248 | 18,686~27,574.39 | 4.44~5.15 | 26.15~44.62 | 66.73~77.65 | |
| PYL24 | 202 | 22,653.53 | 4.7 | 47.65 | 74.75 | |
| PYL14 | 168~381 | 18,895.38~43,039.84 | 4.46~4.8 | 25.8~31.73 | 72.66~78.99 | |
| PYL21 | 188 | 21,413.6 | 8.44 | 34.03 | 81.38 | |
| PYL19 | 108~143 | 12,205.97~16,086.06 | 4.39~4.59 | 40.25~43.88 | 76.36~92.96 | |
| PYL15 | 169~329 | 18,665.8~36,424.99 | 4.71~5.55 | 29.9~42.91 | 72.12~86.8 | |
| PYL18 | 102~115 | 11,253.82~12,760.6 | 5.57~5.86 | 26.7~36.85 | 77.91~85 | |
| PYL12 | 103~163 | 11,430.91~18,163.62 | 4.46~4.9 | 23.22~45.19 | 77.04~93.74 | |
| PYL29 | 163 | 18,053.43 | 4.64 | 29.94 | 86.01 | |
| PYL26 | 186 | 20,986.81 | 6.44 | 39.15 | 84.25 | |
| PYL8 | 615 | 67,740.22 | 7.23 | 38.38 | 93.07 | |
| PYL30 | 190 | 21,527.63 | 6.06 | 38.55 | 91.63 | |
| PYL25 | 190 | 21,487.52 | 6.06 | 40.01 | 89.05 | |
| PYL27 | 190 | 21,463.46 | 6.06 | 40.31 | 88.53 | |
| PYL23 | 220 | 24,606.14 | 5.62 | 43.17 | 92 | |
| PYL13 | 191~229 | 21,355.14~25,681.5 | 5.78~6.11 | 40.64~48.96 | 94.29~98.25 | |
| PYL28 | 191 | 21,369.16 | 5.78 | 48.96 | 94.29 | |
| PYL16 | 134~282 | 15,264.5~32,181.38 | 5.55~8.03 | 38.94~44.91 | 95.9~100.9 | |
| II | PYL6 | 186~223 | 20,570.31~24,181.23 | 8.5~9.21 | 42.26~46.91 | 80.59~85.02 |
| PYL17 | 214~222 | 23,268.13~24,100.99 | 7.66~8.26 | 50.16~54.14 | 79.41~80.56 | |
| PYL22 | 306 | 34,088.89 | 6.3 | 46.78 | 93.24 | |
| PYL10 | 214~238 | 23,103.88~26,319.76 | 7.11~9.22 | 38.54~51.13 | 78.35~85.47 | |
| PYL11 | 214 | 23,086.16~23,310.38 | 7.1~7.68 | 49.26~54.69 | 86.82~90.93 | |
| III | PYL5 | 124~191 | 13,517.37~21,341.06 | 5.2~7.01 | 2.05~51.39 | 82.43~89.69 |
| PYL9 | 203~210 | 22,319.97~23,190 | 4.93~5.53 | 32.74~42.8 | 82.1~86.85 | |
| PYL7 | 201~259 | 22,392.17~28,658.25 | 5.08~5.62 | 29.23~34.58 | 79.57~84.33 | |
| PYL20 | 169 | 18,961.56~18,961.58 | 5.94 | 38.88~40.02 | 85.27~85.86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, X.; Qiu, C.; Gai, Z.; Zhai, J.; Song, J.; Sun, J.; Li, Z. Pan-Genome-Based Characterization of the PYL Transcription Factor Family in Populus. Plants 2025, 14, 2541. https://doi.org/10.3390/plants14162541
Han X, Qiu C, Gai Z, Zhai J, Song J, Sun J, Li Z. Pan-Genome-Based Characterization of the PYL Transcription Factor Family in Populus. Plants. 2025; 14(16):2541. https://doi.org/10.3390/plants14162541
Chicago/Turabian StyleHan, Xiaoli, Chen Qiu, Zhongshuai Gai, Juntuan Zhai, Jia Song, Jianhao Sun, and Zhijun Li. 2025. "Pan-Genome-Based Characterization of the PYL Transcription Factor Family in Populus" Plants 14, no. 16: 2541. https://doi.org/10.3390/plants14162541
APA StyleHan, X., Qiu, C., Gai, Z., Zhai, J., Song, J., Sun, J., & Li, Z. (2025). Pan-Genome-Based Characterization of the PYL Transcription Factor Family in Populus. Plants, 14(16), 2541. https://doi.org/10.3390/plants14162541






