Identification and Characterization of the StCPAI Gene Family in Potato
Abstract
1. Introduction
2. Results
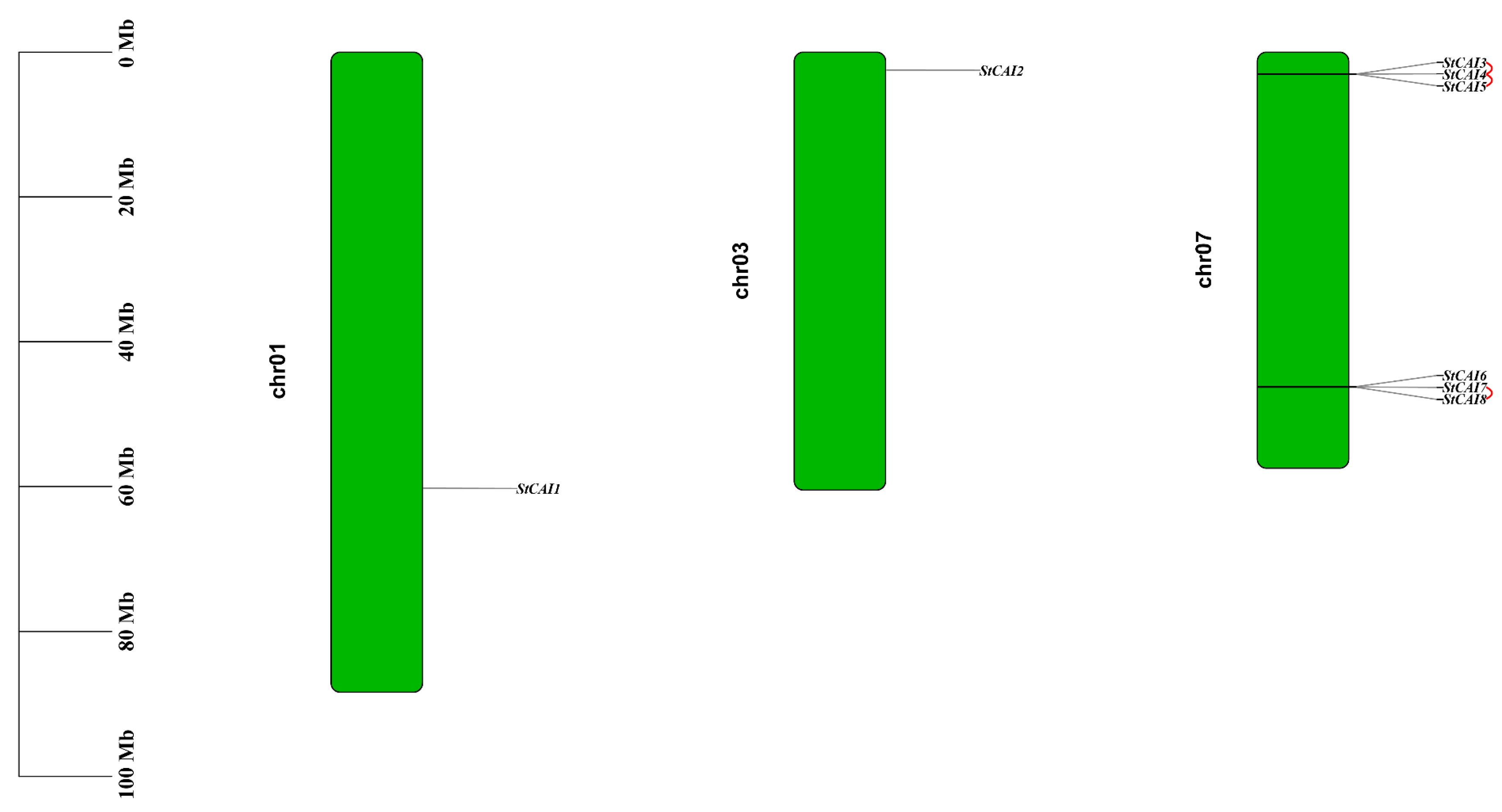
2.1. Identification and Chromosomal Distribution of the StCPAI Gene Family
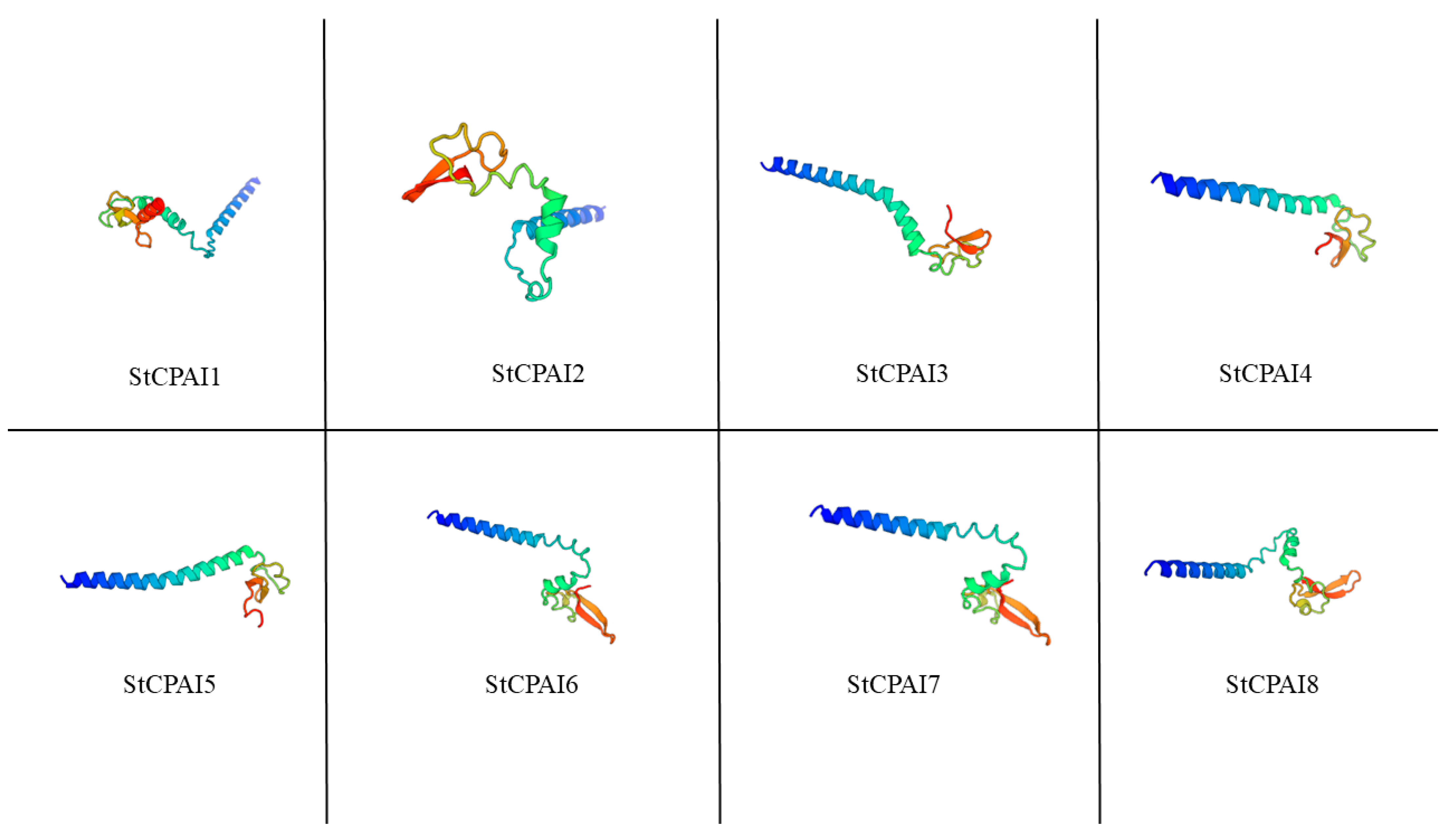
2.2. Physicochemical Properties, Subcellular Localization, and Protein Secondary/Tertiary Structure Prediction of StCPAIs
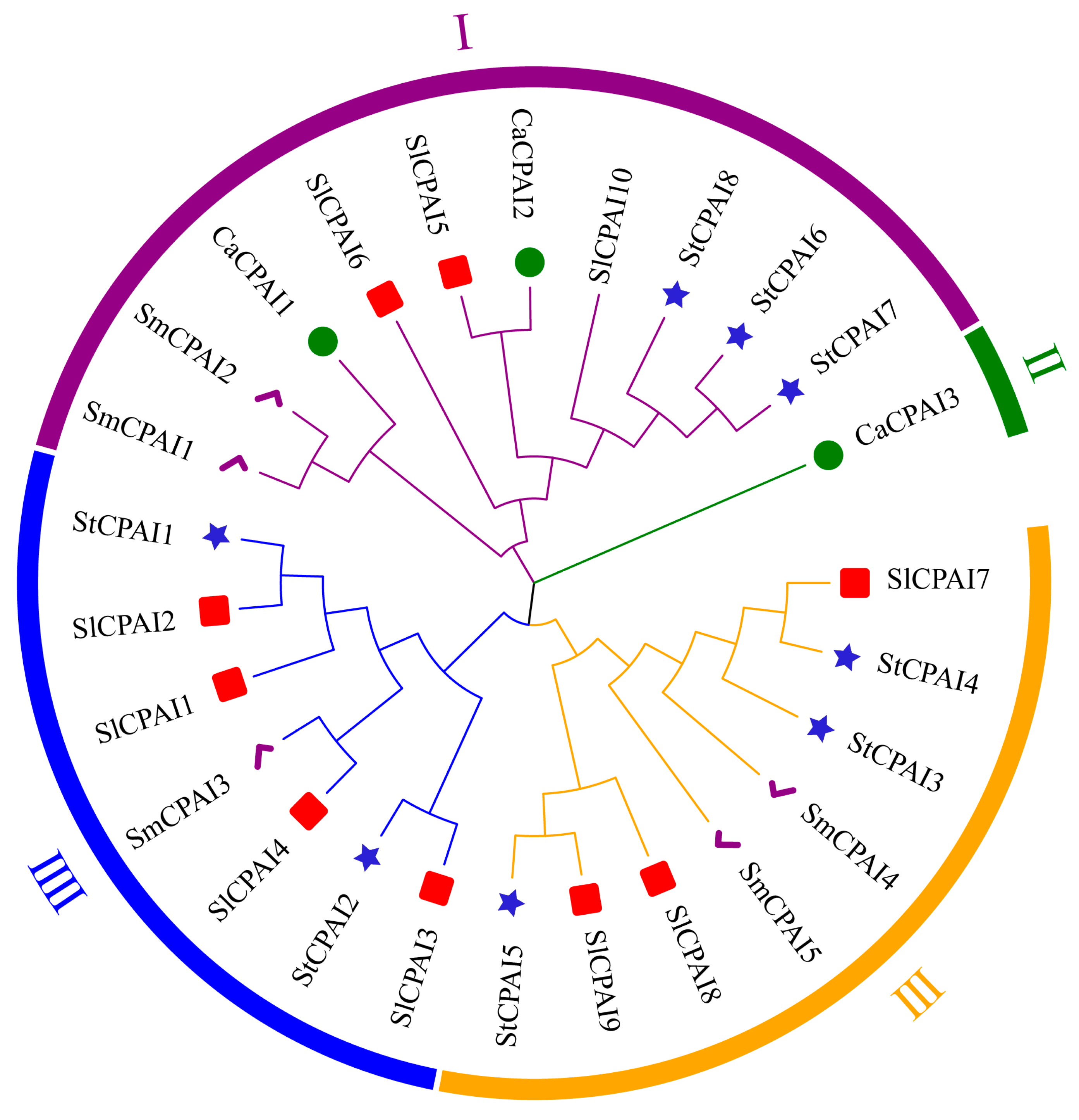
2.3. Phylogenetic Analysis of StCPAI
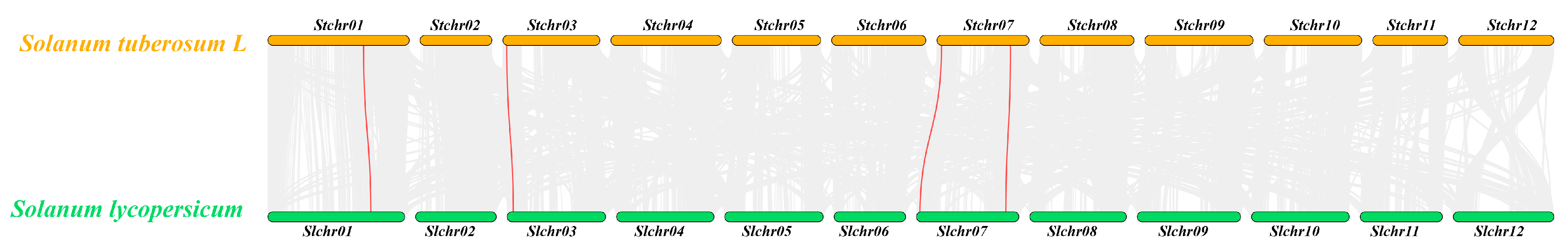
2.4. Collinearity Analysis Between StCPAI and Tomato SlCPAI Gene Families
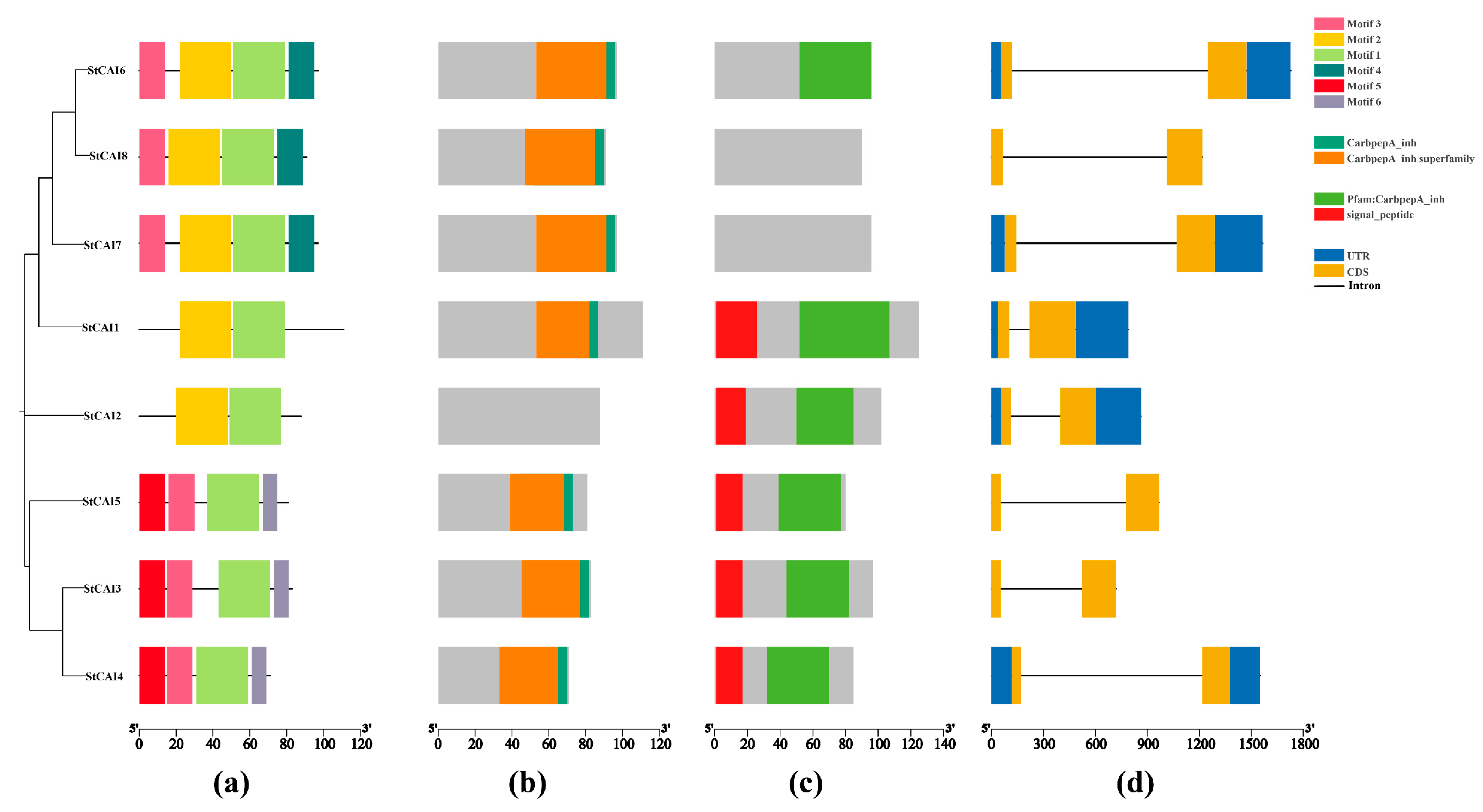
2.5. Conserved Motifs and Gene Structure Analysis of StCPAI Family Members
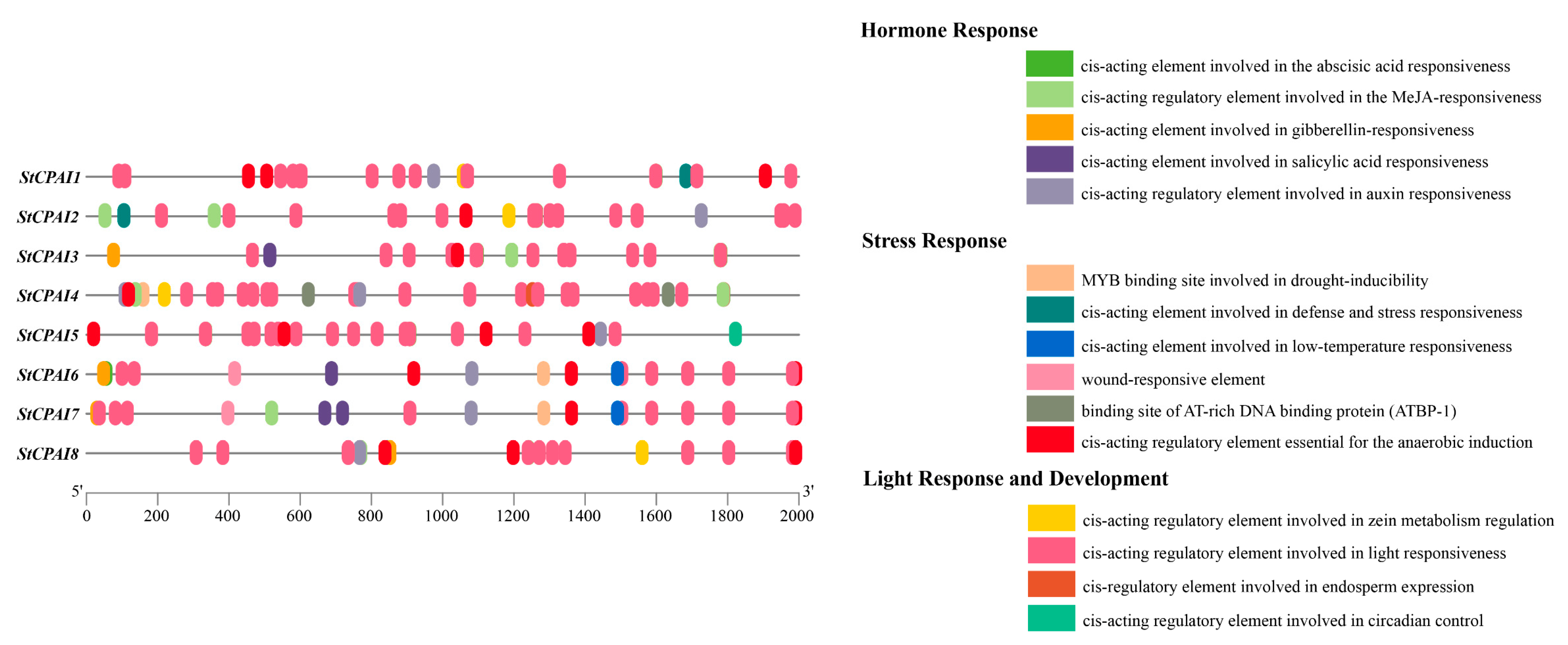
2.6. Analysis of Cis-Acting Elements in Promoters of StCPAI Gene Family Members
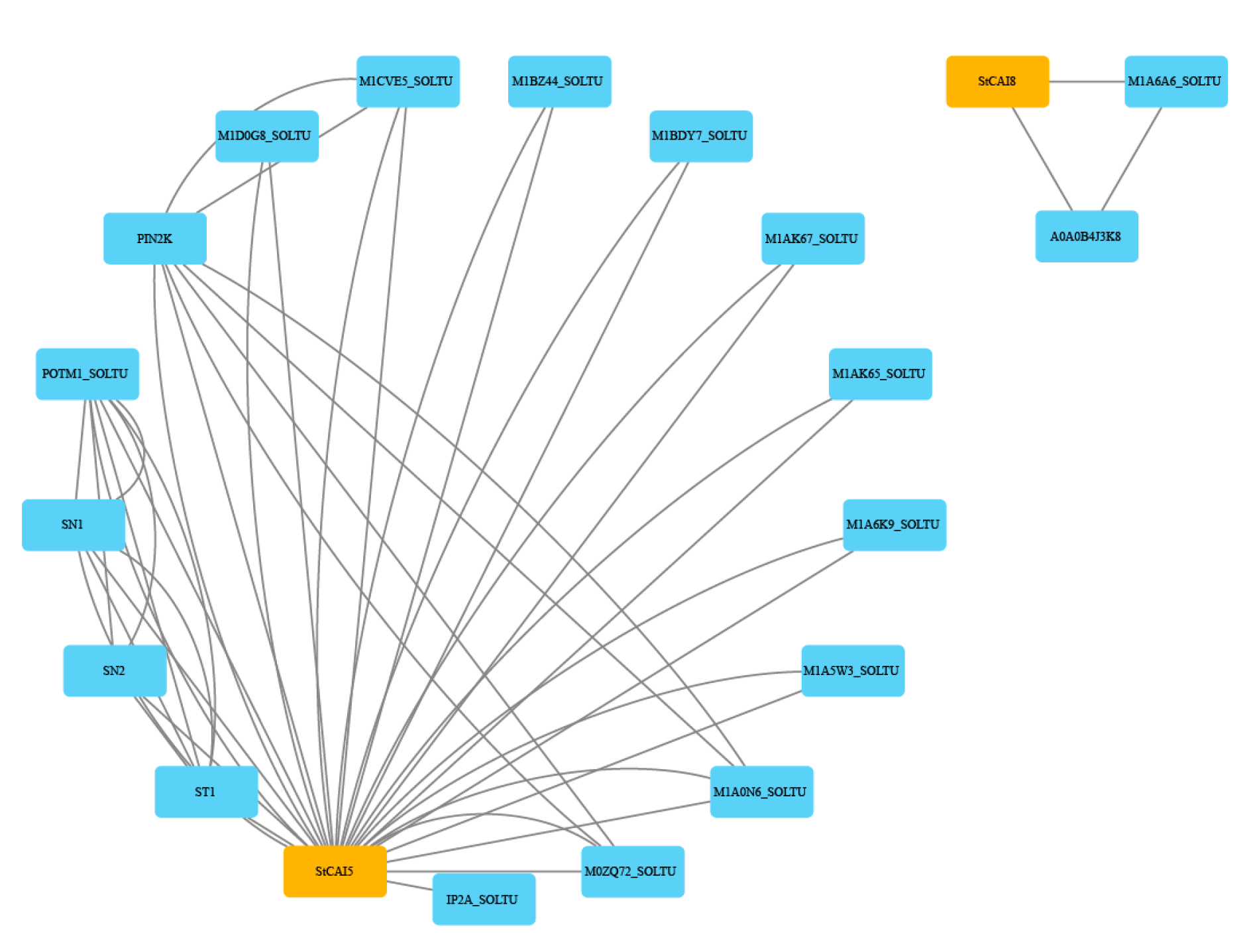
2.7. Analysis of the StCPAI Protein Interaction Network
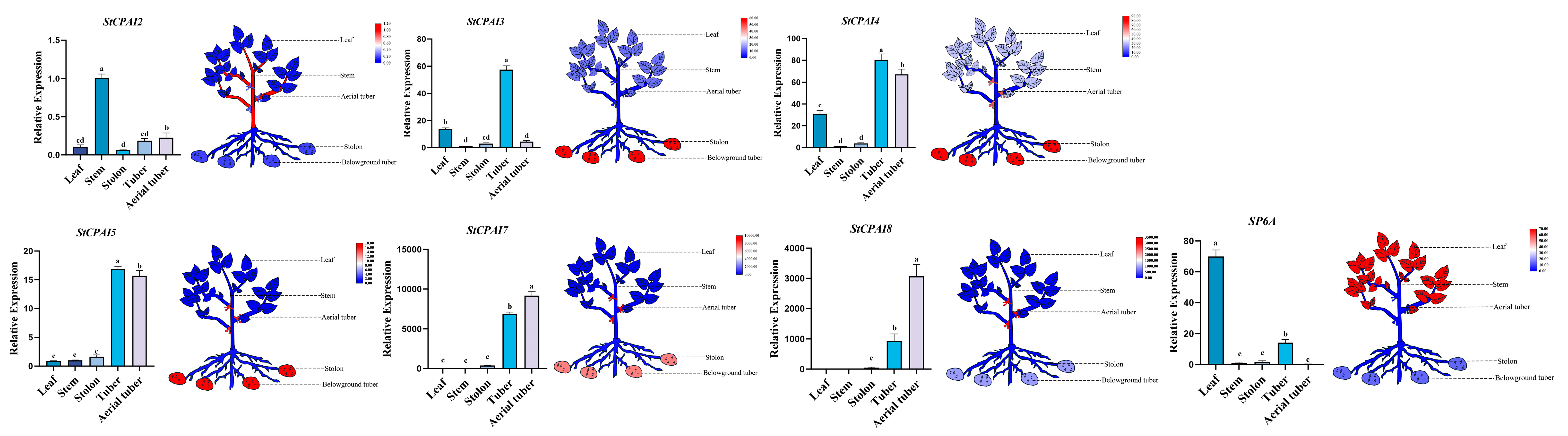
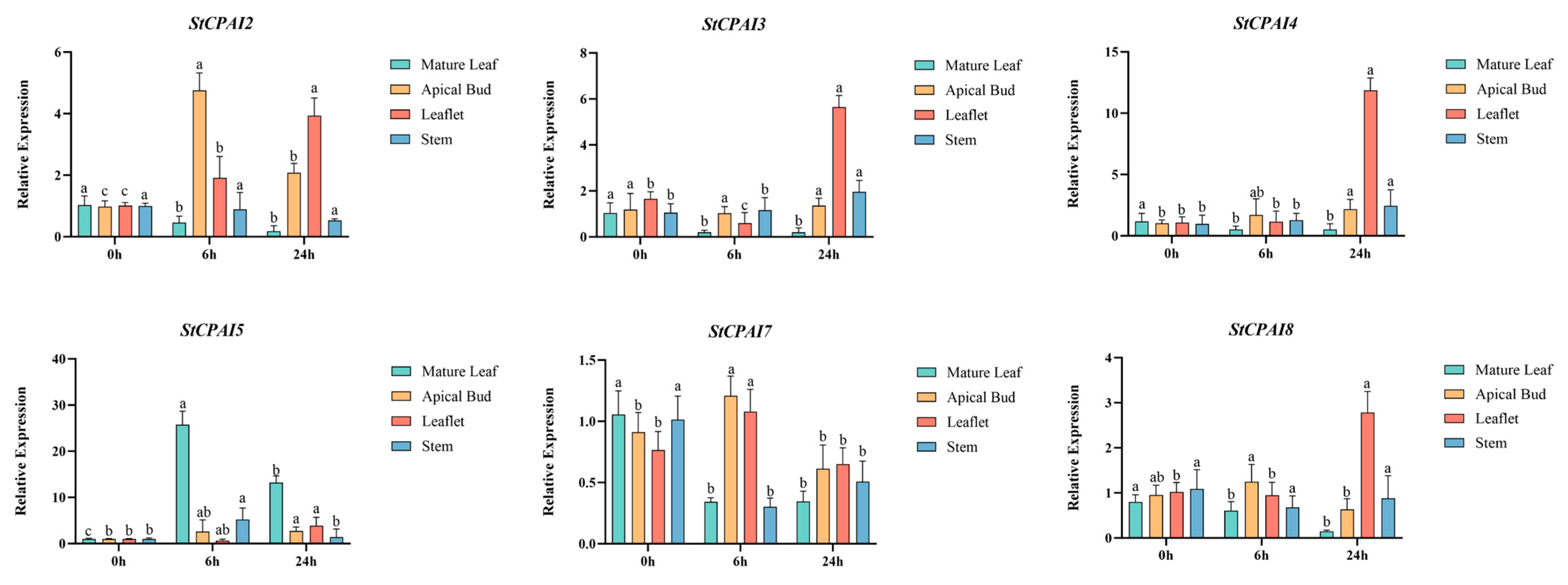
2.8. Expression Pattern Analysis of StCPAI Gene Family Members in Different Organs and Under Auxin Treatment
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of StCPAI Gene Family Members
4.3. Chromosomal Localization and Tandem Repeats of StCPAI Gene
4.4. Analysis of Physicochemical Properties, Subcellular Localization Prediction, Secondary and Tertiary Structures of StCPAI Protein
4.5. Phylogenetic Tree Analysis of StCPAI
4.6. Interspecies Synteny Analysis of StCPAI Family Members
4.7. Conserved Motif and Gene Structure Analysis of the StCPAI Family
4.8. Cis-Acting Element Analysis in StCPAI Promoters
4.9. StCPAI Protein Interaction Network Analysis
4.10. Quantitative Real-Time PCR (qRT-PCR)
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Camire, M.E.; Kubow, S.; Donnelly, D.J. Potatoes and Human Health. Crit. Rev. Food Sci. Nutr. 2009, 49, 823–840. [Google Scholar] [CrossRef] [PubMed]
- Burlingame, B.; Mouillé, B.; Charrondière, R. Nutrients, bioactive non-nutrients and anti-nutrients in potatoes. J. Food Compos. Anal. 2009, 22, 494–502. [Google Scholar] [CrossRef]
- Landa, B.B.; Cachinero-Díaz, J.M.; Lemanceau, P.; Jiménez-Díaz, R.M.; Alabouvette, C. Effect of fusaric acid and phytoanticipins on growth of rhizobacteria andFusarium oxysporum. Can. J. Microbiol. 2002, 48, 971–985. [Google Scholar] [CrossRef]
- Sels, J.; Mathys, J.; Coninck, B.D.; Cammue, B.; Bolle, M.D.D. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef]
- García-Olmedo, F.; Rodríguez-Palenzuela, P.; Molina, A.; Alamillo, J.M.; López-Solanilla, E.; Berrocal-Lobo, M.; Poza-Carrión, C. Antibiotic activities of peptides, hydrogen peroxide and peroxynitrite in plant defence. FEBS Lett. 2001, 498, 219–222. [Google Scholar] [CrossRef]
- Li, J.; Hu, S.; Jian, W.; Xie, C.; Yang, X. Plant antimicrobial peptides: Structures, functions, and applications. Bot. Stud. 2021, 62, 5. [Google Scholar] [CrossRef]
- Mazurkiewicz-Pisarek, A.; Baran, J.; Ciach, T. Antimicrobial Peptides: Challenging Journey to the Pharmaceutical, Biomedical, and Cosmeceutical Use. Int. J. Mol. Sci. 2023, 24, 9031. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.S.; Ryan, C.A. Accumulation of a metallo-carboxypeptidase inhibitor in leaves of wounded potato plants. Biochem. Biophys. Res. Commun. 1981, 101, 1164–1170. [Google Scholar] [CrossRef]
- Villanueva, J.; Canals, F.; Prat, S.; Ludevid, D.; Querol, E.; Avilés, F.X. Characterization of the wound-induced metallocarboxypeptidase inhibitor from potato1. FEBS Lett. 1998, 440, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Bayés, A.; Comellas-Bigler, M.; de la Vega, M.R.; Maskos, K.; Bode, W.; Aviles, F.X.; Jongsma, M.A.; Beekwilder, J.; Vendrell, J. Structural basis of the resistance of an insect carboxypeptidase to plant protease inhibitors. Proc. Natl. Acad. Sci. USA 2005, 102, 16602–16607. [Google Scholar] [CrossRef]
- Bayés, À.; Vega, M.R.d.l.; Vendrell, J.; Avilés, F.; Jongsma, M.; Beekwilder, J. Response of the digestive system of Helicoverpa zea to ingestion of potato carboxypeptidase inhibitor and characterization of an uninhibited carboxypeptidase B. Insect Biochem. Mol. Biol. 2006, 36, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Quilis, J.; Meynard, D.; Vila, L.; Avilés, F.X.; Guiderdoni, E.; San Segundo, B. A potato carboxypeptidase inhibitor gene provides pathogen resistance in transgenic rice. Plant Biotechnol. J. 2007, 5, 537–553. [Google Scholar] [CrossRef] [PubMed]
- Treggiari, D.; Zoccatelli, G.; Chignola, R.; Molesini, B.; Minuz, P.; Pandolfini, T. Tomato cystine-knot miniproteins possessing anti-angiogenic activity exhibit in vitro gastrointestinal stability, intestinal absorption and resistance to food industrial processing. Food Chem. 2017, 221, 1346–1353. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wen, J.; Liu, L.; Chen, J.; Wang, C.; Li, Z.; Wang, G.; Pichersky, E.; Xu, H. Engineering of tomato type VI glandular trichomes for trans-chrysanthemic acid biosynthesis, the acid moiety of natural pyrethrin insecticides. Metab. Eng. 2022, 72, 188–199. [Google Scholar] [CrossRef]
- Pease, J.B.; Guerrero, R.F.; Sherman, N.A.; Hahn, M.W.; Moyle, L.C. Molecular mechanisms of postmating prezygotic reproductive isolation uncovered by transcriptome analysis. Mol. Ecol. 2016, 25, 2592–2608. [Google Scholar] [CrossRef]
- Manara, A.; Fasani, E.; Molesini, B.; DalCorso, G.; Pennisi, F.; Pandolfini, T.; Furini, A. The Tomato Metallocarboxypeptidase Inhibitor I, which Interacts with a Heavy Metal-Associated Isoprenylated Protein, Is Implicated in Plant Response to Cadmium. Molecules 2020, 25, 700. [Google Scholar] [CrossRef]
- Harada, E.; Kim, J.-A.; Meyer, A.J.; Hell, R.; Clemens, S.; Choi, Y.-E. Expression Profiling of Tobacco Leaf Trichomes Identifies Genes for Biotic and Abiotic Stresses. Plant Cell Physiol. 2010, 51, 1627–1637. [Google Scholar] [CrossRef]
- Guruprasad, K.; Reddy, B.V.B.; Pandit, M.W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. Des. Sel. 1990, 4, 155–161. [Google Scholar] [CrossRef]
- Schilmiller, A.L.; Miner, D.P.; Larson, M.; McDowell, E.; Gang, D.R.; Wilkerson, C.; Last, R.L. Studies of a Biochemical Factory: Tomato Trichome Deep Expressed Sequence Tag Sequencing and Proteomics. Plant Physiol. 2010, 153, 1212–1223. [Google Scholar] [CrossRef]
- Ruggieri, V.; Francese, G.; Sacco, A.; D’Alessandro, A.; Rigano, M.M.; Parisi, M.; Milone, M.; Cardi, T.; Mennella, G.; Barone, A. An association mapping approach to identify favourable alleles for tomato fruit quality breeding. BMC Plant Biol. 2014, 14, 337. [Google Scholar] [CrossRef] [PubMed]
- D’Esposito, D.; Manzo, D.; Ricciardi, A.; Garonna, A.P.; De Natale, A.; Frusciante, L.; Pennacchio, F.; Ercolano, M.R. Tomato transcriptomic response to Tuta absoluta infestation. BMC Plant Biol. 2021, 21, 358. [Google Scholar] [CrossRef]
- Aliberti, A.; Olivieri, F.; Graci, S.; Rigano, M.M.; Barone, A.; Ruggieri, V. Genomic Dissection of a Wild Region in a Superior Solanum pennellii Introgression Sub-Line with High Ascorbic Acid Accumulation in Tomato Fruit. Genes 2020, 11, 847. [Google Scholar] [CrossRef] [PubMed]
- Navarro, C.; Abelenda, J.A.; Cruz-Oró, E.; Cuéllar, C.A.; Tamaki, S.; Silva, J.; Shimamoto, K.; Prat, S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 2011, 478, 119–122. [Google Scholar] [CrossRef]
- Shibagaki, N.; Grossman, A.R. The Role of the STAS Domain in the Function and Biogenesis of a Sulfate Transporter as Probed by Random Mutagenesis. J. Biol. Chem. 2006, 281, 22964–22973. [Google Scholar] [CrossRef]
- Oliveira-Lima, M.; Benko-Iseppon, A.; Neto, J.; Rodriguez-Decuadro, S.; Kido, E.; Crovella, S.; Pandolfi, V. Snakin: Structure, Roles and Applications of a Plant Antimicrobial Peptide. Curr. Protein Pept. Sci. 2017, 18, 368–374. [Google Scholar] [CrossRef]
- Berrocal-Lobo, M.; Segura, A.; Moreno, M.; López, G.; García-Olmedo, F.; Molina, A. Snakin-2, an Antimicrobial Peptide from Potato Whose Gene Is Locally Induced by Wounding and Responds to Pathogen Infection. Plant Physiol. 2002, 128, 951–961. [Google Scholar] [CrossRef]
- Rehman, S.; Aziz, E.; Akhtar, W.; Ilyas, M.; Mahmood, T. Structural and functional characteristics of plant proteinase inhibitor-II (PI-II) family. Biotechnol. Lett. 2017, 39, 647–666. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, J.; Zhang, Y.; Cai, X.; Gong, P.; Zhang, J.; Wang, T.; Li, H.; Ye, Z. Overexpression of SlGMEs leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Cell Rep. 2010, 30, 389–398. [Google Scholar] [CrossRef]
- Ma, L.; Wang, Y.; Liu, W.; Liu, Z. Overexpression of an alfalfa GDP-mannose 3, 5-epimerase gene enhances acid, drought and salt tolerance in transgenic Arabidopsis by increasing ascorbate accumulation. Biotechnol. Lett. 2014, 36, 2331–2341. [Google Scholar] [CrossRef]
- Zhang, G.-Y.; Liu, R.-R.; Zhang, C.-Q.; Tang, K.-X.; Sun, M.-F.; Yan, G.-H.; Liu, Q.-Q. Manipulation of the Rice L-Galactose Pathway: Evaluation of the Effects of Transgene Overexpression on Ascorbate Accumulation and Abiotic Stress Tolerance. PLoS ONE 2015, 10, e0125870. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.K.; Lin, T.; Hannapel, D.J. Untranslated Regions of a Mobile Transcript Mediate RNA Metabolism. Plant Physiol. 2009, 151, 1831–1843. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | Transcript ID | Molecular Mass (kDa) | Acid Amino Length | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Localization | Protein Secondary Structure | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alpha Helix | Beta Turn | Random Coil | |||||||||
| StCPAI1 | Soltu.DM.01G022210.1 | 13.93 | 125aa | 5.65 | 50.04 | 73.36 | 0.021 | Extracellular | 38 | 0 | 54 |
| StCPAI2 | Soltu.DM.03G002590.1 | 11.5 | 102aa | 7.59 | 21.65 | 83.14 | 0.215 | Extracellular | 37 | 0 | 42 |
| StCPAI3 | Soltu.DM.07G002650.1 | 11.24 | 97aa | 5.38 | 41.39 | 73.4 | −0.105 | Extracellular | 31 | 0 | 39 |
| StCPAI4 | Soltu.DM.07G002660.1 | 9.68 | 85aa | 5.32 | 41.83 | 70 | 0.004 | Extracellular | 30 | 0 | 32 |
| StCPAI5 | Soltu.DM.07G002670.1 | 9.03 | 80aa | 7.64 | 43.51 | 74.5 | −0.116 | Extracellular | 36 | 0 | 38 |
| StCPAI6 | Soltu.DM.07G016200.1 | 10.64 | 96aa | 8.58 | 56.51 | 94.48 | 0.362 | Extracellular | 36 | 0 | 44 |
| StCPAI7 | Soltu.DM.07G016210.1 | 10.57 | 96aa | 8.58 | 58.52 | 81.25 | 0.27 | Extracellular | 38 | 0 | 50 |
| StCPAI8 | Soltu.DM.07G016220.1 | 9.99 | 90aa | 8.25 | 43.23 | 87.78 | 0.296 | Extracellular | 39 | 0 | 34 |
| Gene | Forward Primer Sequence (5′→3′) | Reverse Primer Sequence (5′→3′) | Amplicon Length |
|---|---|---|---|
| qStCPAI2 | ATTGTGACTTGCGGGCATCGTTG | CGCAATGAGAAACCGCAGACCCA | 100 bp |
| qStCPAI3 | GGCCCAAGAAGATATACAACAAAT | CAAGTTTGTCTGAAATTCCAACAG | 157 bp |
| qStCPAI4 | GGCCCAAGAAGATCAAGATCCAAT | CAAGTTTGTCTGAAATTCCAACAG | 105 bp |
| qStCPAI5 | CAAGATGTGACGAAACTTTTTCAGGA | TTATTGAAAACTCTGGCGCCGC | 147 bp |
| qStCPAI7 | TCCCCCAGTGGTGGTACATACAG | CGCACGCACATAAACACACATAGA | 94 bp |
| qStCPAI8 | CCGCTTTAGTTAATATGCAAGTG | CAATCGGCGTTTGTGTTGCA | 130 bp |
| qACTIN11 | GGATCTTGCTGGTCGTGATTTAAC | CATAGGCAAGCTTTTCCTTCATGT | 122 bp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Wu, W.; Liu, T.; Shi, W.; Ma, K.; He, Z.; Chen, L.; Du, C.; Wang, C.; Yang, Z. Identification and Characterization of the StCPAI Gene Family in Potato. Plants 2025, 14, 2472. https://doi.org/10.3390/plants14162472
Wang Z, Wu W, Liu T, Shi W, Ma K, He Z, Chen L, Du C, Wang C, Yang Z. Identification and Characterization of the StCPAI Gene Family in Potato. Plants. 2025; 14(16):2472. https://doi.org/10.3390/plants14162472
Chicago/Turabian StyleWang, Zhiqi, Wenbo Wu, Tao Liu, Wenting Shi, Kai Ma, Zhouwen He, Lixuan Chen, Chong Du, Chaonan Wang, and Zhongmin Yang. 2025. "Identification and Characterization of the StCPAI Gene Family in Potato" Plants 14, no. 16: 2472. https://doi.org/10.3390/plants14162472
APA StyleWang, Z., Wu, W., Liu, T., Shi, W., Ma, K., He, Z., Chen, L., Du, C., Wang, C., & Yang, Z. (2025). Identification and Characterization of the StCPAI Gene Family in Potato. Plants, 14(16), 2472. https://doi.org/10.3390/plants14162472






