Genome-Wide Association Study and Candidate Gene Identification for Girth Traits in Rubber Tree
Abstract
1. Introduction
2. Results
2.1. Phenotypic Data Analysis
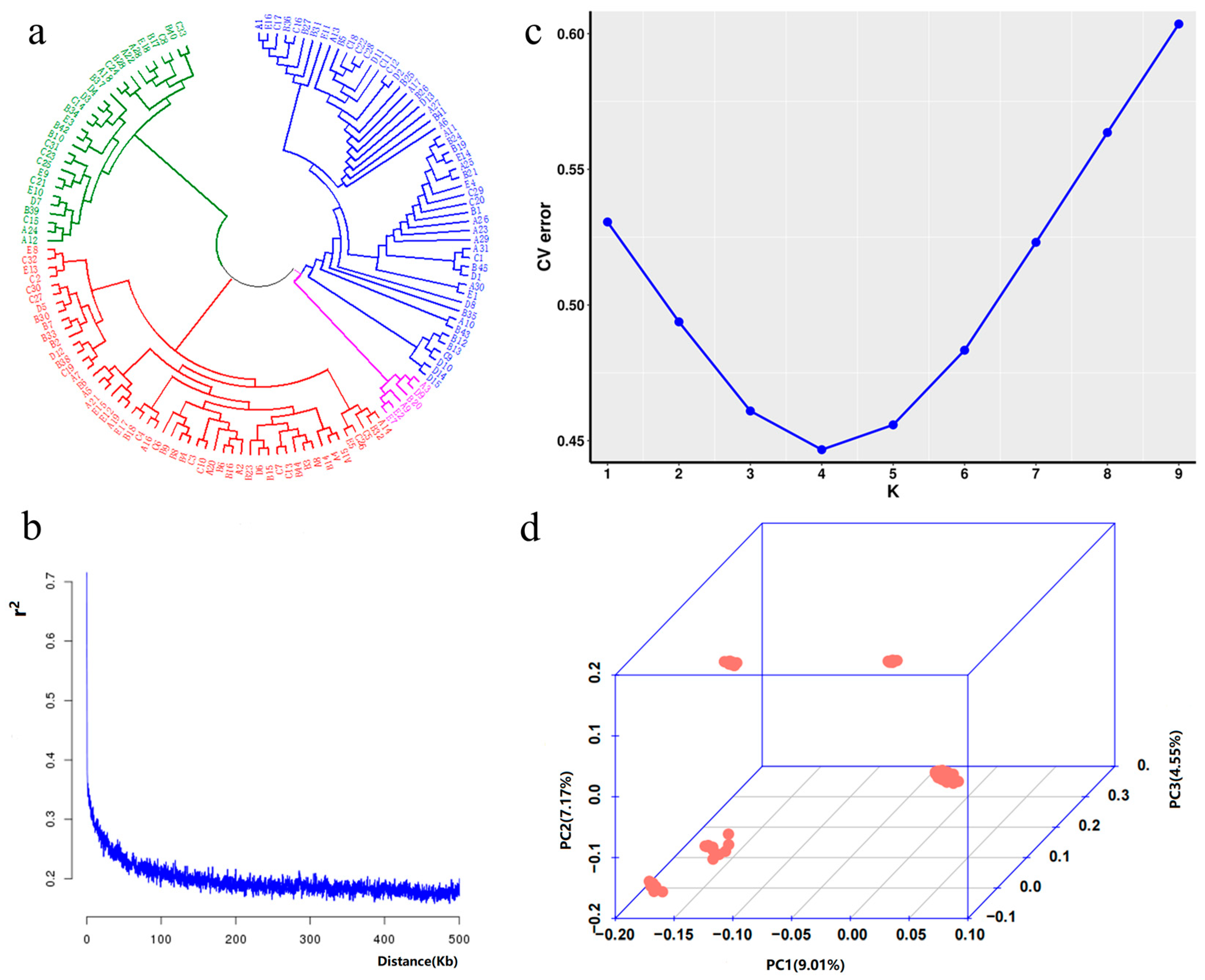
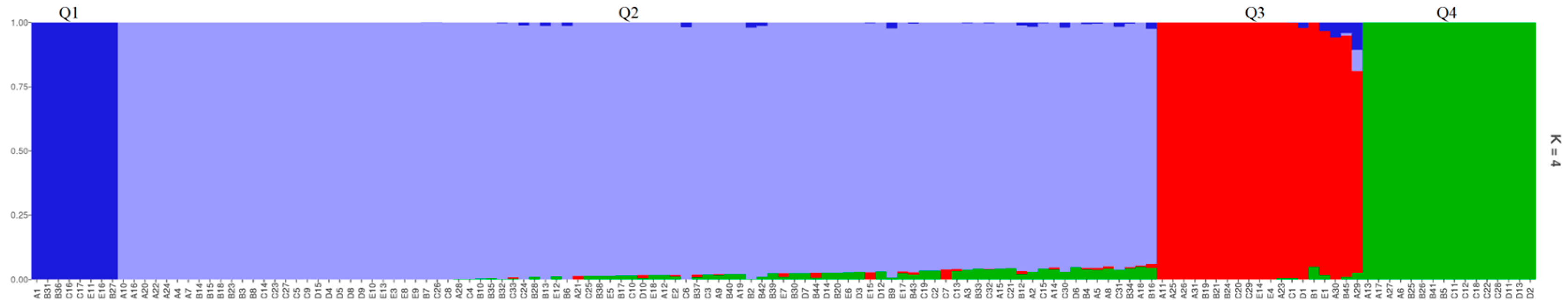
2.2. Population Stratification Analysis
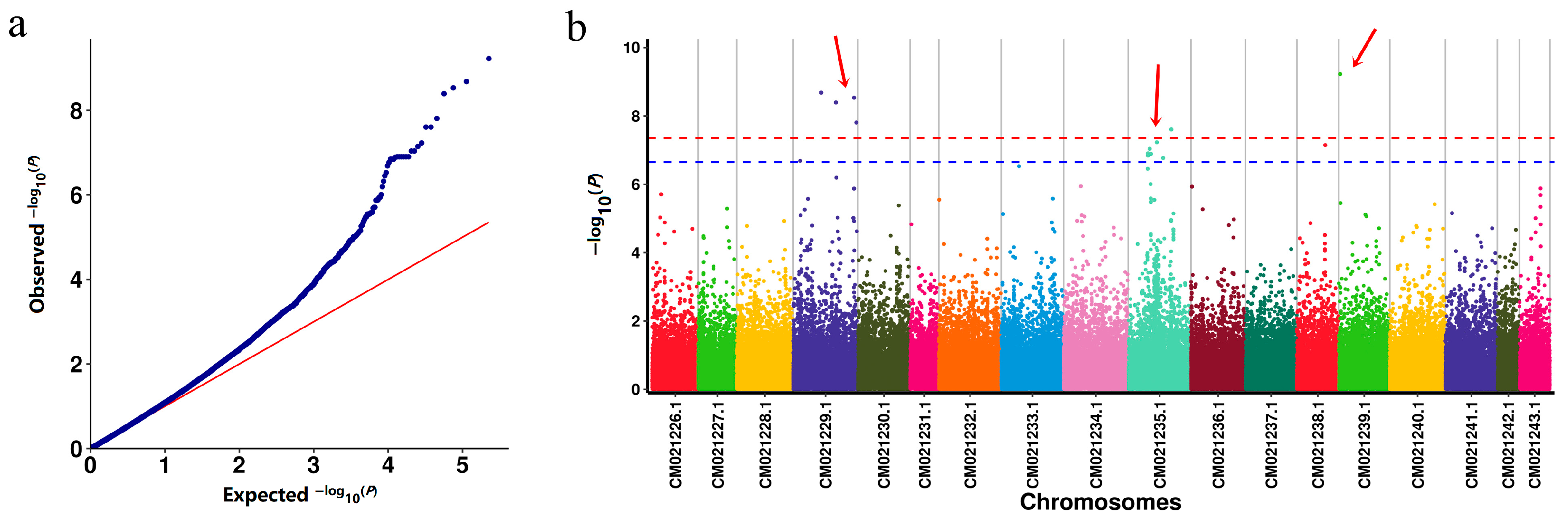
2.3. Genome-Wide Association Analysis
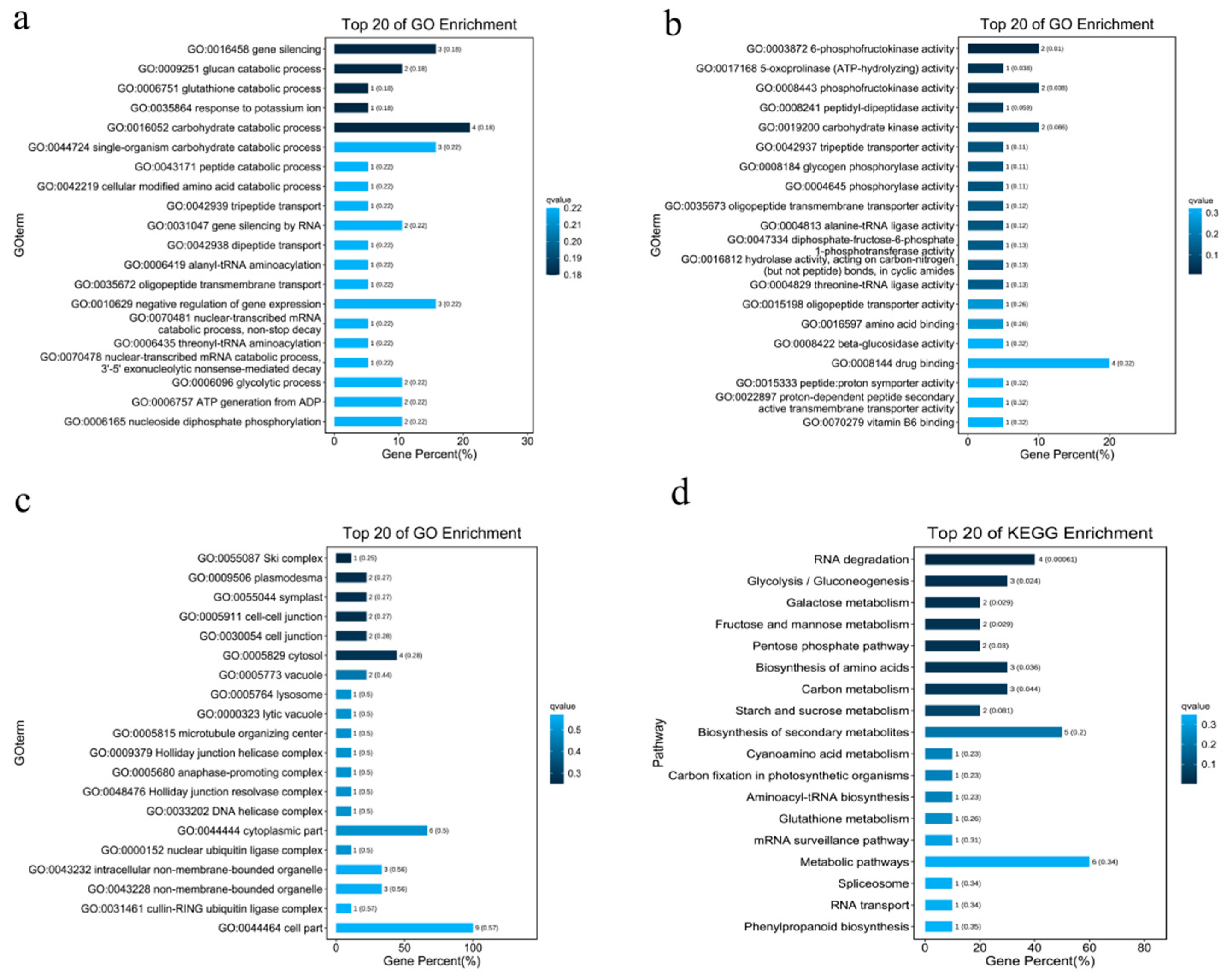
2.4. Candidate Gene Identification and Enrichment Analysis
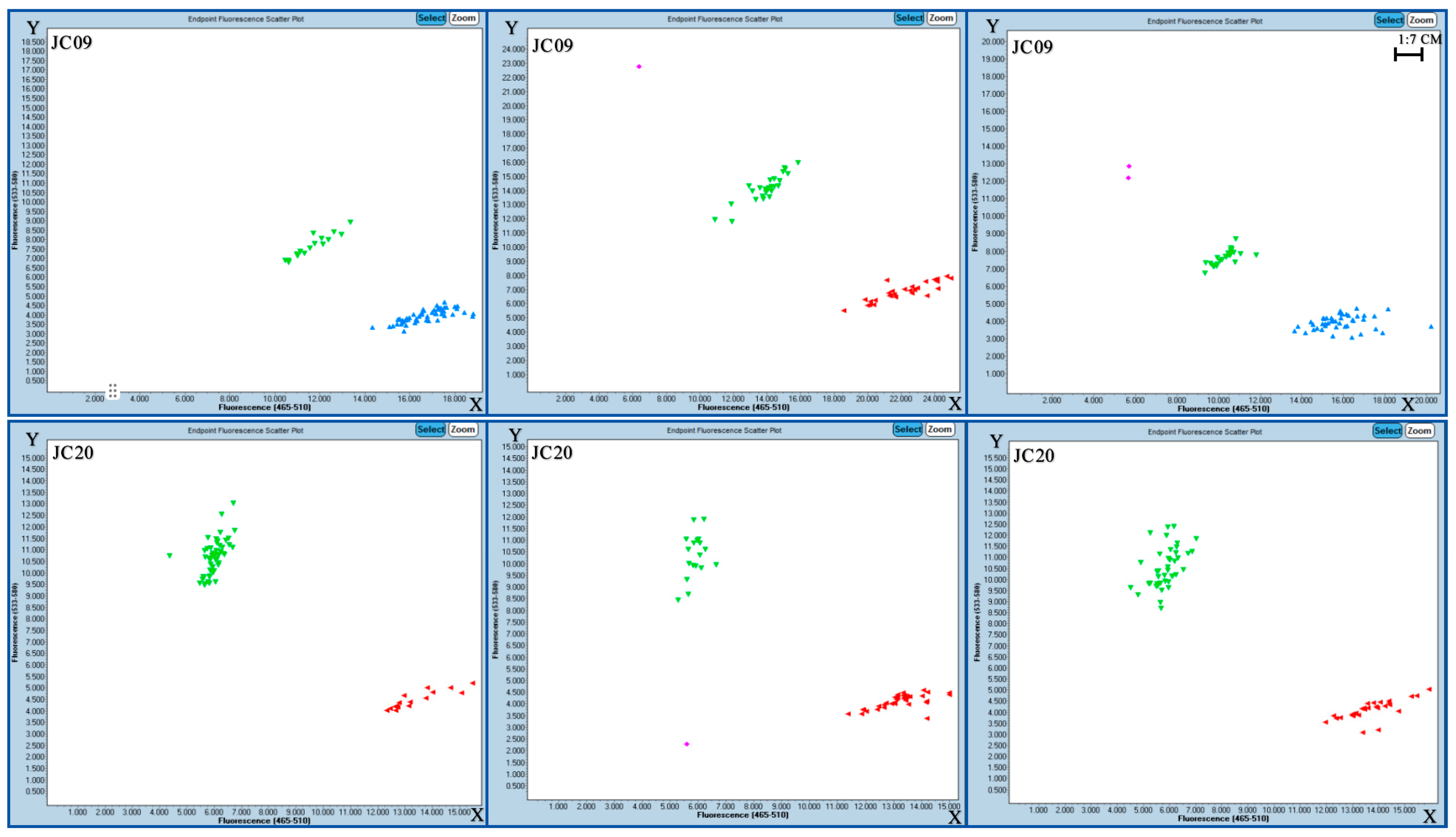
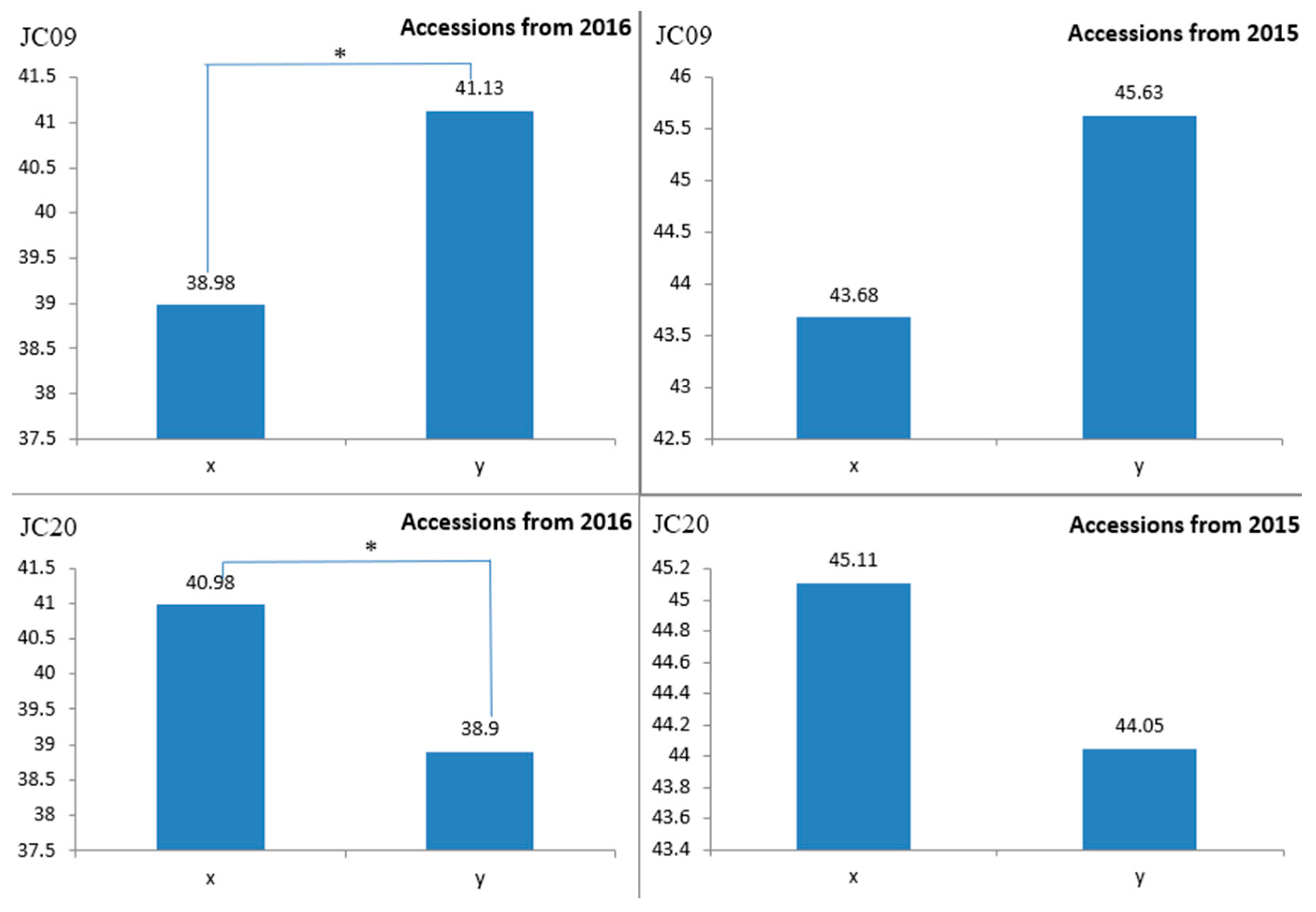
2.5. Verification of SNP Molecular Markers
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Phenotypic Data Collection and Analysis
4.3. Genotyping by Sequencing
4.3.1. DNA Extraction and Quality Testing
- Sample processing: Grind 1–2 g of fresh leaves to a powder with liquid nitrogen.
- Lysis and purification: Add CTAB extraction solution (65 °C water bath for 1 h), then extract with chloroform-isoamyl alcohol (24:1) to remove protein.
- Precipitation and washing: Precipitate DNA with isopropanol and wash three times with 75% ethanol.
- Dissolution and quality testing: Dissolve the DNA precipitate in ddH2O. Check the concentration and purity (OD260/OD280 = 1.8 ± 0.2) with a BioPhotometer and confirm integrity via 1% agarose gel electrophoresis.
4.3.2. Library Construction
4.3.3. Genotyping by Sequencing
4.4. Population Stratification Analysis
4.4.1. Systematically Developed Tree
4.4.2. Linkage Disequilibrium (LD) Decay Analysis
4.4.3. Population Structure Analysis
4.4.4. Principal Component Analysis (PCA) and Kinship Analysis
4.5. Genome-Wide Association Study (GWAS)
4.6. Enrichment Analysis Method
4.6.1. Analysis Tools and Databases
4.6.2. Statistical Testing and Significance Assessment
4.6.3. Visualization
4.7. Validation of SNP Molecular Markers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GWAS | Genome-wide association analysis |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LD | Linkage disequilibrium |
| QTL | Quantitative trait locus |
| SD | Standard deviation |
| SE | Standard error |
| CV | Coefficient of variation |
| GDI | Genetic diversity index |
| MLM | Mixed linear model |
References
- Yang, B.; Huang, X.; Zhang, Y.; Gao, X.; Ding, S.; Qi, J.; Wang, X. Genome-Wide Identification of Rubber Tree SCAMP Genes and Functional Characterization of HbSCAMP3. Plants 2024, 13, 2729. [Google Scholar] [CrossRef]
- Chao, J.; Wu, S.; Shi, M.; Xu, X.; Gao, Q.; Du, H.; Gao, B.; Guo, D.; Yang, S.; Zhang, S.; et al. Genomic Insight into Domestication of Rubber Tree. Nat. Commun. 2023, 14, 4651. [Google Scholar] [CrossRef]
- Chanroj, V.; Rattanawong, R.; Phumichai, T.; Tangphatsornruang, S.; Ukoskit, K. Genome-Wide Association Mapping of Latex Yield and Girth in Amazonian Accessions of Hevea brasiliensis Grown in a Suboptimal Climate Zone. Genomics 2017, 109, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tan, Y.; Xing, J.; Liu, X.; Zeng, X.; Li, W.; Wu, Y.; Zhang, S.; Qi, J.; Li, Z.; et al. Resequencing-Based QTL Mapping of Girth and Rubber Yield Traits in a Full-Sib Rubber Tree Population GT1 × CATAS8-79. Ind. Crops Prod. 2024, 222, 119867. [Google Scholar] [CrossRef]
- Bhusudsawang, G.; Rattanawong, R.; Phumichai, T.; Pootakham, W.; Tangphatsornruang, S.; Ukoskit, K. Identification of Candidate Gene-Based Markers for Girth Growth in Rubber Trees. Plants 2021, 10, 1440. [Google Scholar] [CrossRef] [PubMed]
- Francisco, F.R.; Aono, A.H.; da Silva, C.C.; Gonçalves, P.S.; Junior, E.J.S.; Le Guen, V.; Fritsche-Neto, R.; Souza, L.M.; de Souza, A.P. Unravelling Rubber Tree Growth by Integrating GWAS and Biological Network-Based Approaches. Front. Plant Sci. 2021, 12, 768589. [Google Scholar] [CrossRef]
- Souza, L.M.; Gazaffi, R.; Mantello, C.C.; Silva, C.C.; Garcia, D.; Le Guen, V.; Cardoso, S.E.A.; Garcia, A.A.F.; Souza, A.P. QTL Mapping of Growth-Related Traits in a Full-Sib Family of Rubber Tree (Hevea brasiliensis) Evaluated in a Sub-Tropical Climate. PLoS ONE 2013, 8, e61238. [Google Scholar] [CrossRef]
- Conson, A.R.O.; Taniguti, C.H.; Amadeu, R.R.; Andreotti, I.A.A.; de Souza, L.M.; Dos Santos, L.H.B.; Rosa, J.R.B.F.; Mantello, C.C.; da Silva, C.C.; Junior, E.J.S.; et al. High-Resolution Genetic Map and QTL Analysis of Growth-Related Traits of Hevea brasiliensis Cultivated Under Suboptimal Temperature and Humidity Conditions. Front. Plant Sci. 2018, 9, 1255. [Google Scholar] [CrossRef]
- Liang, C.; Zhang, Y.; Liu, X.; Tan, Y.; Xu, T.; Zheng, C.; Zhang, S.; Qi, J.; Liu, X.; Zeng, X. Genome-Wide Association Study Identifies QTL for Girth and Dry Rubber Yield in a Progeny Population of Whickham Hevea Germplasms. Ind. Crops Prod. 2024, 216, 118749. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The Advantages and Limitations of Trait Analysis with GWAS: A Review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef]
- Hart, A.; Kranzler, H. Alcohol Dependence Genetics: Lessons Learned from Genome-Wide Association Studies (GWAS) and Post-GWAS Analyses. Alcohol. Clin. Exp. Res. 2015, 39, 1312–1327. [Google Scholar] [CrossRef]
- Tibbs Cortes, L.; Zhang, Z.; Yu, J. Status and Prospects of Genome-wide Association Studies in Plants. Plant Genome 2021, 14, e20077. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and Limitations of Genome-Wide Association Studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Wang, L.; Qu, L.; Liu, H.; Sun, Y.; Le, M.; Wang, Q.; Wei, S.; Zheng, Y.; Lin, W.; et al. Genomic Evolution and Insights into Agronomic Trait Innovations of Sesamum Species. Plant Commun. 2024, 5, 100729. [Google Scholar] [CrossRef]
- Shang, Y.; Ma, Y.; Zhou, Y.; Zhang, H.; Duan, L.; Chen, H.; Zeng, J.; Zhou, Q.; Wang, S.; Gu, W.; et al. Plant Science. Biosynthesis, Regulation, and Domestication of Bitterness in Cucumber. Science 2014, 346, 1084–1088. [Google Scholar] [CrossRef] [PubMed]
- Mokrzycka, M.; Stojałowski, S.; Tyrka, M.; Matysik, P.; Żmijewska, B.; Marcinkowski, R.; Woźna-Pawlak, U.; Martofel, R.; Rokicki, M.; Rakoczy-Trojanowska, M.; et al. Genome-Wide Association Analysis for Hybrid Breeding in Wheat. Int. J. Mol. Sci. 2022, 23, 15321. [Google Scholar] [CrossRef] [PubMed]
- Warraich, A.S.; Krishnamurthy, S.L.; Sooch, B.S.; Vinaykumar, N.M.; Dushyanthkumar, B.M.; Bose, J.; Sharma, P.C. Rice GWAS Reveals Key Genomic Regions Essential for Salinity Tolerance at Reproductive Stage. Acta Physiol. Plant. 2020, 42, 134. [Google Scholar] [CrossRef]
- Yan, Y.; Liang, C.; Liu, X.; Tan, Y.; Lu, Y.; Zhang, Y.; Luo, H.; He, C.; Cao, J.; Tang, C.; et al. Genome-Wide Association Study Identifies Candidate Genes Responsible for Inorganic Phosphorus and Sucrose Content in Rubber Tree Latex. Trop. Plants 2023, 2. [Google Scholar] [CrossRef]
- Cheng, H.; Song, X.; Hu, Y.; Wu, T.; Yang, Q.; An, Z.; Feng, S.; Deng, Z.; Wu, W.; Zeng, X.; et al. Chromosome-Level Wild Hevea brasiliensis Genome Provides New Tools for Genomic-Assisted Breeding and Valuable Loci to Elevate Rubber Yield. Plant Biotechnol. J. 2023, 21, 1058–1072. [Google Scholar] [CrossRef]
- Cun, Z.; Li, X.; Zhang, J.-Y.; Hong, J.; Gao, L.-L.; Yang, J.; Ma, S.-Y.; Chen, J.-W. Identification of Candidate Genes and Residues for Improving Nitrogen Use Efficiency in the N-Sensitive Medicinal Plant Panax Notoginseng. BMC Plant Biol. 2024, 24, 105. [Google Scholar] [CrossRef]
- Ohkama-Ohtsu, N.; Oikawa, A.; Zhao, P.; Xiang, C.; Saito, K.; Oliver, D.J. A Gamma-Glutamyl Transpeptidase-Independent Pathway of Glutathione Catabolism to Glutamate via 5-Oxoproline in Arabidopsis. Plant Physiol. 2008, 148, 1603–1613. [Google Scholar] [CrossRef]
- Perby, L.K.; Richter, S.; Weber, K.; Hieber, A.J.; Hess, N.; Crocoll, C.; Mogensen, H.K.; Pribil, M.; Burow, M.; Nielsen, T.H.; et al. Cytosolic Phosphofructokinases Are Important for Sugar Homeostasis in Leaves of Arabidopsis thaliana. Ann. Bot. 2022, 129, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Schopper, S.; Mühlenbock, P.; Sörensson, C.; Hellborg, L.; Lenman, M.; Widell, S.; Fettke, J.; Andreasson, E. Arabidopsis Cytosolic Alpha-Glycan Phosphorylase, PHS2, Is Important During Carbohydrate Imbalanced Conditions. Plant Biol. Stuttg. Ger. 2015, 17, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Arisha, M.H.; Zhang, Z.; Yan, H.; Kou, M.; Song, W.; Li, C.; Gao, R.; Ma, M.; Wang, X.; et al. Comparative Transcriptomic and Proteomic Analysis Reveals Common Molecular Factors Responsive to Heat and Drought Stresses in Sweetpotaoto (Ipomoea batatas). Front. Plant Sci. 2022, 13, 1081948. [Google Scholar] [CrossRef]
- Ayaz, A.; Saqib, S.; Huang, H.; Zaman, W.; Lü, S.; Zhao, H. Genome-wide comparative analysis of long-chain acyl-CoA synthetases (LACSs) gene family: A focus on identification, evolution and expression profiling related to lipid synthesis. Plant Physiol. Biochem. 2021, 161, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Saqib, S.; Ullah, F.; Omollo, W.; Liu, Y.; Tao, H.; Zaman, W.; Temur, A.; Liu, B.; Lai, Y.; Chen, Z.; et al. Identifying hotspots and climate drivers of alien plant species for conservation prioritization across the Pan-Himalaya. Biol. Conserv. 2025, 302, 110994. [Google Scholar] [CrossRef]
- McCarthy, M.; Abecasis, G.; Cardon, L.; Goldstein, D.; Little, J.; Ioannidis, J.; Hirschhorn, J. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat. Rev. Genet. 2008, 9, 356–369. [Google Scholar] [CrossRef]
- Barbeira, A.; Dickinson, S.; Bonazzola, R.; Zheng, J.; Wheeler, H.; Torres, J.; Torstenson, E.; Shah, K.; Garcia, T.; Edwards, T.; et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018, 9, 18–25. [Google Scholar] [CrossRef]
- Pasaniuc, B.; Price, A. Dissecting the genetics of complex traits using summary association statistics. Nat. Rev. Genet. 2017, 18, 117–127. [Google Scholar] [CrossRef]
- Gilkerson, J.; Perez-Ruiz, J.; Chory, J.; Callis, J. The Plastid-Localized PfkB-Type Carbohydrate Kinases FRUCTOKINASE-LIKE 1 and 2 Are Essential for Growth and Development of Arabidopsis thaliana. BMC Plant Biol. 2012, 12, 102. [Google Scholar] [CrossRef]
- Li, Z.; Liu, P.; Zhang, X.; Zhang, Y.; Ma, L.; Liu, M.; Guan, Z.; Zhang, Y.; Li, P.; Zou, C.; et al. Genome-wide association studies and QTL mapping uncover the genetic architecture of ear tip-barrenness in maize. Physiol. Plant. 2020, 170, 27–39. [Google Scholar] [CrossRef]
- Spencer, C.; Su, Z.; Donnelly, P.; Marchini, J. Designing genome-wide association studies: Sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009, 5, e1000477. [Google Scholar] [CrossRef] [PubMed]
- Zewei, A.; Huasun, H. A Method for Genomic DNA Extraction from Leaves of Rubber Tree (Hevea brasiliensis Muell. Arg.). Plant Physiol. Commun. 2005, 41, 513–515. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Dong, S.; Xu, J.; He, W.; Yang, T. PopLDdecay: A Fast and Effective Tool for Linkage Disequilibrium Decay Analysis Based on Variant Call Format Files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Chang, C.; Chow, C.; Tellier, L.; Vattikuti, S.; Purcell, S.; Lee, J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, 47–48. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast Model-Based Estimation of Ancestry in Unrelated Individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.; Visscher, P. GCTA: A Tool for Genome-Wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-Wide Efficient Mixed-Model Analysis for Association Studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Ibrahim, S.; Li, K.; Ahmad, N.; Kuang, L.; Sadau, S.B.; Tian, Z.; Huang, L.; Wang, X.; Dun, X.; Wang, H. Genetic Dissection of Mature Root Characteristics by Genome-Wide Association Studies in Rapeseed (Brassica napus L.). Plants 2021, 10, 2569. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chen, B.; Xu, K.; Wu, J.; Song, W.; Bancroft, I.; Harper, A.L.; Trick, M.; Liu, S.; Gao, G.; et al. Genome-Wide Association Study Dissects the Genetic Architecture of Seed Weight and Seed Quality in Rapeseed (Brassica napus L.). DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2014, 21, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Luo, P.; Tian, M.; He, J.; Zhang, H.; Yan, Q.; Ye, Y. A SNP Locus and Primer for Rubber Tree Germplasm Typing and Its Applications. 2023. Available online: https://d.wanfangdata.com.cn/patent/ChhQYXRlbnROZXdTMjAyNTAzMTgwODIxNTASEENOMjAyMzExNTA5MDc0LjMaCGFkaDNobzQ0 (accessed on 6 February 2024). (In Chinese).
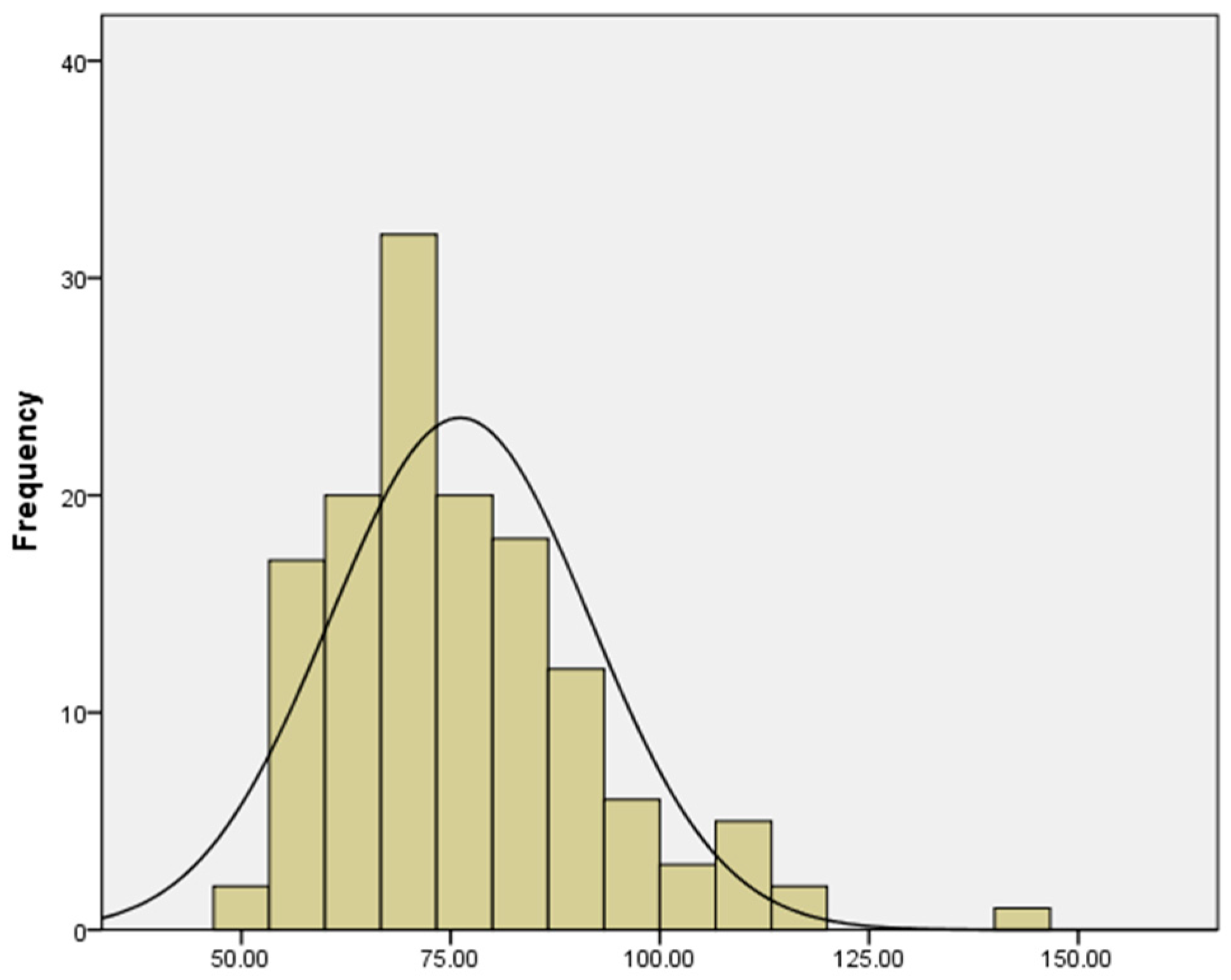
| Trait | Sample | Mean (cm) | SD | SE | Min (cm) | Max (cm) | Range (cm) | Skew | Kurtosis | CV | GDI | KS Test |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| girth | 138 | 76.13 | 15.57 | 1.33 | 48.5 | 142 | 93.5 | 1.05 | 1.62 | 0.205 | 4.907 | 0.143 |
| Chromosomes | Position | p | R2 | Genes |
|---|---|---|---|---|
| CM021229.1 | 44994744 | 2.08 × 10−9 | 0.254 | GH714_028664; GH714_028663; GH714_028652 |
| CM021229.1 | 68430381 | 4.03 × 10−9 | 0.299 | GH714_033616; GH714_033625; GH714_033634 |
| CM021229.1 | 97573289 | 2.93 × 10−9 | 0.580 | GH714_018846; GH714_018861 |
| CM021229.1 | 101378399 | 1.55 × 10−8 | 0.279 | GH714_020134; GH714_020127; GH714_020118; GH714_020116; GH714_020113; GH714_020079; GH714_020074; GH714_020070; GH714_020064; GH714_020061 |
| CM021235.1 | 69150797/69150814 | 2.48 × 10−8 | 0.165 | GH714_002023; GH714_002035; GH714_002048 |
| CM021239.1 | 2245924 | 5.94 × 10−10 | 0.235 | GH714_035167; GH714_035169; GH714_035172; GH714_035173; GH714_035177; GH714_035184 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Zhang, Z.; Ouyang, H.; Zhang, H.; Cheng, H.; Zhang, X.; Gao, X.; He, J.; Yan, Q.; Ye, Y.; et al. Genome-Wide Association Study and Candidate Gene Identification for Girth Traits in Rubber Tree. Plants 2025, 14, 2460. https://doi.org/10.3390/plants14162460
Li W, Zhang Z, Ouyang H, Zhang H, Cheng H, Zhang X, Gao X, He J, Yan Q, Ye Y, et al. Genome-Wide Association Study and Candidate Gene Identification for Girth Traits in Rubber Tree. Plants. 2025; 14(16):2460. https://doi.org/10.3390/plants14162460
Chicago/Turabian StyleLi, Wenxiu, Zishan Zhang, Huan Ouyang, Hualin Zhang, Han Cheng, Xiaofei Zhang, Xinsheng Gao, Junjun He, Qing Yan, Yana Ye, and et al. 2025. "Genome-Wide Association Study and Candidate Gene Identification for Girth Traits in Rubber Tree" Plants 14, no. 16: 2460. https://doi.org/10.3390/plants14162460
APA StyleLi, W., Zhang, Z., Ouyang, H., Zhang, H., Cheng, H., Zhang, X., Gao, X., He, J., Yan, Q., Ye, Y., Yi, Y., Li, P., Luo, P., & Xie, R. (2025). Genome-Wide Association Study and Candidate Gene Identification for Girth Traits in Rubber Tree. Plants, 14(16), 2460. https://doi.org/10.3390/plants14162460






