Screening of qPCR Reference Genes in Quinoa Under Cold, Heat, and Drought Gradient Stress
Abstract
1. Introduction
2. Results
2.1. Primer Specificity and Amplification Efficiency of Candidate Reference Genes
2.2. Expression Levels of Candidate Reference Genes
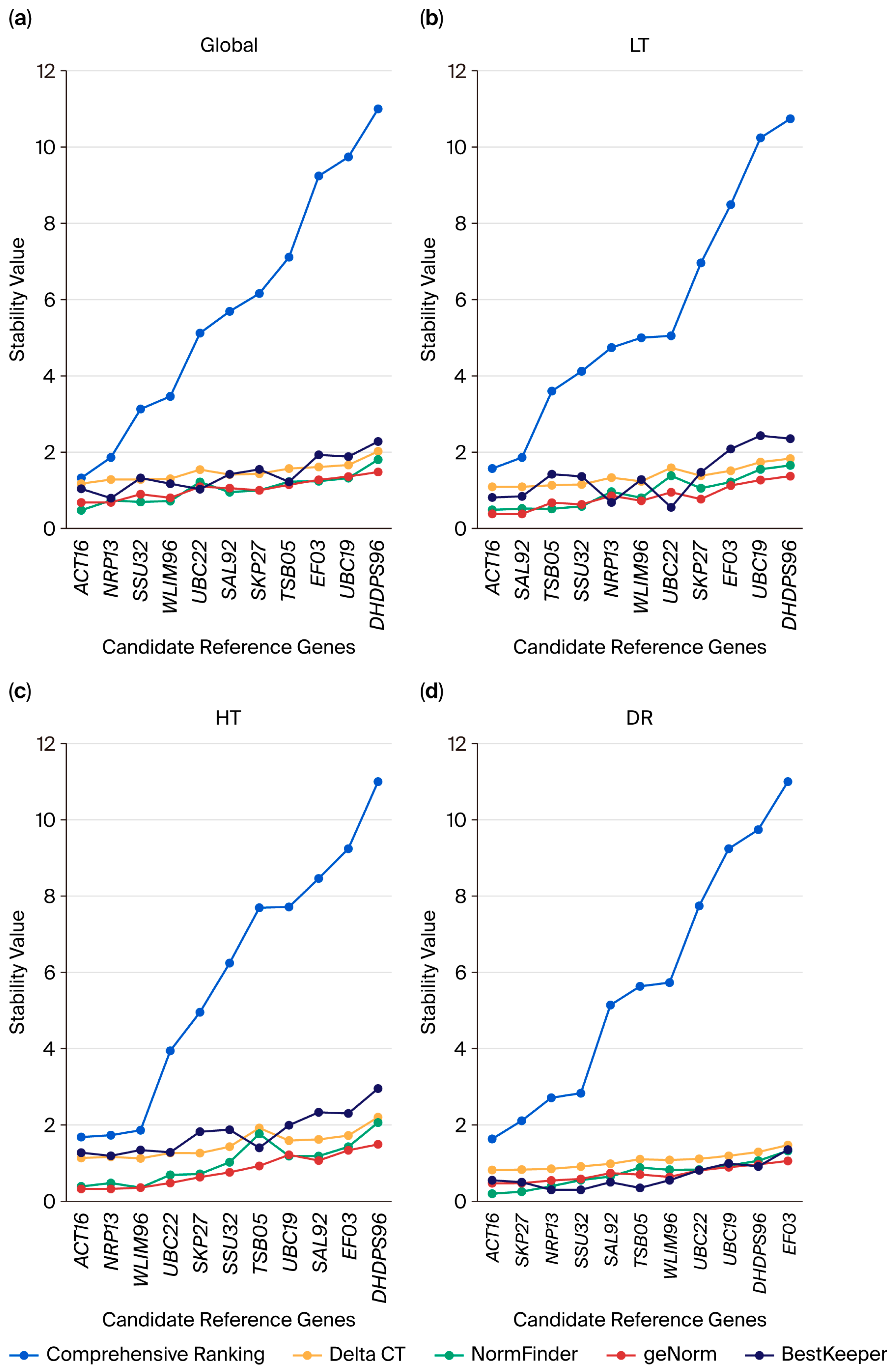
2.3. Analysis of the Expression Stability of Candidate Reference Genes
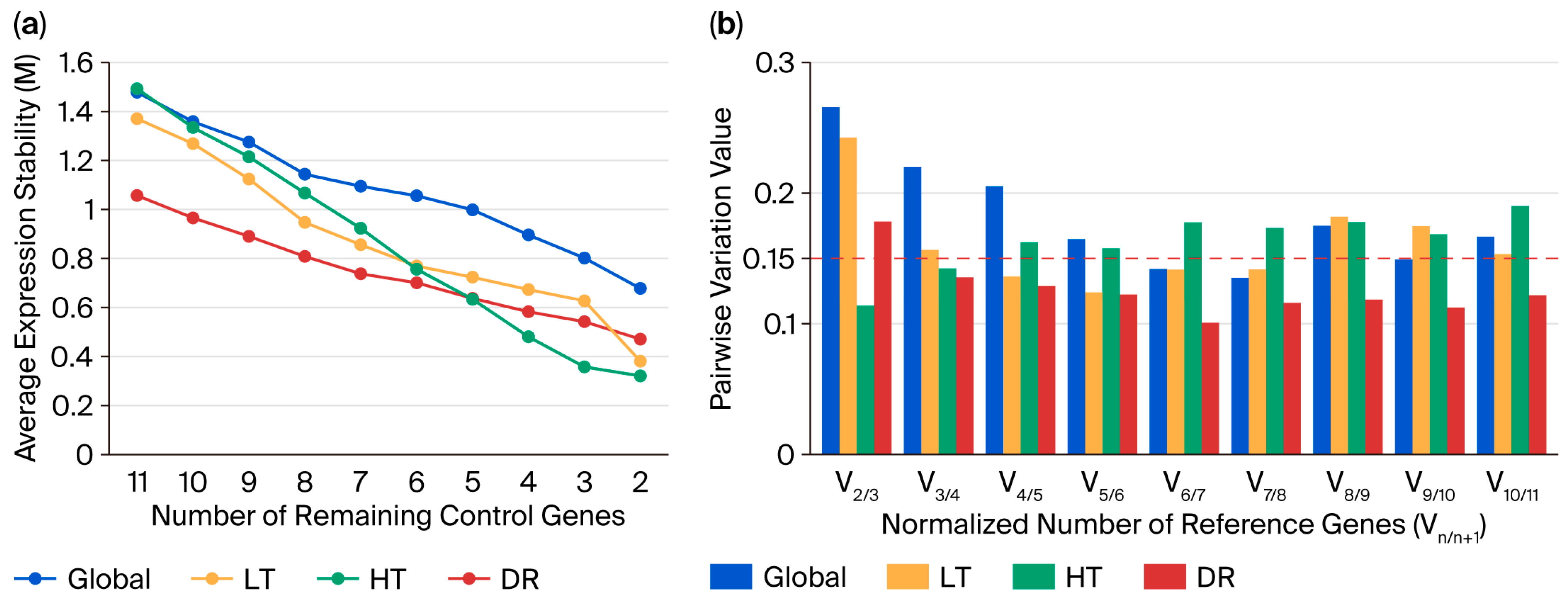
2.4. Analysis of Candidate Reference Gene Expression Stability Using GeNorm
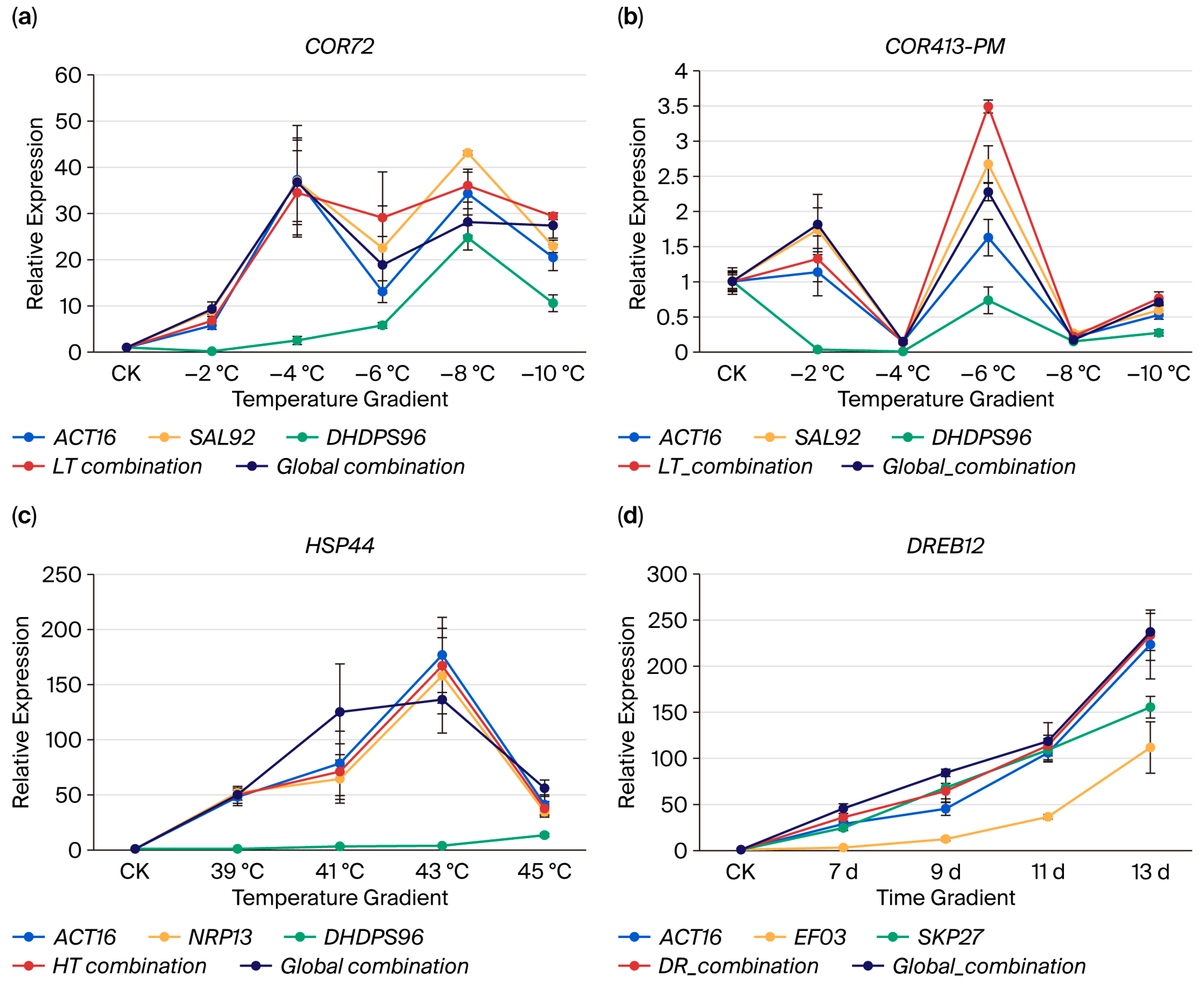
2.5. Validation of Candidate Reference Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Extraction and cDNA Synthesis
4.3. Candidate Gene Selection and Primer Design
4.4. Primer Specificity Verification and qPCR Analysis
4.5. Primer Amplification Efficiency Analysis
4.6. RT-qPCR Data Analysis of Reference Gene Stability
4.7. Normalization of Stress-Response Gene Expression in Chenopodium quinoa by RT-qPCR Analysis
4.8. Data Processing and Accessibility
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Saharan, B.S.; Brar, B.; Duhan, J.S.; Kumar, R.; Marwaha, S.; Rajput, V.D.; Minkina, T. Molecular and physiological mechanisms to mitigate abiotic stress conditions in plants. Life 2022, 12, 1634. [Google Scholar] [CrossRef]
- Ghadirnezhad, S.S.R.; Rahimi, R.; Zand-Silakhoor, A.; Fathi, A.; Fazeli, A.; Radicetti, E.; Mancinelli, R. Enhancing seed germination under abiotic stress: Exploring the potential of nano-fertilization. J. Soil Sci. Plant Nutr. 2024, 24, 5319–5341. [Google Scholar] [CrossRef]
- Alvar-Beltrán, J.; Verdi, L.; Marta, A.D.; Dao, A.; Vivoli, R.; Sanou, J.; Orlandini, S. The effect of heat stress on quinoa (cv. Titicaca) under controlled climatic conditions. J. Agric. Sci. 2020, 158, 255–261. [Google Scholar] [CrossRef]
- Abugoch, L.E.; Romero, N.; Tapia, C.A.; Silva, J.; Rivera, M. Study of some physicochemical and functional properties of quinoa (Chenopodium quinoa Willd) protein isolates. J. Agric. Food Chem. 2008, 56, 4745–4750. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmöckel, S.M.; Li, B.; Borm, T.J.A.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N.; et al. The genome of Chenopodium quinoa. Nature 2017, 542, 307–312. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, X.; Gao, A.; Cao, M.; Yang, D.; An, K.; Guo, S.; Yin, H. Genome-wide identification and expression analysis of 1-Aminocyclopropane-1-Carboxylate Synthase (ACS) gene family in Chenopodium quinoa. Plants 2023, 12, 4021. [Google Scholar] [CrossRef]
- Li, T.; Zhang, M.; Li, M.; Wang, X.; Xing, S. Molecular characterization and expression analysis of YABBY genes in Chenopodium quinoa. Genes 2023, 14, 2103. [Google Scholar] [CrossRef]
- De Magalhães, J.P.; Finch, C.E.; Janssens, G. Next-generation sequencing in aging research: Emerging applications, problems, pitfalls and possible solutions. Ageing Res. Rev. 2010, 9, 315–323. [Google Scholar] [CrossRef]
- Wong, M.L.; Medrano, J.F. Real-time PCR for mRNA quantitation. Biotechniques 2005, 39, 75–85. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Wen, L.; Tan, B.; Guo, W. Estimating transgene copy number in precocious trifoliate orange by TaqMan real-time PCR. Plant Cell Tissue Organ Cult. 2012, 109, 363–371. [Google Scholar] [CrossRef]
- Dheda, K.; Huggett, J.F.; Chang, J.S.; Kim, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005, 344, 141–143. [Google Scholar] [CrossRef]
- Klumb, E.K.; Rickes, L.N.; Braga, E.J.B.; Bianchi, V.J. Evaluation of stability and validation of reference genes for real time PCR expression studies in leaves and roots of Prunus spp. rootstocks under flooding. Sci. Hortic. 2019, 247, 310–319. [Google Scholar] [CrossRef]
- Bustin, S.; Nolan, T. Talking the talk, but not walking the walk: RT-qPCR as a paradigm for the lack of reproducibility in molecular research. Eur. J. Clin. Investig. 2017, 47, 756–774. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: Amodel-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determina tion of stable housekeeping genes, differentially regulat ed target genes and sample integrity: BestKeeper--Excel-based tool using pairwise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Roy, N.V.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulo cytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genomics 2023, 23, 125. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Sgamma, T.; Pape, J.; Massiah, A.; Jackson, S. Selection of reference genes for diurnal and developmental time-course real-time PCR expression analyses in lettuce. Plant Methods 2016, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Knopkiewicz, M.; Wojtaszek, P. Validation of reference genes for gene expression analysis using quantitative polymerase chain reaction in pea lines (Pisum sativum) with different lodging susceptibility. Ann. Appl. Biol. 2019, 174, 86–91. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Q.; Jiang, Y.; Li, Y.; Zhang, H.; Li, R. Reference genes identification for normalization of qPCR under multiple stresses in Hordeum brevisubulatum. Plant Methods 2018, 14, 110. [Google Scholar] [CrossRef]
- Zhang, J.; Feng, Y.; Yang, M.; Xiao, Y.; Liu, Y.; Yuan, Y.; Li, Z.; Zhang, Y.; Zhuo, M.; Zhang, J.; et al. Systematic screening and validation of reliable reference genes for qRT-PCR analysis in Okra (Abelmoschus esculentus L.). Sci. Rep. 2022, 12, 12913. [Google Scholar] [CrossRef]
- Chen, H.; Hu, B.; Zhao, L.; Shi, D.; She, Z.; Huang, X.; Priyadarshani, S.V.G.N.; Niu, X.; Qin, Y. Differential expression analysis of reference genes in pineapple (Ananas comosus L.) during reproductive development and response to abiotic stress, hormonal stimuli. Trop. Plant Biol. 2019, 12, 67–77. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, C.; Lan, H.; Gao, S.; Liu, H.; Liu, J.; Cao, M.; Pan, G.; Rong, T.; Zhang, S. Validation of potential reference genes for qPCR in maize across abiotic stresses, hormone treatments, and tissue types. PLoS ONE 2014, 9, e95445. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-R.; Nam, H.-Y.; Kim, S.-U.; Kim, S.-I.; Chang, Y.-J. Normalization of reverse transcription quantitativePCR with housekeeping genes in rice. Biotechnol. Lett. 2003, 25, 1869–1872. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Xue, Y.; He, Y.; Zhou, X.; Hu, H.; Liu, J.; Feng, L.; Yang, W.; Luo, J.; Zhang, H.; et al. Validation of reference genes for quantitative real-time PCR normalization in Ananas comosus var. bracteatus during chimeric leaf development and response to hormone stimuli. Front. Genet. 2021, 12, 716137. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, C.; Yang, S.; Zhong, Q.; Tian, J. Screening and verification of reference genes for analysis of gene expression in garlic (Allium sativum L.) under cold and drought stress. Plants 2023, 12, 763. [Google Scholar] [CrossRef]
- Wang, X.; Shu, X.; Su, X.; Xiong, Y.; Xiong, Y.; Chen, M.; Tong, Q.; Ma, X.; Zhang, J.; Zhao, J. Selection of suitable reference genes for RT-qPCR gene expression analysis in centipedegrass under different abiotic stress. Genes 2023, 14, 1874. [Google Scholar] [CrossRef]
- Zhu, J. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, S. Selection of the reference gene for expression normalization in Salsola ferganica under abiotic stress. Genes 2022, 13, 571. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, B.; Wang, X.; Wei, X. Screening of stable internal reference gene of quinoa under hormone treatment and abiotic stress. Physiol. Mol. Biol. Plants 2021, 27, 2459–2470. [Google Scholar] [CrossRef]
- Contreras, E.; Martín-Fernández, L.; Manaa, A.; Vicente-Carbajosa, J.; Iglesias-Fernández, R. Identification of reference genes for precise expression analysis during germination in Chenopodium quinoa seeds under salt stress. Int. J. Mol. Sci. 2023, 24, 15878. [Google Scholar] [CrossRef]
- Maldonado-Taipe, N.; Patirange, D.S.R.; Schmockel, S.M.; Jung, C.; Emrani, N. Validation of suitable genes for normalization of diurnal gene expression studies in Chenopodium quinoa. PLoS ONE 2021, 16, e0233821. [Google Scholar] [CrossRef]
- Xie, Y.; Xue, J.; Jiang, X.; Yin, H.; Zhao, X.; Li, X. Screening of reference genes in Chenopodium quinoa under Peronospora variabilis stress and verification of their stability. J. Fujian Agric. For. Univ. 2024, 53, 191–198. [Google Scholar]
- Zhu, X.; Wang, B.; Liu, W.; Wei, X.; Wang, X.; Du, X.; Liu, H. Genome-wide analysis of AP2/ERF gene and functional analysis of CqERF24 gene in drought stress in quinoa. Int. J. Biol. Macromol. 2023, 253, 127582. [Google Scholar] [CrossRef]
- Xie, H.; Wang, Q.; Zhang, P.; Zhang, X.; Huang, T.; Guo, Y.; Liu, J.; Li, L.; Li, H.; Qin, P. Transcriptomic and metabolomic analysis of the response of quinoa seedlings to low temperatures. Biomolecules 2022, 12, 977. [Google Scholar] [CrossRef]
- Zhuang, H.; Fu, Y.; He, W.; Wang, L.; Wei, Y. Selection of appropriate reference genes for quantitative real-time PCR in Oxytropis ochrocephala Bunge using transcriptome datasets under abiotic stress treatments. Front. Plant Sci. 2015, 6, 475. [Google Scholar] [CrossRef]
- Fan, C.; Ma, J.; Guo, Q.; Li, X.; Wang, H.; Lu, M. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS ONE 2013, 8, e56573. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, J.; Huang, S.; Guo, T.; Deng, L.; Hua, W. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene 2014, 538, 113–122. [Google Scholar] [CrossRef]
- Zhou, A.; Liu, E.; Li, H.; Li, Y.; Feng, S.; Gong, S.; Wang, J. PsCor413pm2, a plasma membrane-localized, cold-regulated protein from Phlox subulata, confers low temperature tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2579. [Google Scholar] [CrossRef]
- Deng, Y.; Lin, Y.; Wei, G.; Hu, X.; Zheng, Y.; Ma, J. Overexpression of the CpCOR413PM1 gene from wintersweet (Chimonanthus praecox) enhances cold and drought tolerance in Arabidopsis. Horticulturae 2024, 10, 599. [Google Scholar] [CrossRef]
- Chaudhary, R.; Baranwal, V.K.; Kumar, R.; Sircar, D.; Chauhan, H. Genome-wide identification and expression analysis of Hsp70, Hsp90, and Hsp100 heat shock protein genes in barley under stress conditions and reproductive development. Funct. Integr. Genom. 2019, 19, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yue, J.; Wu, X.; Xu, C.; Yu, J. An ABA-responsive DRE-binding protein gene from Setaria italica, SiARDP, the target gene of SiAREB, plays a critical role under drought stress. J. Exp. Bot. 2014, 65, 5415–5427. [Google Scholar] [CrossRef]
- Zhou, M.; Ma, J.; Pang, J.; Zhang, Z.; Tang, Y.; Wu, Y. Regulation of plant stress response by dehydration responsive element binding (DREB) transcription factors. Afr. J. Biotechnol. 2010, 9, 9255–9269. [Google Scholar]
- Xu, L.; Xu, H.; Cao, Y.; Yang, P.; Feng, Y.; Tang, Y.; Yuan, S.; Ming, J. Validation of reference genes for quantitative real-time PCR during bicolor tepal development in Asiatic hybrid lilies (Lilium spp.). Front. Plant Sci. 2017, 8, 669. [Google Scholar] [CrossRef]
- Wang, X.; Wu, Z.; Bao, W.; Hu, H.; Chen, M.; Chai, T.; Wang, H. Identification and evalution of reference genes for quantitative real-time PCR analysis in Polygonum cuspidatum based on transcriptome data. BMC Plant Biol. 2019, 19, 498. [Google Scholar] [CrossRef]
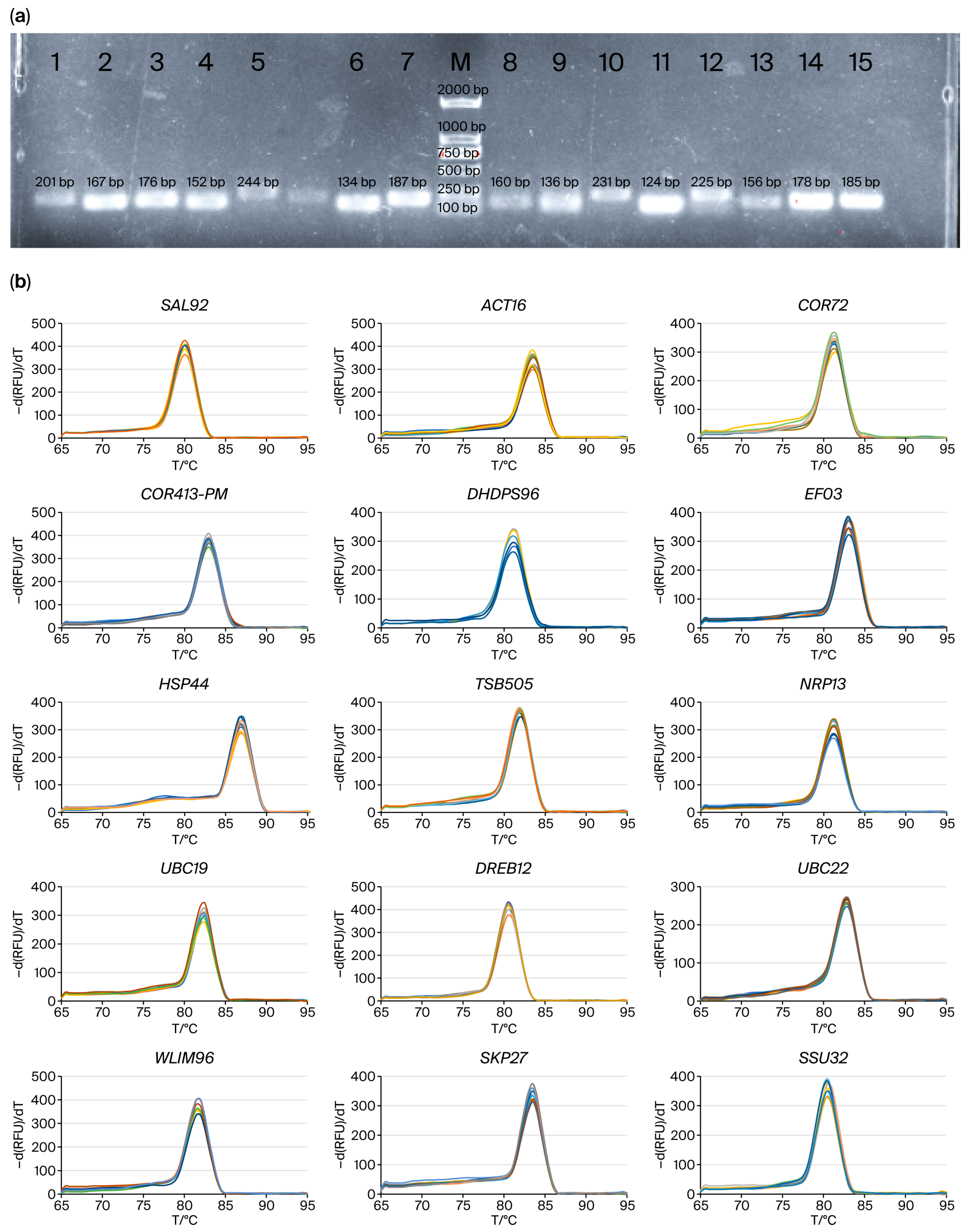

| Gene | Coefficient of Variation (CV) | Amplification Efficiency/% | Linear Correlation Coefficient (R2) |
|---|---|---|---|
| SAL92 | 0.0208 (LT) | 99.02% | 0.9881 |
| ACT16 | 0.0411 (LT) | 96.66% | 0.9891 |
| DHDPS96 | 0.0412 (HT) | 94.93% | 0.9877 |
| EF03 | 0.0876 (HT) | 104.03% | 0.9961 |
| TSB05 | 0.0417 (HT) | 102.34% | 0.9986 |
| NPR13 | 0.0784 (DR) | 104.36% | 0.9911 |
| UBC19 | 0.0463 (DR) | 102.83% | 0.9978 |
| UBC22 | 0.0853 (Global) | 96.97% | 0.9967 |
| WLIM96 | 0.0806 (Global) | 98.04% | 0.9837 |
| SKP27 | 0.0829 (Global) | 108.03% | 0.9879 |
| SSU32 | 0.0891 (Global) | 97.33% | 0.9901 |
| COR72 | 1.2999 (Global) | 98.72% | 0.9835 |
| COR413-PM | 0.5632 (Global) | 98.49% | 0.9919 |
| HSP44 | 0.9073 (Global) | 102.31% | 0.9880 |
| DREB12 | 0.6486 (Global) | 99.30% | 0.9900 |
| Group | Experiment Treatments (Leaves Were Collected from Six-Week-Old Quinoa) |
|---|---|
| Normal treatment (CK) | Plants were continuously maintained under normal growth conditions (24 °C, 12 h light/12 h dark cycle, with regular watering) in the growth chamber. |
| Low-temperature treatment (LT) | Five temperature gradients were set: −2 °C, −4 °C, −6 °C, −8 °C, and −10 °C. Upon reaching the target temperature, plants were transferred from the growth conditions (24 °C) to a low-temperature incubator. Plants were subjected to stress treatment for 6 h at a constant temperature. |
| Heat treatment (HT) | Four temperature gradients were set: 39 °C, 41 °C, 43 °C, and 45 °C. Plants were transferred directly from growth conditions (24 °C) into a growth chamber that had been pre-set and stabilized at the target temperature. Plants were subjected to stress treatment for 6 h at a constant temperature. |
| Drought treatment (DR) | Four stress gradients were set based on the duration after watering cessation: 7, 9, 11, and 13 days. The day when watering was stopped was denoted as day 0 of drought treatment. Drought treatment was conducted within the growth chamber (24 °C, 12 h light/12 h dark). |
| Gene ID | Gene | Description | Function | Primer Sequence (5′~3′) | Product Size |
|---|---|---|---|---|---|
| AUR62038592 | SAL92 | IT4 phosphatase-associated protein | It is required for SIT4’s role in G1 cyclin transcription and for bud formation. | F: GAACACTCACATAGCACCTT R: CGAACCAACACCTCCATA | 167 bp |
| AUR62019116 | ACT16 | Actin | Actin is a ubiquitous protein involved in the formation of filaments that are major components of the cytoskeleton. | F: TTGTGCTCAGTGGTGGTA R: CATCTGTTGGAAGGTGCT | 176 bp |
| AUR62003513 | NRP13 | Asparagine-rich protein | It plays a role in phytohormone response, embryo development and programmed cell death by pathogens or ozone. | F: GAACAAGCCGGAATGTAA R: AAATAAACCCAAGCCAGA | 152 bp |
| AUR62036119 | UBC19 | Ubiquitin-conjugating enzyme E2 | It acts as a ubiquitin-binding enzyme | F: ATTGATAAGCTAGGGAGG R: AGAGGGTAAAGTTGTTGC | 244 bp |
| AUR62021096 | DHDPS96 | Dihydrodipicolinate synthase 2 | Key enzymes of lysine biosynthesis pathway | F: CTTTACAAACGCCACCAT R: GAGAAGCAGAGCGAGGAC | 201 bp |
| AUR62026903 | EF03 | Translation elongation factor | Catalyzes the GTP-dependent ribosomal translocation step during translation elongation | F: CCGCACTGTGATGAGCAA R: TGGAACGAACCTTGGGAT | 134 bp |
| AUR62012196 | WLIM96 | LIM domain-containing protein | The exact function is unknown | F: ACAAGGTCGCCAAGCAAA R: TTCCATCAAGGGCAGCAT | 187 bp |
| AUR62013027 | SKP27 | S-phase kinase-associated protein | Participate in the ubiquitination of target proteins and subsequent proteasomal degradation | F: TTTTGGCTGCTAACTACCT R: TTCTCCCTCCTAACCTCC | 160 bp |
| AUR62012972 | COR72 | Cold-regulated protein | Participate in low temperature response | F: GGTAGACAAGGCAGAGGA R: TGTAGGCTGATGATGGTTAT | 136 bp |
| AUR62021644 | HSP44 | Heat shock protein | Molecular chaperones that suppress protein aggregation and protect against cell stress | F: CCTCGCACAGTCCCATAC R: CAACTCAGCCTTCGCATC | 231 bp |
| AUR62016670 | COR413-PM | Cold-regulated 413-plasma membrane protein | Participate in low temperature response | F: AGCATCCTATGTCCGTGGTG R: CCCGTTAGCCCTTGTGAA | 124 bp |
| AUR62012312 | DREB12 | Dehydration response element binding protein | Participate in plant drought stress | F: ACTTGCCGCATTACCCAG R: GCATCATCGCAGCATTTT | 225 bp |
| AUR62020505 | TSB05 | Tryptophan synthase beta-subunit 2 | Catalyzes the final step in the biosynthesis of L-tryptophan | F: TCTGAAAGACTTGGGACG R: TTCGGAAGAGTTGGACAC | 156 bp |
| AUR62036432 | SSU32 | Ssu72-like family protein | It has intrinsic phosphatase activity and plays an essential role in the transcription cycle | F: CCTCAACGCTGGCAAGAT R: CACCAATAGCCGCCTCCT | 178 bp |
| AUR62028822 | UBC22 | Ubiquitin-conjugating enzyme E2 | It acts as a ubiquitin-binding enzyme | F: AAGAGGTTGATGAGGGAT R: GGAGGCTTATTTGGGTAG | 185 bp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Q.; Wang, X.; Dong, S.; Fu, J.; Lin, Y.; Zhang, Y.; Zhao, B.; Guo, F. Screening of qPCR Reference Genes in Quinoa Under Cold, Heat, and Drought Gradient Stress. Plants 2025, 14, 2434. https://doi.org/10.3390/plants14152434
Lu Q, Wang X, Dong S, Fu J, Lin Y, Zhang Y, Zhao B, Guo F. Screening of qPCR Reference Genes in Quinoa Under Cold, Heat, and Drought Gradient Stress. Plants. 2025; 14(15):2434. https://doi.org/10.3390/plants14152434
Chicago/Turabian StyleLu, Qiuwei, Xueying Wang, Suxuan Dong, Jinghan Fu, Yiqing Lin, Ying Zhang, Bo Zhao, and Fuye Guo. 2025. "Screening of qPCR Reference Genes in Quinoa Under Cold, Heat, and Drought Gradient Stress" Plants 14, no. 15: 2434. https://doi.org/10.3390/plants14152434
APA StyleLu, Q., Wang, X., Dong, S., Fu, J., Lin, Y., Zhang, Y., Zhao, B., & Guo, F. (2025). Screening of qPCR Reference Genes in Quinoa Under Cold, Heat, and Drought Gradient Stress. Plants, 14(15), 2434. https://doi.org/10.3390/plants14152434






