Accumulating Heterozygous Deleterious Mutations in Conserved Soybean Germplasm over Successive Regenerations
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection and Acquisition
2.2. RNA-Seq Analysis
2.3. SNP Calling
2.4. Identification of Deleterious SNPs
2.5. Gene Ontology (GO) and Expression Analysis
2.6. Mutation Burden Estimation and Its Association with Conservation-Related Features
2.7. Additional Association Analyses
2.8. Comparison of Seed-Based and Leaf-Based RNA-Seq Analyses
3. Results
3.1. SNP Identification and Annotation
3.2. Deleterious Mutation
3.3. Ontology of the Associated Genes
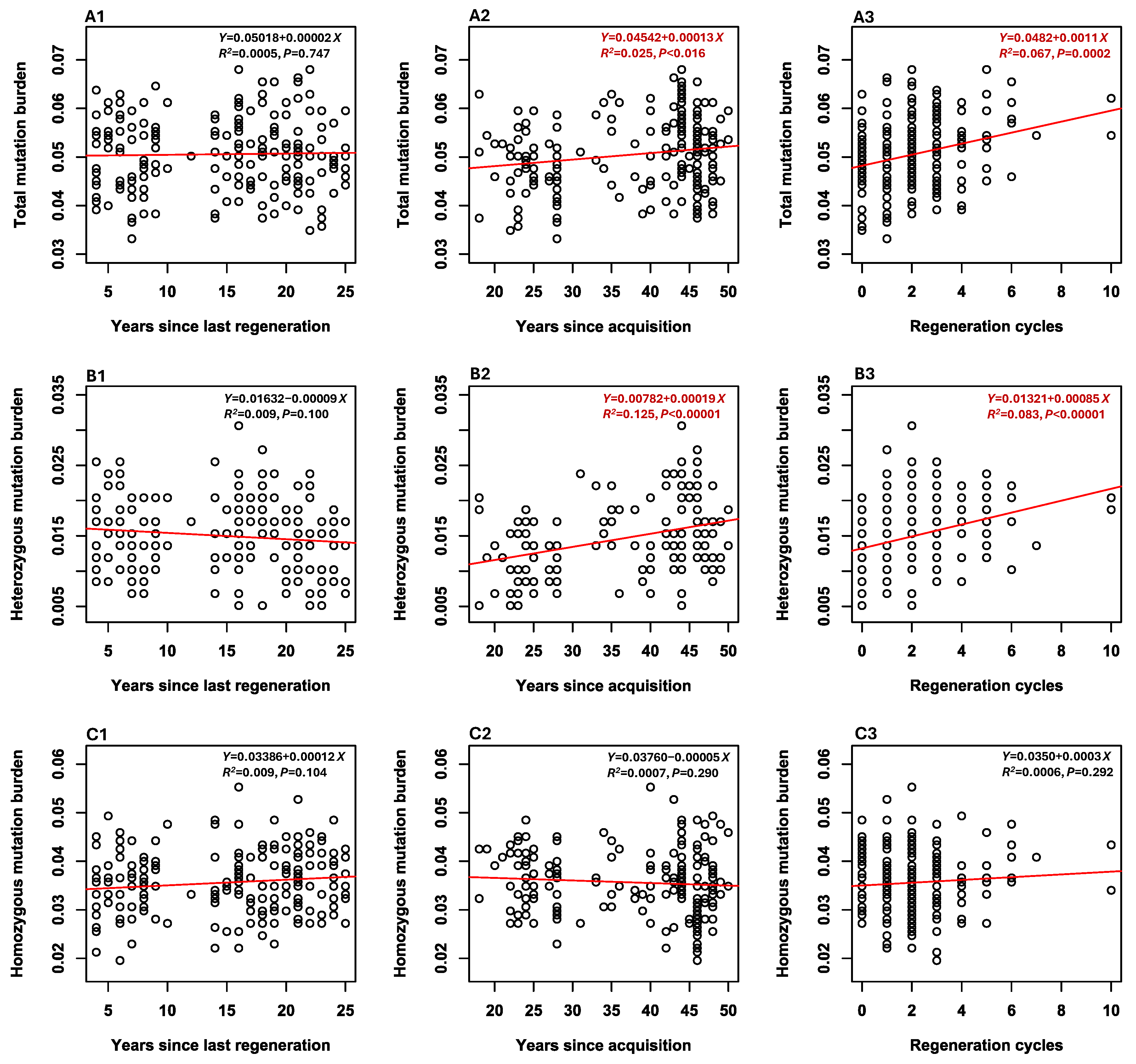
3.4. Mutation Burden
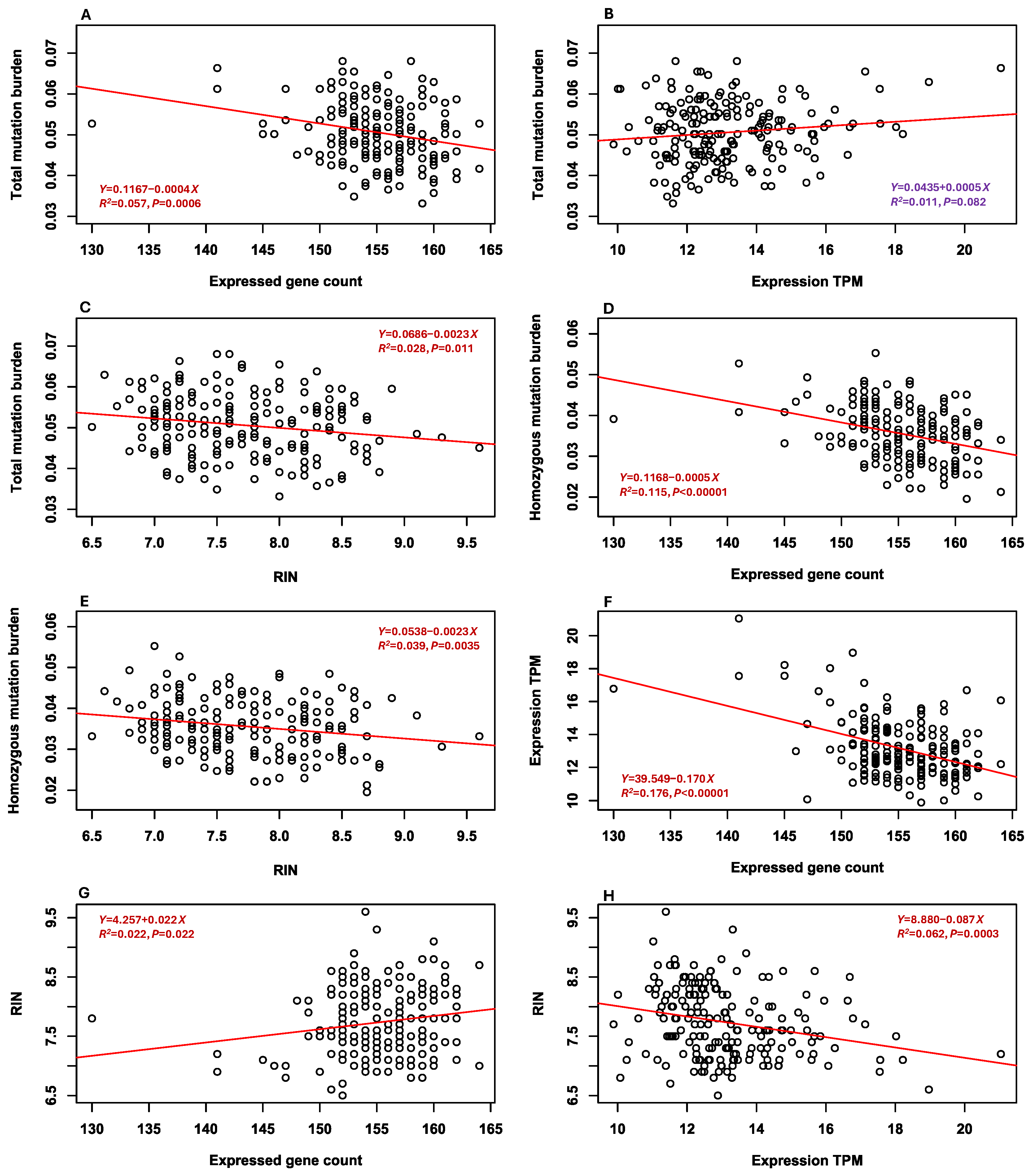
3.5. Associations Between Mutation Burdens and Sample Features
3.6. Three Other Mutation Estimates
3.7. Associations Between Mutation Burdens and Other Mutation Estimates
3.8. Associations Between Three Other Mutation Estimates and Sample Features
3.9. Comparisons Between Seed-Based and Leaf-Based Estimates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The Third Report on the State of the World’s Plant Genetic Resources for Food and Agriculture; FAO Commission on Genetic Resources for Food and Agriculture Assessments: Rome, Italy, 2025. [Google Scholar]
- Engels, J.M.M.; Ebert, A.W. How Can We Strengthen the Global Genetic Resources’ Conservation and Use System? Plants 2024, 13, 702. [Google Scholar] [CrossRef]
- Ashton, T. Genetical aspects of seed storage. In The Storage of Seeds for Maintenance of Viability; Owen, E.B., Ed.; Commonwealth Agricultural Bureaux: Farnham Royal, UK, 1956; pp. 34–38. [Google Scholar]
- Roberts, E.H. Mutations During Seed Storage. Acta Hortic. 1978, 83, 279–282. [Google Scholar] [CrossRef]
- Dourado, A.M.; Roberts, E.H. Phenotypic Mutations Induced during Storage in Barley and Pea Seeds. Ann. Bot. 1984, 54, 781–790. [Google Scholar] [CrossRef]
- Schoen, D.J.; David, J.L.; Bataillou, T.M. Deleterious Mutation Accumulation and the Regeneration of Genetic Resources. Proc. Natl. Acad. Sci. USA 1998, 95, 394–399. [Google Scholar] [CrossRef]
- Hoffman, P.D.; Leonard, J.M.; Lindberg, G.E.; Bollmann, S.R.; Hays, J.B. Rapid Accumulation of Mutations During Seed-to-seed Propagation of Mismatch-repair-defective Arabidopsis. Genes Dev. 2004, 18, 2676–2685. [Google Scholar] [CrossRef] [PubMed]
- Moyers, B.T.; Morrell, P.L.; McKay, J.K. Genetic Costs of Domestication and Improvement. J. Hered. 2018, 109, 103–116. [Google Scholar] [CrossRef]
- Kono, T.J.Y.; Liu, C.; Vonderharr, E.E.; Koenig, D.; Fay, J.C.; Smith, K.P.; Morrell, P.L. The Fate of Deleterious Variants in a Barley Genomic Prediction Population. Genetics 2019, 213, 1531–1544. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, S.L.; Heslop-Harrison, P.; Spillane, C.; McKeown, P.C.; Edwards, D.; Goldman, I.; Ortiz, R. Evolutionary Dynamics and Adaptive Benefits of Deleterious Mutations in Crop Gene Pools. Trends Plant Sci. 2023, 28, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.B. Will a Plant Germplasm Accession Conserved in a Genebank Change Genetically over Time? Front. Plant Sci. 2024, 15, 1437541. [Google Scholar] [CrossRef]
- Cooper, G.M.; Stone, E.A.; Asimenos, G.; Green, E.D.; Batzoglou, S.; Sidow, A. Distribution and Intensity of Constraint in Mammalian Genomic Sequence. Genome Res. 2005, 15, 901–913. [Google Scholar] [CrossRef]
- Henn, B.M.; Botigué, L.R.; Bustamante, C.D.; Clark, A.G.; Gravel, S. Estimating the Mutation Load in Human Genomes. Nat. Rev. Genet. 2015, 16, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Henikoff, S. SIFT: Predicting Amino Acid Changes That Affect Protein Function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Mezmouk, S.; Ross-Ibarra, J. The Pattern and Distribution of Deleterious Mutations in Maize. G3 Genes Genomes Genet. 2014, 4, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Renaut, S.; Rieseberg, L.H. The Accumulation of Deleterious Mutations as a Consequence of Domestication and Improvement in Sunflowers and Other Compositae Crops. Mol. Biol. Evol. 2015, 32, 2273–2283. [Google Scholar] [CrossRef]
- Kono, T.J.Y.; Fu, F.; Mohammadi, M.; Hoffman, P.J.; Liu, C.; Stupar, R.M.; Smith, K.P.; Tiffin, P.; Fay, J.C.; Morrell, P.L. The Role of Deleterious Substitutions in Crop Genomes. Mol. Biol. Evol. 2016, 33, 2307–2317. [Google Scholar] [CrossRef]
- Liu, Q.; Zhou, Y.; Morrell, P.L.; Gaut, B.S. Deleterious Variants in Asian Rice and the Potential Cost of Domestication. Mol. Biol. Evol. 2017, 34, 908–924. [Google Scholar] [CrossRef] [PubMed]
- Ramu, P.; Esuma, W.; Kawuki, R.; Rabbi, I.Y.; Egesi, C.; Bredeson, J.V.; Bart, R.S.; Verma, J.; Buckler, E.S.; Lu, F. Cassava Haplotype Map Highlights Fixation of Deleterious Mutations During Clonal Propagation. Nat. Genet. 2017, 49, 959–963. [Google Scholar] [CrossRef]
- Valluru, R.; Gazave, E.E.; Fernandes, S.B.; Ferguson, J.N.; Lozano, R.; Hirannaiah, P.; Zuo, T.; Brown, P.J.; Leakey, A.D.; Gore, M.A.; et al. Deleterious Mutation Burden and Its Association with Complex Traits in Sorghum (Sorghum bicolor). Genetics 2019, 211, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Lozano, R.; Gazave, E.; dos Santos, J.P.R.; Stetter, M.G.; Valluru, R.; Bandillo, N.; Fernandes, S.B.; Brown, P.J.; Shakoor, N.; Mockler, T.C.; et al. Comparative Evolutionary Genetics of Deleterious Load in Sorghum and Maize. Nat. Plants 2021, 7, 17–24. [Google Scholar] [CrossRef]
- Fu, Y.B.; Peterson, G.W.; Horbach, C. Deleterious and Adaptive Mutations in Plant Germplasm Conserved Ex Situ. Mol. Biol. Evol. 2023, 40, msad238. [Google Scholar] [CrossRef]
- Sun, S.; Wang, B.; Li, C.; Xu, G.; Yang, J.; Hufford, M.B.; Ross-Ibarra, J.; Wang, H.; Wang, L. Unraveling Prevalence and Effects of Deleterious Mutations in Maize Elite Lines Across Decades of Modern Breeding. Mol. Biol. Evol. 2023, 40, msad170. [Google Scholar] [CrossRef]
- Wu, Y.; Li, D.; Hu, Y.; Li, H.; Ramstein, G.P.; Zhou, S.; Zhang, X.; Bao, Z.; Zhang, Y.; Song, B.; et al. Phylogenomic Discovery of Deleterious Mutations Facilitates Hybrid Potato Breeding. Cell 2023, 186, 2313–2328. [Google Scholar] [CrossRef]
- Plekhanova, E.; Nuzhdin, S.V.; Utkin, L.V.; Samsonova, M.G. Prediction of Deleterious Mutations in Coding Regions of Mammals with Transfer Learning. Evol. Appl. 2019, 12, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Bertorelle, G.; Raffini, F.; Bosse, M.; Bortoluzzi, C.; Iannucci, A.; Trucchi, E.; Morales, H.; van Oosterhout, C. Genetic Load: Genomic Estimates and Applications in Non-Model Animals. Nat. Rev. Genet. 2022, 23, 492–503. [Google Scholar] [CrossRef]
- Fu, Y.B.; Cober, E.R.; Morrison, M.J.; Marsolais, F.; Peterson, G.W.; Horbach, C. Patterns of Genetic Variation in a Soybean Germplasm Collection as Characterized with Genotyping-by-Sequencing. Plants 2021, 10, 1611. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.B.; Cober, E.R.; Morrison, M.J.; Marsolais, F.; Zhou, R.; Xu, N.; Gahagan, A.C.; Horbach, C. Variability in Maturity, Oil and Protein Concentration, and Genetic Distinctness among Soybean Accessions Conserved at Plant Gene Resources of Canada. Plants 2022, 11, 3525. [Google Scholar] [CrossRef] [PubMed]
- Soy Canada. Statistics At a Glance. Available online: http://soycanada.ca/statistics/at-a-glance/ (accessed on 16 June 2025).
- MacMillan, K.P.; Gulden, R.H. Effect of Seeding Date, Environment and Cultivar on Soybean Seed Yield, Yield Components, and Seed Quality in the Northern Great Plains. Agron. J. 2020, 112, 1666–1678. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- FAO. Genebank Standards for Plant Genetic Resources for Food and Agriculture, Rev; FAO: Rome, Italy, 2014. [Google Scholar]
- FAO. Practical Guide for the Application of the Genebank Standards for Plant Genetic Resources for Food and Agriculture: Conservation of Orthodox Seeds in Seed Genebanks; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- Qiagen. TissueLyser Handbook, 2nd ed.; Qiagen: Hilden, Germany, 2010; pp. 18–19. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 April 2025).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome Sequence of the Palaeopolyploid Soybean. Nature 2010, 463, 178–183, Erratum in Nature 2010, 465, 120. [Google Scholar] [CrossRef]
- Song, Q.; Jenkins, J.; Jia, G.; Hyten, D.L.; Pantalone, V.; Jackson, S.A.; Schmutz, J.; Cregan, P.B. Construction of High Resolution Genetic Linkage Maps to Improve the Soybean Genome Sequence Assembly Glyma1.01. BMC Genom. 2016, 17, 33. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Naithani, S.; Geniza, M.; Jaiswal, P. Variant Effect Prediction Analysis Using Resources Available at Gramene Database. In Plant Genomics Databases: Methods and Protocols; van Dijk, A., Ed.; Humana Press: New York, NY, USA, 2017; pp. 270–297. [Google Scholar] [CrossRef]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT Missense Predictions for Genomes. Nat. Protoc. 2015, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Davydov, E.V.; Goode, D.L.; Sirota, M.; Cooper, G.M.; Sidow, A.; Batzoglou, S. Identifying a High Fraction of the Human Genome to be Under Selective Constraint Using GERP++. PLoS Comput. Biol. 2010, 6, e1001025. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://www.R-project.org/ (accessed on 22 September 2024).
- Charlesworth, D.; Morgan, M.T.; Charlesworth, B. Mutation Accumulation in Finite Outbreeding and Inbreeding Populations. Genet. Res. 1993, 61, 39–56. [Google Scholar] [CrossRef]
- Fu, Y.B.; Namkoong, G.; Carlson, J.E. Comparison of Breeding Strategies for Purging Inbreeding Depression via Simulation. Conserv. Biol. 1998, 12, 856–864. [Google Scholar] [CrossRef]
- Fu, Y.B. Patterns of Purging Deleterious Genes of Synergistic Interactions with Different Breeding Schemes. Theor. Appl. Genet. 1999, 98, 337–356. [Google Scholar] [CrossRef]
- Fleming, M.B.; Richards, C.M.; Walters, C. Decline in RNA Integrity of Dry-Stored Soybean Seeds Correlates with Loss of Germination Potential. J. Exp. Bot. 2017, 68, 2219–2230. [Google Scholar] [CrossRef]
- Fleming, M.B.; Hill, L.M.; Walters, C. The Kinetics of Ageing in Dry Stored Seeds: A Comparison of Viability Loss and RNA Degradation in Unique Legacy Seed Collections. Ann. Bot. 2019, 123, 1133–1146. [Google Scholar] [CrossRef]
- Walters, C.; Fleming, M.B.; Hill, L.M.; Dorr, E.J.; Richards, C.M. Stress–Response Relationships Related to Ageing and Death of Orthodox Seeds: A Study Comparing Viability and RNA Integrity in Soya Bean (Glycine max) cv. Williams 82. Seed Sci. Res. 2020, 30, 161–172. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, S.; Fu, Y.B.; Wang, H. Arabidopsis Seed Stored mRNAs Are Degraded Constantly over Aging Time, as Revealed by New Quantification Methods. Front. Plant Sci. 2020, 10, 1764. [Google Scholar] [CrossRef] [PubMed]
- Tetreault, H.; Fleming, M.; Hill, L.; Dorr, E.; Yeater, K.; Richards, C.; Walters, C. A Power Analysis for Detecting Aging of Dry-Stored Soybean Seeds: Germination versus RNA Integrity Assessments. Crop Sci. 2023, 63, 1481–1493. [Google Scholar] [CrossRef]
- Tetreault, H.M.; Zingerman, Z.; Hill, L.; Ibrahim, S.; Maschinski, J.; Heineman, K.D.; Walters, C. Assessing the RNA Integrity in Dry Seeds Collected from Diverse Endangered Species Native to the USA. Front. Plant Sci. 2025, 16, 1585631. [Google Scholar] [CrossRef]
- Fu, Y.B. Variability in Predicted Deleterious Mutations among Barley Accessions Conserved Ex Situ. Crop Sci. 2024, 64, 3372–3380. [Google Scholar] [CrossRef]
- Schoen, D.J.; Brown, A.H.D. The Conservation of Wild Plant Species in Seed Banks. BioScience 2001, 51, 960–966. [Google Scholar] [CrossRef]
- Redden, R.; Partington, D. Gene Bank Scheduling of Seed Regeneration: Internal Report on a Long Term Storage Study. J. Integr. Agric. 2018, 17, 60345–60347. [Google Scholar] [CrossRef]
- Richards, C.M.; Lockwood, D.R.; Volk, G.M.; Walters, C. Modeling Demographics and Genetic Diversity in Ex Situ Collections During Seed Storage and Regeneration. Crop Sci. 2010, 50, 2440–2447. [Google Scholar] [CrossRef]
- Hay, F.R.; Whitehouse, K.J. Rethinking the Approach to Viability Monitoring in Seed Genebanks. Conserv. Physiol. 2017, 5, cox009. [Google Scholar] [CrossRef] [PubMed]
- Hay, F.R.; Whitehouse, K.J.; Ellis, R.H.; Sackville Hamilton, N.R.; Lusty, C.; Ndjiondjop, M.N.; Tia, D.; Wenzl, P.; Santos, L.G.; Yazbek, M.; et al. CGIAR Genebank Viability Data Reveal Inconsistencies in Seed Collection Management. Glob. Food Sec. 2021, 30, 100557. [Google Scholar] [CrossRef]
- Roles, A.J.; Conner, J.K. Fitness Effects of Mutation Accumulation in a Natural Outbred Population of Wild Radish (Raphanus raphanistrum): Comparison of Field and Greenhouse Environments. Evolution 2008, 62, 1066–1075. [Google Scholar] [CrossRef] [PubMed]

| Variant | Count/Proportion | Variant | Count/Proportion |
|---|---|---|---|
| SNP calling and filtering | Loss-of-function variant ** | ||
| Total SNPs without missing values | 58,548 | Total count | 869 |
| SNP annotation with VEP * | Proportion | 0.015 | |
| Missense_variant (MV) | 19,847 | SIFT analysis with CT *** | |
| Proportion of MV in total SNPs | 0.339 | SIFT-deleterious SNPs (SDS) | 5912 |
| Synonymous_variant (SV) | 20,478 | Proportion of SDS in total SNPs | 0.101 |
| Proportion of SV in total SNPs | 0.350 | Deleterious_low_confidence SNPs | 941 |
| Splice_acceptor_variant | 45 | Tolerated SNPs | 34,076 |
| Splice_donor_variant | 50 | Deleterious SNPs by SIFT+RS | |
| Stop_gained | 450 | SDS+RS-filtered SNPs (RSD) | 588 |
| Stop_lost | 57 | Proportion of RSD in total SNPs | 0.010 |
| Start_lost | 45 | Fixed RSD | 1 |
| Splice_region_variant | 194 | Proportion of fixed RSD in total SNPs | 0.00002 |
| Stop_retained_variant | 28 | Weakly deleterious with RS < 1 | 45 |
| 5_prime_UTR_variant | 5354 | Proportion of RSD | 0.077 |
| 3_prime_UTR_variant | 7068 | Mildly deleterious with RS of 1–3 | 477 |
| Non_coding_transcript_exon_variant | 208 | Proportion of RSD | 0.811 |
| Intron_variant | 1131 | Highly deleterious with RS > 3 | 66 |
| Upstream_gene_variant | 1584 | Proportion of RSD | 0.112 |
| Downstream_gene_variant | 863 | ||
| Intergenic_variant | 1146 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© His Majesty the King in Right of Canada, 2025. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Y.-B.; Horbach, C. Accumulating Heterozygous Deleterious Mutations in Conserved Soybean Germplasm over Successive Regenerations. Plants 2025, 14, 2429. https://doi.org/10.3390/plants14152429
Fu Y-B, Horbach C. Accumulating Heterozygous Deleterious Mutations in Conserved Soybean Germplasm over Successive Regenerations. Plants. 2025; 14(15):2429. https://doi.org/10.3390/plants14152429
Chicago/Turabian StyleFu, Yong-Bi, and Carolee Horbach. 2025. "Accumulating Heterozygous Deleterious Mutations in Conserved Soybean Germplasm over Successive Regenerations" Plants 14, no. 15: 2429. https://doi.org/10.3390/plants14152429
APA StyleFu, Y.-B., & Horbach, C. (2025). Accumulating Heterozygous Deleterious Mutations in Conserved Soybean Germplasm over Successive Regenerations. Plants, 14(15), 2429. https://doi.org/10.3390/plants14152429







