Identification and Characterization of NAC Transcription Factors Involved in Pine Wilt Nematode Resistance in Pinus massoniana
Abstract
1. Introduction
2. Results
2.1. Biophysical Properties Analysis of P. massoniana NAC Proteins
2.2. Multiple Sequence Alignment Analysis of the P. massoniana NAC Family Proteins
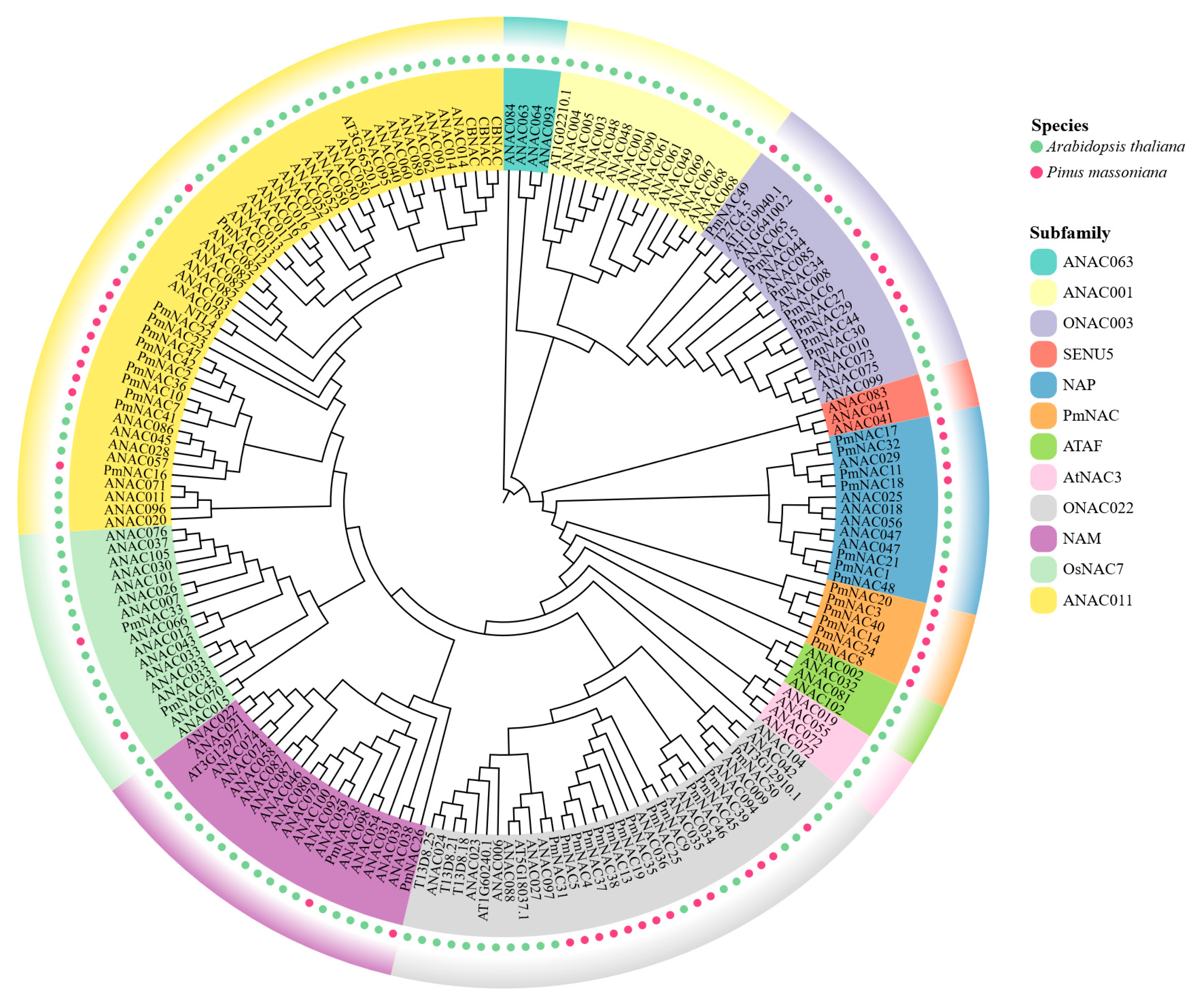
2.3. Classification and Phylogenetic Analysis of PmNACs
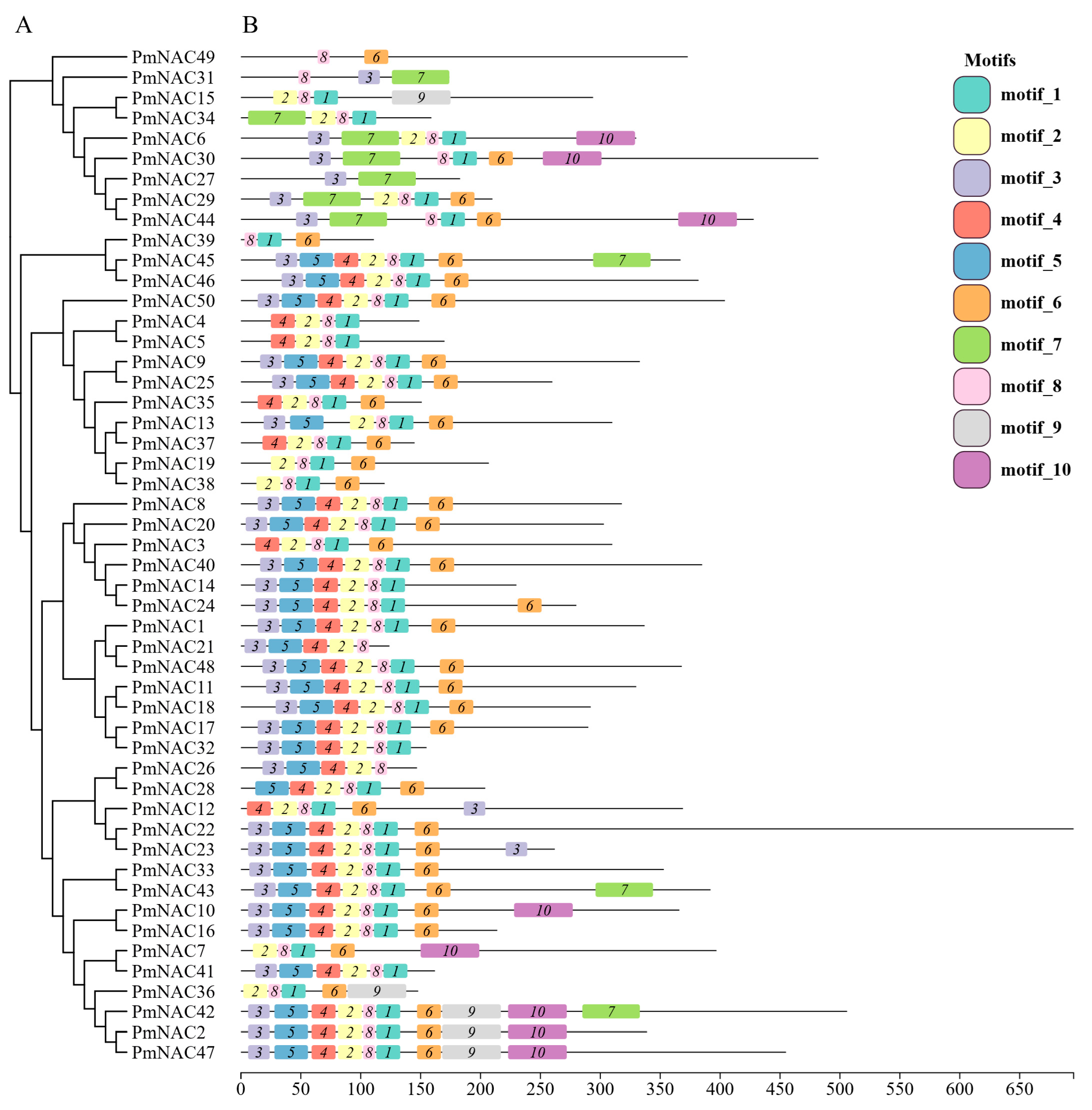
2.4. Conserved Motif Analysis of P. massoniana NAC Family Proteins
2.5. Three-Dimensional Structure Model of PmNAC Protein
2.6. Tissue-Specific Expression Profiles of PmNAC Family Genes
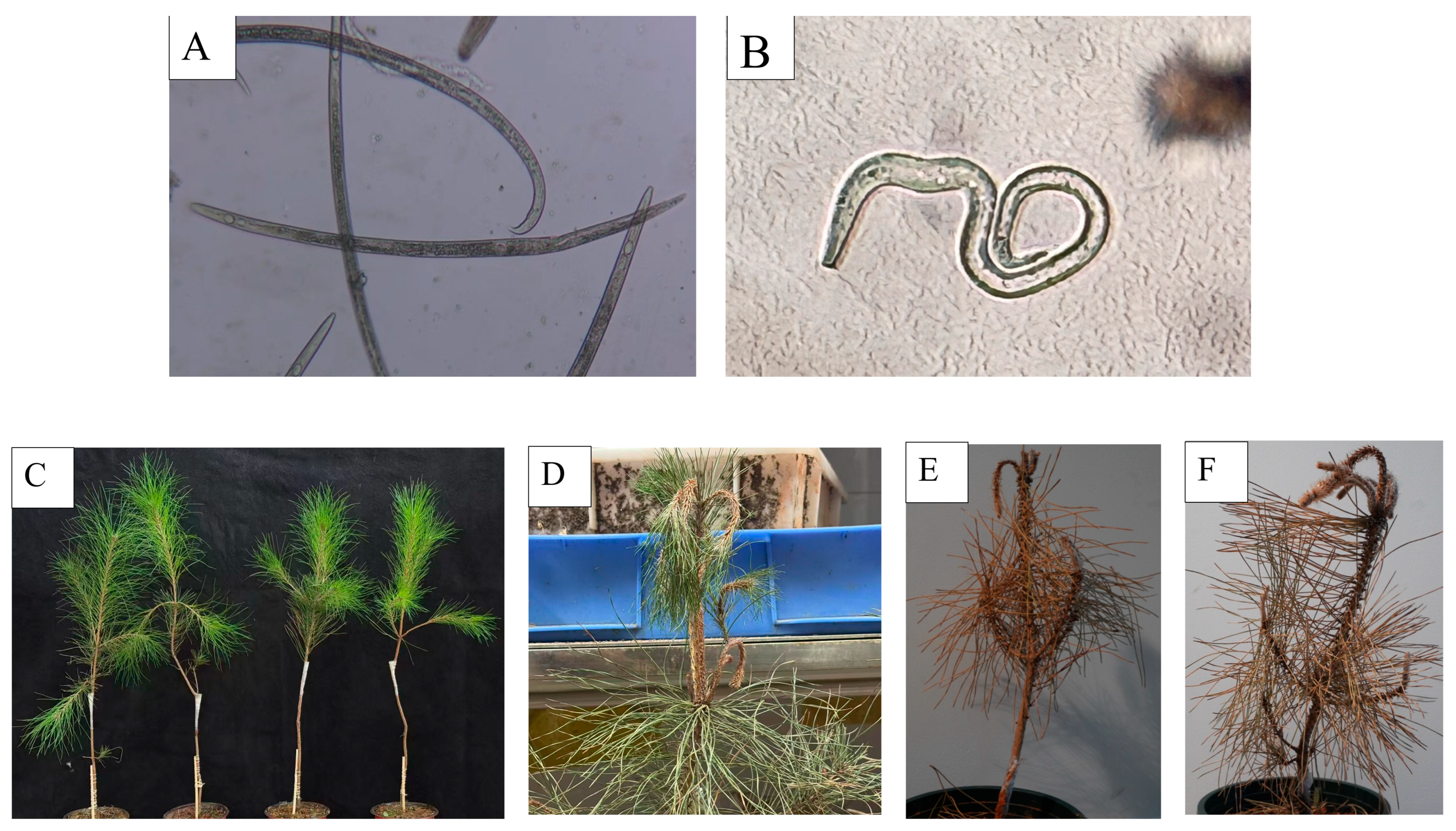
2.7. Isolation and Identification of Nematodes and Inoculation of B. xylophilus on P. massoniana
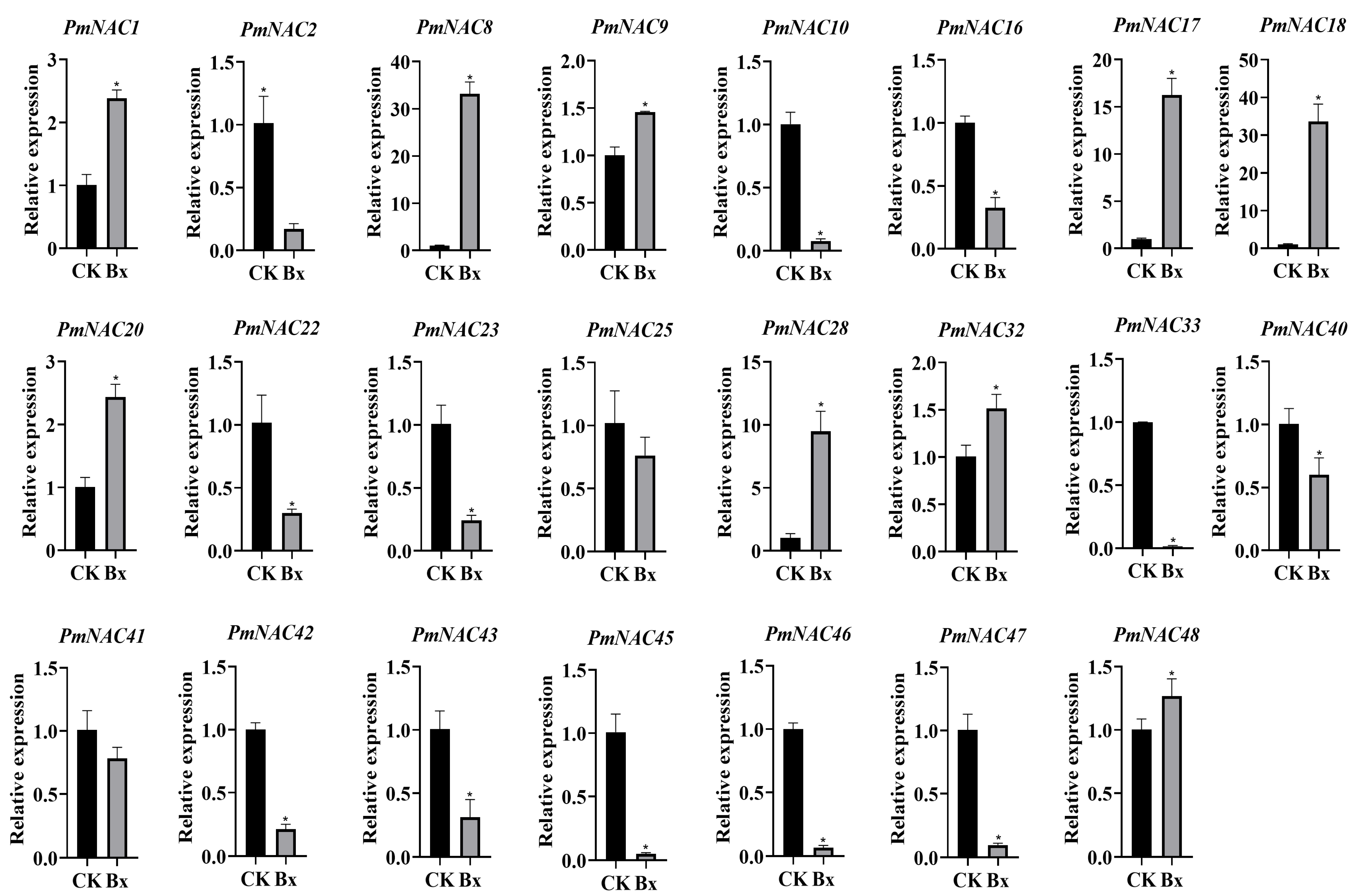
2.8. Response of PmNAC Family Genes to Nematode Infection
2.9. Response of PmNAC Genes to Exogenous MeJA and SA
2.10. Auto-Activation Activity of PmNAC8
2.11. Subcellular Localization of PmNAC8
2.12. PmNAC8 Promotes Reactive Oxygen Species (ROS) Accumulation in Plants and Induces the Expression of Pathogenesis-Related (PR) Genes
3. Discussion
4. Materials and Methods
4.1. Physicochemical Property Analysis of the NAC Family in P. massoniana
4.2. Sequence Alignment and Phylogenetic Analysis
4.3. Conserved Motif Prediction and Subdomain Analysis
4.4. Tertiary Structure Modeling
4.5. Healthy Plant Materials
4.6. Isolation and Identification of PWN
4.7. Inoculation of the PWN and Sample Preparation
4.8. Verification of Pathogenicity of PWN
4.9. SA and MeJA Processing and Material Preparation
4.10. RNA Isolation and qRT-PCR
4.11. Yeast Self-Activation Assay
4.12. Subcellular Localization Analysis
4.13. Transient Overexpression of PmNAC8 in N. benthamiana
4.14. Detection of Reactive Oxygen Species (ROS)
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, J.-Q.; Zhang, L.-Z.; Ma, J.; Li, C.-J.; Zang, Y.-D.; Sun, H.; Zhang, D.-M. Hepatoprotective Diterpenes from the Nodes of Pinus Massoniana. J. Asian Nat. Prod. Res. 2025, 27, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Brugger, D.; Wilhelm, B.; Schusser, B.; Gisch, N.; Matthes, J.; Zhao, J.; Windisch, W. Masson Pine Pollen (Pinus Massoniana) Activate HD11 Chicken Macrophages in Vitro. J. Ethnopharmacol. 2024, 325, 117870. [Google Scholar] [CrossRef]
- Tang, F.; Zhou, Y.; Feng, J.; Li, J.; Feng, J.; Lv, Y.; Mao, Y.; Wang, Y.; Deng, P.; Bai, Y. Important Role of Pinus Massoniana Mixed Forests in Enhancing Soil Carbon Stocks in Degraded Forests in Southern China. CATENA 2025, 250, 108792. [Google Scholar] [CrossRef]
- Xu, S.; Huang, W.; Wang, D.; Zhang, B.; Sun, H.; Yan, J.; Ding, J.; Ma, X. Risk Assessment of Carbon Stock Loss in Chinese Forests Due to Pine Wood Nematode Invasion. Forests 2025, 16, 315. [Google Scholar] [CrossRef]
- Modesto, I.; Mendes, A.; Carrasquinho, I.; Miguel, C.M. Molecular Defense Response of Pine Trees (Pinus Spp.) to the Parasitic Nematode Bursaphelenchus Xylophilus. Cells 2022, 11, 3208. [Google Scholar] [CrossRef]
- Akbulut, S.; Stamps, W.T. Insect Vectors of the Pinewood Nematode: A Review of the Biology and Ecology of Monochamus Species. For. Pathol. 2012, 42, 89–99. [Google Scholar] [CrossRef]
- Song, L.; Liu, W.; Ma, Y.; Zhao, Y.; Zhao, R.; Sun, J.; Zhang, B. Comprehensive Insights Into Pinewood Nematode Effectors: Evolutionary Diversity, Functional Roles, and Implications for Pine Wilt Disease Management. Plant Cell Environ. 2025. [Google Scholar] [CrossRef]
- Zhang, Y.; Du, G.; Guo, Q.; Dong, G.; Wang, M.; Zhang, T.; Li, R. Transcriptome Sequencing and Analysis of Genes Related to Disease Resistance in Pinus Thunbergii. Forests 2023, 14, 650. [Google Scholar] [CrossRef]
- Wang, X.-Y.; Wu, X.-Q.; Wen, T.-Y.; Feng, Y.-Q.; Zhang, Y. Terpene Production Varies in Pinus Thunbergii Parl. with Different Levels of Resistance, with Potential Effects on Pinewood Nematode Behavior. Forests 2022, 13, 1140. [Google Scholar] [CrossRef]
- Back, M.A.; Bonifacio, L.; Inacio, M.L.; Mota, M.; Boa, E. Pine Wilt Disease: A Global Threat to Forestry. Plant Pathol. 2024, 73, 1026–1041. [Google Scholar] [CrossRef]
- Naidoo, S.; Slippers, B.; Plett, J.M.; Coles, D.; Oates, C.N. The Road to Resistance in Forest Trees. Front. Plant Sci. 2019, 10, 273. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, H.; Wang, J.; Li, D.; Wang, H.; Jiang, Y.; Fei, X.; Sun, L.; Li, F. Research Progress on the Resistance Mechanism of Host Pine to Pine Wilt Disease. Plant Pathol. 2024, 73, 469–477. [Google Scholar] [CrossRef]
- Modesto, I.; Sterck, L.; Arbona, V.; Gomez-Cadenas, A.; Carrasquinho, I.; Van de Peer, Y.; Miguel, C.M. Insights Into the Mechanisms Implicated in Pinus Pinaster Resistance to Pinewood Nematode. Front. Plant Sci. 2021, 12, 690857. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.M.; Langer, S.; Carrasquinho, I.; Bergström, E.; Larson, T.; Thomas-Oates, J.; António, C. Pinus Pinaster Early Hormonal Defence Responses to Pinewood Nematode (Bursaphelenchus Xylophilus) Infection. Metabolites 2021, 11, 227. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Chen, J.; Li, Y.; Liu, X.; Li, Y.; Wang, B.; Cao, J.; Gu, Y.; Ma, W.; Ma, L. Molecular Defense Response of Bursaphelenchus Xylophilus to the Nematophagous Fungus Arthrobotrys Robusta. Cells 2023, 12, 543. [Google Scholar] [CrossRef]
- Couto, D.; Zipfel, C. Regulation of Pattern Recognition Receptor Signalling in Plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef]
- Liu, Q.; Wei, Y.; Xu, L.; Hao, Y.; Chen, X.; Zhou, Z. Transcriptomic Profiling Reveals Differentially Expressed Genes Associated with Pine Wood Nematode Resistance in Masson Pine (Pinus Massoniana Lamb.). Sci. Rep. 2017, 7, 4693. [Google Scholar] [CrossRef] [PubMed]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes Involved in Organ Separation in Arabidopsis: An Analysis of the Cup-Shaped Cotyledon Mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; He, H.; Chang, Y.; Miao, B.; Liu, Z.; Wang, Q.; Dong, F.; Xiong, L. Multiple Roles of NAC Transcription Factors in Plant Development and Stress Responses. J. Integr. Plant Biol. 2025, 67, 510–538. [Google Scholar] [CrossRef]
- Pereira-Santana, A.; Alcaraz, L.D.; Castaño, E.; Sanchez-Calderon, L.; Sanchez-Teyer, F.; Rodriguez-Zapata, L. Comparative Genomics of NAC Transcriptional Factors in Angiosperms: Implications for the Adaptation and Diversification of Flowering Plants. PLoS ONE 2015, 10, e0141866. [Google Scholar] [CrossRef]
- Ernst, H.A.; Nina Olsen, A.; Skriver, K.; Larsen, S.; Lo Leggio, L. Structure of the Conserved Domain of ANAC, a Member of the NAC Family of Transcription Factors. EMBO Rep. 2004, 5, 297–303. [Google Scholar] [CrossRef]
- Zheng, C.; Yang, Q.; Wang, X.; Chen, Y.; He, R.; Li, X.; Pan, H.; Zhuo, R.; Qu, T.; Qiu, W. Transcription Factors Involved in Plant Stress and Growth and Development: NAC. Agronomy 2025, 15, 949. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The Role of WRKY Transcription Factors in Plant Abiotic Stresses. Biochim. Biophys. Acta 2012, 1819, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Han, G.H.; Huang, R.N.; Hong, L.H.; Xu, J.X.; Hong, Y.G.; Wu, Y.H.; Chen, W.W. The Transcription Factor NAC102 Confers Cadmium Tolerance by Regulating WAKL11 Expression and Cell Wall Pectin Metabolism in Arabidopsis. J. Integr. Plant Biol. 2023, 65, 2262–2278. [Google Scholar] [CrossRef] [PubMed]
- Goverse, A.; Smant, G. The Activation and Suppression of Plant Innate Immunity by Parasitic Nematodes. Annu. Rev. Phytopathol. 2014, 52, 243–265. [Google Scholar] [CrossRef]
- Xu, P.; Ma, W.; Hu, J.; Cai, W. The Nitrate-Inducible NAC Transcription Factor NAC056 Controls Nitrate Assimilation and Promotes Lateral Root Growth in Arabidopsis Thaliana. PLoS Genet. 2022, 18, e1010090. [Google Scholar] [CrossRef]
- Shu, L.; Li, L.; Jiang, Y.-Q.; Yan, J. Advances in Membrane-Tethered NAC Transcription Factors in Plants. Plant Sci. 2024, 342, 112034. [Google Scholar] [CrossRef]
- Ooka, H. Comprehensive Analysis of NAC Family Genes in Oryza Sativa and Arabidopsis Thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Wu, Y.; Deng, Z.; Lai, J.; Zhang, Y.; Yang, C.; Yin, B.; Zhao, Q.; Zhang, L.; Li, Y.; Yang, C.; et al. Dual Function of Arabidopsis ATAF1 in Abiotic and Biotic Stress Responses. Cell Res. 2009, 19, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Staskawicz, B.J.; Dangl, J.L. The Plant Immune System: From Discovery to Deployment. Cell 2024, 187, 2095–2116. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The Plant Immune System. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Sun, J.-Q.; Jiang, H.-L.; Li, C.-Y. Systemin/Jasmonate-Mediated Systemic Defense Signaling in Tomato. Mol. Plant 2011, 4, 607–615. [Google Scholar] [CrossRef]
- Ren, T.; Wang, J.; Zhao, M.; Gong, X.; Wang, S.; Wang, G.; Zhou, C. Involvement of NAC Transcription Factor SiNAC1 in a Positive Feedback Loop via ABA Biosynthesis and Leaf Senescence in Foxtail Millet. Planta 2018, 247, 53–68. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-Recognition Receptors Are Required for NLR-Mediated Plant Immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the Functional Annotation of Proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The Proteomics Server for in-Depth Protein Knowledge and Analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A Web Application for Visualizing, Modifying and Annotating Phylogenetic Trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Tintori, S.C.; Sloat, S.A.; Rockman, M.V. Rapid Isolation of Wild Nematodes by Baermann Funnel. J. Vis. Exp. 2022, e63287. [Google Scholar] [CrossRef]
- Futai, K. Pine Wood Nematode, Bursaphelenchus Xylophilus. Annu. Rev. Phytopathol. 2013, 51, 61–83. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, F.; Ye, J.; Fang, G.; Pan, H. Classification of Parasitic Nematodes in Pine Trees in China. J. Nanjing For. Univ. 2012, 36, 137. [Google Scholar] [CrossRef]
- Wang, M.; Zhu, S.; Luo, K.; Zhou, L.; Zhao, W. Studies on real--time fluorescent PCR with TaqMan probe for rDNA--ITS2 of pine wood nematode (Bursaphelenchus xylophilus). Sci. Silvae Sin. 2005, 41, 82–85. [Google Scholar]
- Parija, S.C.; Khurana, S. Revisiting Koch’s Postulates: A Tailored Approach for Clinical Parasitology. Trop. Parasitol. 2025, 15, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Huang, W.; Cui, X.; Dong, Y.; Shi, Y.; Ma, H.; Liu, L. Identification of Wheat Yellow Rust Using Optimal Three-Band Spectral Indices in Different Growth Stages. Sensors 2019, 19, 35. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 Transcription Factor: A Node of Convergence for Jasmonate-Mediated and Salicylate-Mediated Signals in Plant Defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef]
- Chen, H.; Yang, Z.; Hu, Y.; Tan, J.; Jia, J.; Xu, H.; Chen, X. Reference Genes Selection for Quantitative Gene Expression Studies in Pinus Massoniana L. Trees 2016, 30, 685–696. Trees 2016, 30, 685–696. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Cesarz, S.; Schulz, A.E.; Beugnon, R.; Eisenhauer, N. Testing Soil Nematode Extraction Efficiency Using Different Variations of the Baermann-Funnel Method. Soil. Org. 2020, 91, 61–72. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Lei, J.; Zhang, M.; Li, J.; Pi, C.; Yu, J.; Yan, X.; Luo, K.; Xia, Y. Identification and Characterization of NAC Transcription Factors Involved in Pine Wilt Nematode Resistance in Pinus massoniana. Plants 2025, 14, 2399. https://doi.org/10.3390/plants14152399
Zhao Z, Lei J, Zhang M, Li J, Pi C, Yu J, Yan X, Luo K, Xia Y. Identification and Characterization of NAC Transcription Factors Involved in Pine Wilt Nematode Resistance in Pinus massoniana. Plants. 2025; 14(15):2399. https://doi.org/10.3390/plants14152399
Chicago/Turabian StyleZhao, Zhengping, Jieyun Lei, Min Zhang, Jiale Li, Chungeng Pi, Jinxiu Yu, Xuewu Yan, Kun Luo, and Yonggang Xia. 2025. "Identification and Characterization of NAC Transcription Factors Involved in Pine Wilt Nematode Resistance in Pinus massoniana" Plants 14, no. 15: 2399. https://doi.org/10.3390/plants14152399
APA StyleZhao, Z., Lei, J., Zhang, M., Li, J., Pi, C., Yu, J., Yan, X., Luo, K., & Xia, Y. (2025). Identification and Characterization of NAC Transcription Factors Involved in Pine Wilt Nematode Resistance in Pinus massoniana. Plants, 14(15), 2399. https://doi.org/10.3390/plants14152399




