Geobotanical Study, DNA Barcoding, and Simple Sequence Repeat (SSR) Marker Analysis to Determine the Population Structure and Genetic Diversity of Rare and Endangered Prunus armeniaca L.
Abstract
1. Introduction
2. Results and Discussion
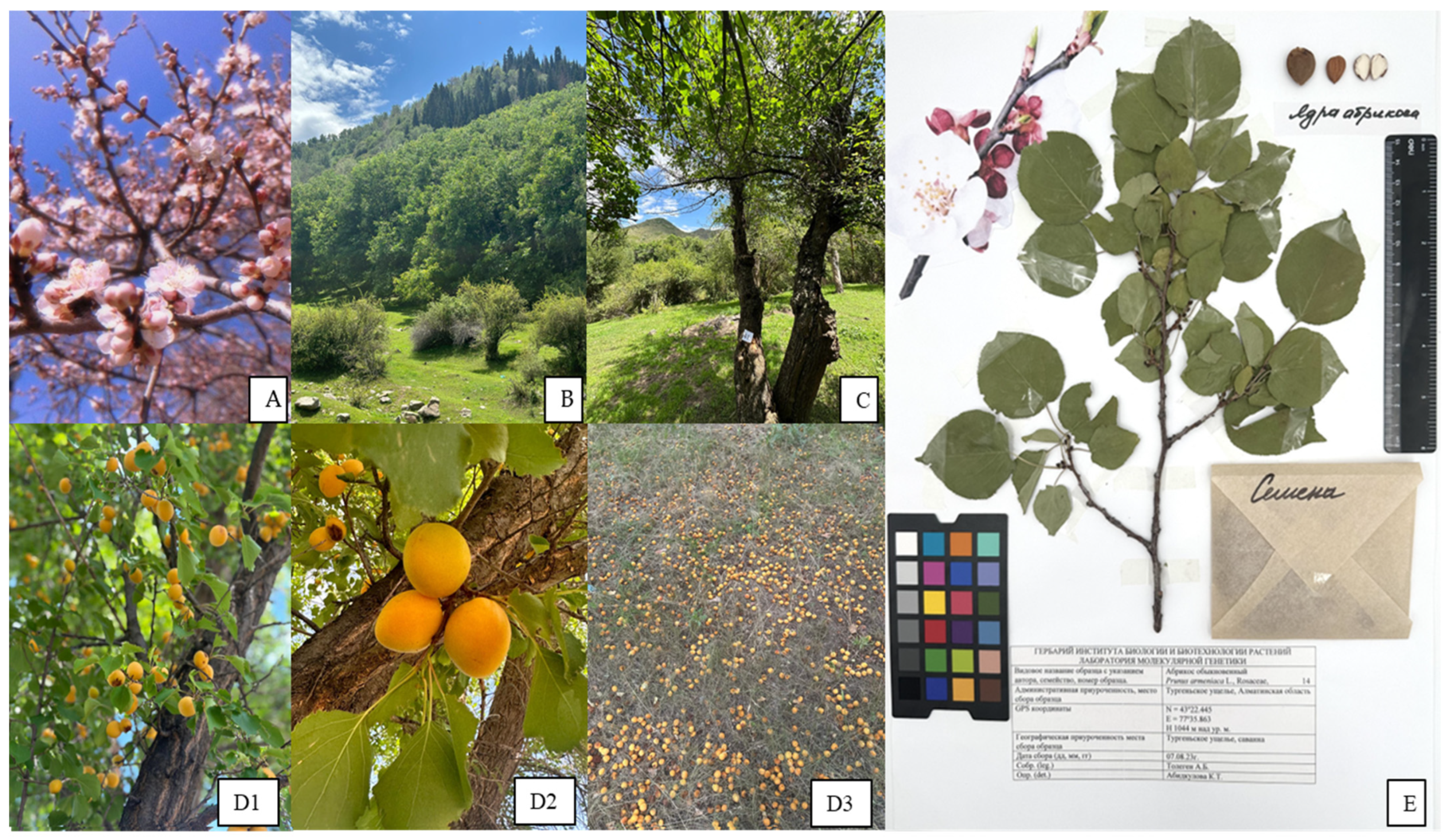
2.1. Description of Populations
2.2. Remote Sensing of Territories for Mapping Natural Populations of P. armeniaca
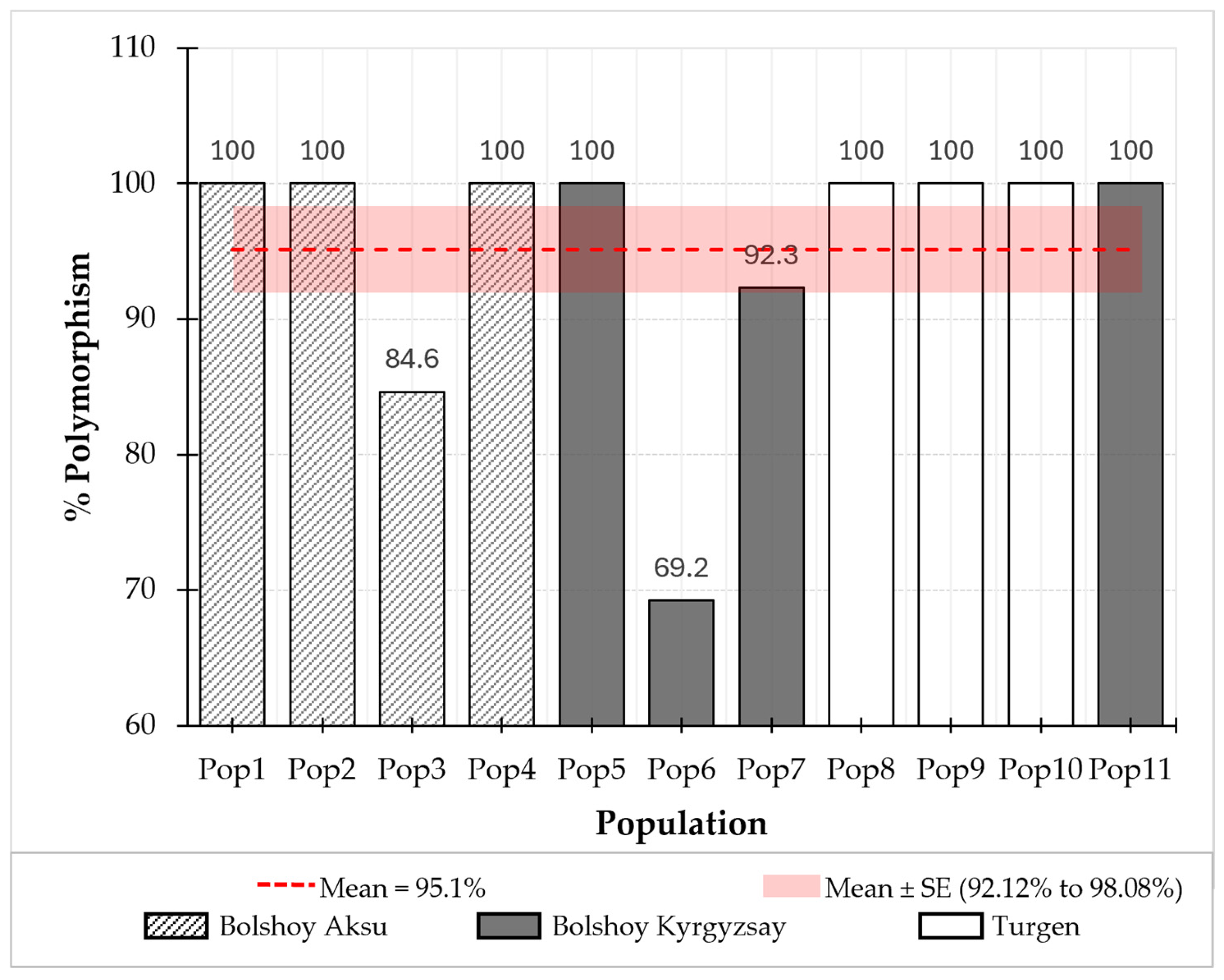
2.3. Population Genetic Diversity of P. armeniaca Using SSR Markers
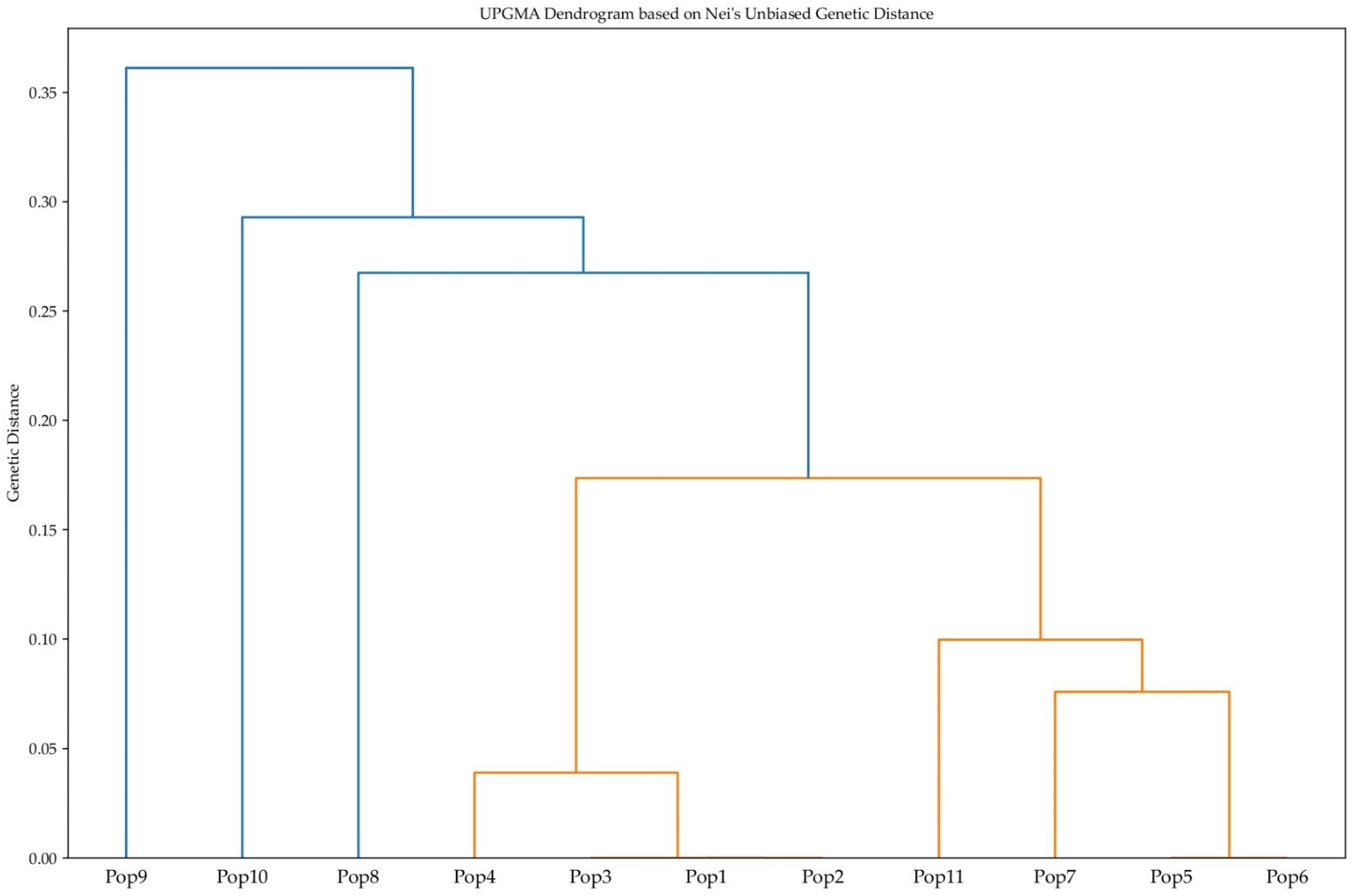
2.4. Population Genetic Structure of P. armeniaca
3. Materials and Methods
3.1. Plant Material and Botanical Description
3.2. Botanical Description
3.3. Herbarium Production
3.4. Remote Sensing of Territories for Mapping Natural Populations of P. armeniaca
3.5. Molecular Analysis
3.5.1. DNA Extraction
3.5.2. PCR Setup
3.5.3. Electrophoretic Accounting of PCR Amplification
3.5.4. Purification of PCR Products and Sequencing
3.5.5. SSR Labeling of the Biodiversity of Rare and Endangered P. armeniaca
3.5.6. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO (Food and Agriculture Organization of the United Nations). Crops and Livestock Products, Primary Apricots. Production in 2023. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 18 April 2025).
- Bourguiba, H.; Scotti, I.; Sauvage, C.; Zhebentyayeva, T.; Ledbetter, C.; Krška, B.; Remay, A.; D’Onofrio, C.; Iketani, H.; Christen, D.; et al. Genetic Structure of a Worldwide Germplasm Collection of Prunus armeniaca L. Reveals Three Major Diffusion Routes for Varieties Coming From the Species’ Center of Origin. Front. Plant Sci. 2020, 11, 638. [Google Scholar] [CrossRef] [PubMed]
- Dzhangaliev, A.D.; Salova, T.N.; Turekhanova, R.M. Wild fruit and nut plants of Kazakhstan. In Horticultural Reviews: Wild Apple and Fruit Trees of Central Asia; Janick, J., Ed.; Wiley: Hoboken, NJ, USA, 2002; Volume 29, pp. 305–371. [Google Scholar]
- Sheikh, Z.N.; Sharma, V.; Shah, R.A.; Raina, S.; Aljabri, M.; Mir, J.I.; AlKenani, N.; Hakeem, K.R. Elucidating Genetic Diversity in Apricot (Prunus armeniaca L.) Cultivated in the North-Western Himalayan Provinces of India Using SSR Markers. Plants 2021, 10, 2668. [Google Scholar] [CrossRef]
- Li, W.; Liu, L.; Wang, Y.; Zhang, Q.; Fan, G.; Zhang, S.; Wang, Y.; Liao, K. Genetic diversity, population structure, and relationships of apricot (Prunus) based on restriction site-associated DNA sequencing. Hortic. Res. 2020, 7, 69. [Google Scholar] [CrossRef] [PubMed]
- Baitulin, I.O. (Ed.) Red Book of Kazakhstan, 2nd ed.; ArtPrintXX1 LLP: Astana, Kazakhstan, 2014; Volume 2, 452p. (In Russian) [Google Scholar]
- Dzhangaliev, A.D.; Salova, T.A.; Salova, T.A.; Turekhanova, P.M. The Wild Fruit and Nut Plants of Kazakhstan. Hortic. Rev. 2003, 29, 305–371. [Google Scholar] [CrossRef]
- Jiang, F.; Zhang, J.; Wang, S.; Yang, L.; Luo, Y.; Gao, S.; Zhang, M.; Wu, S.; Hu, S.; Sun, H.; et al. The apricot (Prunus armeniaca L.) genome elucidates Rosaceae evolution and beta-carotenoid synthesis. Hortic. Res. 2019, 6, 128. [Google Scholar] [CrossRef] [PubMed]
- Shylla, B.; Sharma, D.P.; Handa, A.; Thakur, M.; Sharma, P.; Negi, N. Exploring genetic diversity in apricot (Prunus armeniaca L.) populations using SSR markers. S. Afr. J. Bot. 2025, 177, 50–59. [Google Scholar] [CrossRef]
- Yuan, Z.; Chen, X.; He, T.; Feng, J.; Feng, T.; Zhang, C. Population genetic structure in apricot (Prunus armeniaca L.) cultivars revealed by fluorescent-AFLP markers in southern Xinjiang, China. J. Genet. Genom. 2007, 34, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Zaurov, D.E.; Molnar, T.J.; Eisenman, S.W.; Ford, T.M.; Mavlyanova, R.F.; Capik, J.M.; Reed Funk, S.; Goffreda, J.S. Genetic Resources of apricots (Prunus armeniaca L.) in Central Asia. HortScience 2013, 48, 681–691. [Google Scholar] [CrossRef]
- Groppi, A.; Liu, S.; Cornille, A.; Decroocq, A.; Bui, Q.T.; Tricon, D.; Cruaud, C.; Arribat, S.; Belser, C.; Marande, W.; et al. Population genomics of apricots unravels domestication history and adaptive events. Nat. Commun. 2021, 12, 3956. [Google Scholar] [CrossRef]
- Mukanova, G.S.; Smailova, M.K.; Sankaibaeva, A.G.; Shadmanova, L.S. Analysis of phenological, pomological and biochemical characteristics of wild apricot in Kazakhstan. Acta Hortic. 2024, 1387, 217–224. [Google Scholar] [CrossRef]
- Karatas, N. Evaluation of Nutritional Content in Wild Apricot Fruits for Sustainable Apricot Production. Sustainability 2022, 14, 1063. [Google Scholar] [CrossRef]
- Rivers, M.; Newtown, A.C.; Oldfield, S.; Global Tree Assessment Contributors. Scientists’ warning to humanity on tree extinctions. Plant People Planet 2022, 5, 466–482. [Google Scholar] [CrossRef]
- Bisht, M.; Sekar, K.C.; Mukherjee, S.; Thapliyal, N.; Bahukhandi, A.; Singh, D.; Bhojak, P.; Mehta, P.; Upadhyay, S.; Dey, D. Influence of Anthropogenic Pressure on the Plant Species Richness and Diversity Along the Elevation Gradients of Indian Himalayan High-Altitude Protected Areas. Front. Ecol. Evol. 2022, 10, 751989. [Google Scholar] [CrossRef]
- Fazan, L.; Song, Y.-G.; Kozlowski, G. The Woody Planet: From Past Triumph to Manmade Decline. Plants 2020, 9, 1593. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The biomass distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef]
- Pott, R. Fundamentals and Perspectives of Geobotanical Research in the Twenty-First Century. In Vegetation Structure and Function at Multiple Spatial, Temporal and Conceptual Scales: Geobotany Studies; Box, E., Ed.; Springer: Cham, Switzerland, 2016; pp. 529–548. [Google Scholar]
- Botanic Gardens Conservation International (BGCI). State of the World’s Trees; BGCI: Richmond, UK, 2021; p. 52. [Google Scholar]
- Kushnarenko, S.V.; Romadanova, N.V.; Aralbayeva, M.M. Current state and in vitro conservation of the only endangered population of Corylus avellana in Kazakhstan. Res. Crops 2020, 21, 681–686. [Google Scholar] [CrossRef]
- Romadanova, N.V.; Kushnarenko, S.V. Conservation of plant biodiversity by biotechnology methods. Proc. Appl. Bot. Genet. Breed. 2023, 184, 239–248. [Google Scholar] [CrossRef]
- Bettoni, J.C.; Kretzschmar, A.A.; Bonnart, R.; Shepherd, A.; Volk, G.M. Cryopreservation of 12 Vitis species using apical shoot tipsderived from plants grown in vitro. HortScience 2019, 54, 976–981. [Google Scholar] [CrossRef]
- Bettoni, J.C.; Bonnart, R.; Volk, G.M. Challenges in implementing plant shoot tip cryopreservation technologies. Plant Cell Tissue Organ Cult. 2021, 144, 21–34. [Google Scholar] [CrossRef]
- Bettoni, J.C.; Markovic, Z.; Bi, W.; Volk, G.M.; Matsumoto, T.; Wang, Q.-C. Grapevine shoot tip cryopreservation and cryotherapy: Secure storage of disease-free plants. Plants 2021, 10, 2190. [Google Scholar] [CrossRef]
- Bettoni, J.C.; Wang, M.-R.; Li, J.-W.; Fan, X.; Fazio, G.; Hurtado-Gonzales, O.P.; Volk, G.M.; Wang, Q.-C. Application of biotechniques for in vitro virus and viroid elimination in pome fruit crops. Phytopathology 2024, 114, 930–954. [Google Scholar] [CrossRef]
- Nonić, M.; Šijačić-Nikolić, M. Genetic Diversity: Sources, Threats, and Conservation. In Life on Land: Encyclopedia of the UN Sustainable Development Goals; Leal Filho, W., Azul, A.M., Brandli, L., Lange Salvia, A., Wall, T., Eds.; Springer: Cham, Switzerland, 2021; pp. 421–435. [Google Scholar]
- Woodruff, D.S. Populations, Species, and Conservation Genetics. Encycl. Biodivers. 2001, 28, 811–829. [Google Scholar] [CrossRef]
- Lande, R.; Barrowclough, G.F. Effective population size, genetic variation, and their use in population management. In Viable Populations for Conservation; Soulé, M.E., Ed.; Cambridge University Press: New York, NY, USA, 1987; pp. 87–124. [Google Scholar]
- Lande, R. Genetics and demography in biological conservation. Science 1988, 241, 1455–1460. [Google Scholar] [CrossRef] [PubMed]
- FAO (Food and Agriculture Organization of the United Nations). Crops and Livestock Products. Production in 2022. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 27 February 2025).
- Lateur, M.; Dapena, E.; Szalatnay, D.; Gantar, M.E.; Guyader, A.; Hjalmarsson, I.; Höfer, M.; Ikase, L.; Kelerhals, M.; Lacis, G.; et al. ECPGR Characterization and Evaluation: Descriptors for Apple Genetic Resources; European Cooperative Programme for Plant Genetic Resources: Rome, Italy, 2022; p. 57. [Google Scholar]
- Rotach, P. EUFORGEN Technical Guidelines for Genetic Conservation and Use for Service Tree (Sorbus domestica); International Plant Genetic Resources Institute: Rome, Italy, 2003; p. 6. [Google Scholar]
- Romadanova, N.V.; Zemtsova, A.S.; Altayeva, N.A.; Artimovich, N.A.; Alexandrova, A.M.; Kushnarenko, S.V.; Bettoni, J.C. Geobotanical Study and Preservation of Rare and Endangered Rosaceae Species. Plants 2025, 14, 1526. [Google Scholar] [CrossRef] [PubMed]
- Forsline, P.L.; Aldwinckle, H.S. Evaluation of Malus sieversii seedling populations for disease resistance and horticultural traits. Acta Hort. 2004, 663, 529–534. [Google Scholar] [CrossRef]
- O’Connor, J.R.; Crush, J.R.; Jahufer, Z. Identifying morphological traits associated with vegetative persistence in the perennial ryegrass (Lolium perenne L.) cultivar ‘Grasslands Samson’. J. N. Z. Grassl. 2020, 82, 139–147. [Google Scholar] [CrossRef]
- Pedrotti, F. Plant and Vegetation Mapping. Geobotany Studies, Basics Methods and Case Studies; Springer: Berlin, Germany, 2013; p. 294. [Google Scholar]
- International Board for Plant Genetic Resources (IBPGR); Commission of the European Communities (CEC). Apricot Descriptors (Revised); International Board for Plant Genetic Resources: Rome, Italy, 1991; p. 36. [Google Scholar]
- Lateur, M.; Giovannini, D.; Szalatnay, D.; Flachowsky, H.; Kalmäe, H.; Hudina, M.; Gustavsson, L.; Fernández, F. General Protocols for Using the ECPGR Descriptors for Prunus spp.; ECPGR Working Group on Prunus ECPGR Secretariat: Rome, Italy, 2013; p. 4. [Google Scholar]
- Balenović, I.; Seletković, A.; Pernar, R.; Jazbec, A. Estimation of the mean tree height of forest stands by photo-gram-metric measurement using digital aerial images of high spatial re-solution. Ann. For. Res. 2015, 58, 125–143. [Google Scholar] [CrossRef]
- Balenović, I.; Marjanović, H.; Vuletić, D.; Paladinić, E.; Ostrogović Sever, M.Z.; Indir, K. Quality assessment of high-density digital surface model over different land cover classes. Period. Biol. 2015, 117, 459–470. [Google Scholar] [CrossRef]
- Cienciała, A.; Sobura, S.; Sobolewska-Mikulska, K. Optimising Land Consolidation by Implementing UAV Technology. Sustainability 2022, 14, 441. [Google Scholar] [CrossRef]
- Di Gennaro, S.F.; Dainelli, R.; Palliotti, A.; Toscano, P.; Matese, A. Sentinel-2 Validation for Spatial Variability Assessment in Overhead Trellis System Viticulture Versus UAV and Agronomic Data. Remote Sens. 2019, 11, 2573. [Google Scholar] [CrossRef]
- de Gouw, S.; Morgenroth, J.; Xu, C. An updated survey on the use of geospatial technologies in New Zealand’s plantation forestry sector. N. Z. J. For. Sci. 2020, 50, 8. [Google Scholar] [CrossRef]
- Vélez, S.; Ariza-Sentís, M.; Valente, J. Mapping the Spatial Variability of Botrytis Bunch Rot Risk in Vineyards Using UAV Multispectral Imagery. Eur. J. Agron. 2023, 142, 126691. [Google Scholar] [CrossRef]
- Leeonis, A.N.; Ahmed, M.F.; Mokhtar, M.B.; Lim, C.K.; Halder, B. Challenges of Using a Geographic Information System (GIS) in Managing Flash Floods in Shah Alam, Malaysia. Sustainability 2024, 16, 7528. [Google Scholar] [CrossRef]
- Corbane, C.; Lang, S.; Pipkins, K.; Alleaume, S.; Deshayes, M.; García Millán, V.E.; Strasser, T.; Borre, J.V.; Toon, S.; Michael, F. Remote sensing for mapping natural habitats and their conservation status—New opportunities and challenges. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 7–16. [Google Scholar] [CrossRef]
- Pettorelli, N.; Safi, K.; Turner, W. Satellite remote sensing, biodiversity research and conservation of the future. Philos. Trans. R. Soc. B: Biol. Sci. 2014, 369, 20130190. [Google Scholar] [CrossRef] [PubMed]
- Kocur-Bera, K.; Małek, A. Assessing the feasibility of using remote sensing data and vegetation indices in the estimation of land subject to consolidation. Sensors 2024, 24, 7736. [Google Scholar] [CrossRef]
- Martínez-Casasnovas, J.A.; Llorens, J.; Sandonís, L.; Escolà, A.; Arnó, J. NDVI from satellite images to estimate LiDAR-derived geometric and structural parameters in super-intensive almond orchards”. In Precision Agriculture ’21; Stafford, J.V., Ed.; Wageningen Academic: Leiden, The Netherlands, 2021; pp. 567–573. [Google Scholar]
- Terrence, S.; Siddiq, K.; John, Y.; Ann, F.; Kelley, M.; Don, H.; Richard, E.; Elizabeth, Z. A Preliminary Assessment of Hyperspectral Remote Sensing Technology for Mapping Submerged Aquatic Vegetation in the Upper Delaware River National Parks (USA). ARS 2018, 7, 290–312. [Google Scholar] [CrossRef]
- Twumasi, Y.A.; Merem, E.C.; Ayala-Silva, T.; Osei, A.; Petja, B.M.; Alexander, K. Techniques of Remote Sensing and GIS as Tools for Visualizing Impact of Climate Change-Induced Flood in Southern African Region. Am. J. Clim. Change 2017, 6, 306–327. [Google Scholar] [CrossRef]
- Twumasi, Y.; Merem, E.; Namwamba, J.; Okwemba, R.; Ayala-Silva, T.; Abdollahi, K.; Lukongo, O.; Tate, J.; Cour-Conant, K.; Akinrinwoye, C. Use of GIS and Remote Sensing Technology as a Decision Support Tool in Flood Disaster Management: The Case of Southeast Louisiana, USA. J. Geogr. Inf. Syst. 2020, 12, 141–157. [Google Scholar] [CrossRef]
- Bramel, P.; Volk, G.M. A Global Strategy for the Conservation and Use of Apple Genetic Resources; Crop Trust: Bonn, Germany, 2019; p. 50. [Google Scholar] [CrossRef]
- Bettoni, J.C.; Chen, K.; Volk, G.M. Considerations When Implementing Shoot Tip Cryopreservation. In Training in Plant Genetic Resources: Cryopreservation of Clonal Propagules; Volk, G.M., Ed.; Colorado State University: Fort Collins, CO, USA, 2024; Available online: https://colostate.pressbooks.pub/clonalcryopreservation/chapter/considerations-when-implementing-shoot-tip-cryopreservation/ (accessed on 18 April 2025).
- Wünsch, A.; Hormaza, J. Molecular characterisation of sweet cherry (Prunus avium L.) genotypes using peach [Prunus persica (L.) Batsch] SSR sequences. Heredity 2002, 89, 56–63. [Google Scholar] [CrossRef]
- Zhebentyayeva, T.; Shankar, V.; Scorza, R.; Callahan, A.; Ravelonandro, M.; Castro, S.; DeJong, T.; Saski, C.A.; Dardick, C. Genetic characterization of worldwide Prunus domestica (plum) germplasm using sequence-based genotyping. Hortic. Res. 2019, 6, 12. [Google Scholar] [CrossRef]
- Aranzana, M.J.; Decroocq, V.; Dirlewanger, E.; Eduardo, I.; Gao, Z.S.; Gasic, K.; Iezzoni, A.; Jung, S.; Peace, C.; Prieto, H.; et al. Prunus genetics and applications after de novo genome sequencing: Achievements and prospects. Hortic. Res. 2019, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Batnini, M.A.; Bourguiba, H.; Trifi-Farah, N.; Krichen, L. Molecular diversity and phylogeny of Tunisian Prunus arme-niaca L. by evaluating three candidate barcodes of the chloroplast genome. Sci. Hortic. 2019, 245, 99–106. [Google Scholar] [CrossRef]
- Wu, F.Q.; Shen, S.K.; Zhang, X.J.; Wang, Y.H.; Sun, W.B. Genetic diversity and population structure of an extremely endangered species: The world’s largest Rhododendron. AoB Plants 2014, 7, 10696–10700. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Gostel, M.R.; Kress, W.J. The Expanding Role of DNA Barcodes: Indispensable Tools for Ecology, Evolution, and Conservation. Diversity 2022, 14, 213. [Google Scholar] [CrossRef]
- Letsiou, S.; Madesis, P.; Vasdekis, E.; Montemurro, C.; Grigoriou, M.E.; Skavdis, G.; Moussis, V.; Koutelidakis, A.E.; Tzakos, A.G. DNA Barcoding as a Plant Identification Method. Appl. Sci. 2024, 14, 1415. [Google Scholar] [CrossRef]
- Jiang, K.W.; Zhang, R.; Zhang, Z.F.; Pan, B.; Tian, B. DNA barcoding and molecular phylogeny of Dumasia (Fabaceae: Phaseoleae) reveals a cryptic lineage. Plant Divers. 2020, 42, 376–385. [Google Scholar] [CrossRef]
- Tyagi, K.; Kumar, V.; Kundu, S.; Pakrashi, A.; Prasad, P.; Caleb, J.T.D.; Chandra, K. Identification of Indian Spiders through DNA barcoding: Cryptic species and species complex. Sci. Rep. 2019, 9, 14033. [Google Scholar] [CrossRef]
- Carneiro de Melo Moura, C.; Brambach, F.; Jair Hernandez Bado, K.; Krutovsky, K.V.; Kreft, H.; Tjitrosoedirdjo, S.S.; Siregar, I.Z.; Gailing, O. Integrating DNA Barcoding and Traditional Taxonomy for the Identification of Dipterocarps in Remnant Lowland Forests of Sumatra. Plants 2019, 8, 461. [Google Scholar] [CrossRef]
- Wenne, R. Microsatellites as Molecular Markers with Applications in Exploitation and Conservation of Aquatic Animal Populations. Genes 2023, 14, 808. [Google Scholar] [CrossRef]
- Terrones, A.; van der Bank, M.; Moreno, J.; Juan, A. DNA barcodes and microsatellites: How they complement for species identification in the complex genus Tamarix (Tamaricaceae). J. Syst. Evol. 2022, 60, 1140–1157. [Google Scholar] [CrossRef]
- Meyer, C.P.; Paulay, G. DNA barcoding: Error rates based on comprehensive sampling. PLoS Biol. 2005, 3, e422. [Google Scholar] [CrossRef] [PubMed]
- Shelke, R.G.; Lahane, Y.; Ingle, P.; Pusalkar, P.; Chauhan, S.; Pardhi, P.; Nirmal, R.C.; Prajapati, S.K. Advances in Plant Molecular Taxonomy: Techniques, Challenges, and Applications. Plant Cell Biotechnol. Mol. Biol. 2025, 26, 89–110. [Google Scholar] [CrossRef]
- Volk, G.M.; Henk, A.D.; Richards, C.M.; Forsline, P.L.; Chao, C.T. Malus sieversii: A diverse central Asian apple species in the USDA-ARS National Plant Germplasm System. HortScience 2013, 48, 1440–1444. [Google Scholar] [CrossRef]
- Volk, G.M.; Peace, C.P.; Henk, A.D.; Howard, N.P. DNA profiling with the 20K apple SNP array reveals Malus domestica hybridization and admixture in M. sieversii, M. orientalis, and M. sylvestris genebank accessions. Front. Plant Sci. 2022, 13, 1015658. [Google Scholar] [CrossRef]
- Forsdick, N.J.; Adams, C.I.M.; Alexander, A.; Clark, A.C.; Collier-Anderson, L.; Cubrinovska, I.; Croll Dowgray, M.; Dowle, E.J.; Duntsch, L.; Galla, S.J.; et al. Current Applications and Future Promise of Genetic/Genomic Data for Conservation in an Aotearoa New Zealand Context; Science for Conservation 337; Department of Conservation: Wellington, New Zealand, 2022; p. 57. [Google Scholar]
- Hohenlohe, P.A.; Funk, W.C.; Rajora, O.P. Population genomics for wildlife conservation and management. Mol. Ecol. 2021, 30, 62–82. [Google Scholar] [CrossRef]
- Theissinger, K.; Fernandes, C.; Formenti, G.; Bista, I.; Berg, P.R.; Bleidorn, C.; Bombarely, A.; Crottini, A.; Gallo, G.R.; Godoy, J.A.; et al. How genomics can help biodiversity conservation. Trends Genet. 2023, 39, 545–559. [Google Scholar] [CrossRef]
- Liu, N.; Guan, M.; Ma, B.; Chu, H.; Tian, G.; Zhang, Y.; Li, C.; Zheng, W.; Wang, X. Unraveling genetic mysteries: A comprehensive review of GWAS and DNA insights in animal and plant pathosystems. Int. J. Biol. Macromol. 2025, 285, 138216. [Google Scholar] [CrossRef]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP markers and their impact on plant breeding. Int. J. Plant Genom. 2012, 2012, 728398. [Google Scholar] [CrossRef]
- Messina, R.; Lain, O.; Marrazzo, M.T.; Cipriani, G.; Testolin, R. New set of microsatellite loci isolated in apricot. Mol. Ecol. Notes 2004, 4, 432–434. [Google Scholar] [CrossRef]
- Gao, Z.H.; Shen, Z.J.; Han, Z.H.; Fang, J.G.; Zhang, Y.M.; Zhen, Z. Microsatellite markers and genetic diversity in Japanese apricot (Prunus mume). HortScience 2004, 39, 1571–1574. [Google Scholar] [CrossRef]
- Maghuly, F.; Fernandez, E.B.; Ruthner, S.; Pedryc, A.; Laimer, M. Microsatellite variability in apricots (Prunus armeniaca L.) reflects their geographic origin and breeding history. Tree Genet. Genomes 2005, 1, 151–165. [Google Scholar] [CrossRef]
- Hormaza, J.I. Identification of apricot (Prunus armeniaca L.) genotypes using microsatellite and RAPD markers. Acta Hort. 2001, 546, 209–215. [Google Scholar] [CrossRef]
- Butiuc-Keul, A.; Coste, A.; Postolache, D.; Laslo, V.; Halmagyi, A.; Cristea, V.; Farkas, A. Molecular Characterization of Prunus Cultivars from Romania by Microsatellite Markers. Horticulturae 2022, 8, 291. [Google Scholar] [CrossRef]
- Ayour, J.; Elateri, I.; Allami, M.; Harrak, H.; Alfeddy, M.N.; Audergon, J.-M.; Renard, C.M.G.C.; Benichou, M. Genetic diversity assessment towards core collection construction of Moroccan apricot (Prunus armeniaca L.) germplasm using genomic SSR markers. S. Afr. J. Bot. 2025, 180, 21–34. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Q.; Lu, C.; Liu, Q.; Pan, J.; Zhang, J.; Dong, S. Genetic diversity of Prunus armeniaca L. var. ansu Maxim. germplasm revealed by simple sequence repeat (SSR) markers. PLoS ONE 2022, 17, e0269424. [Google Scholar] [CrossRef]
- Weisskopf, A.; Fuller, D.Q. Apricot: Origins and Development. In Encyclopedia of Global Archaeology; Smith, C., Ed.; Springer: New York, NY, USA, 2014; pp. 294–296. [Google Scholar]
- Mirzaev, M.M.; Kuznetsov, V.V. Apricots in Uzbekistan: Biology, Cultivars, Breeding, Agrotechnology; Fan Publishing: Tashkent, Uzbekistan, 1984; p. 199. (In Russian) [Google Scholar]
- Sitpaeva, G.T.; Turuspekov, Y.; Abugalieva, A.; Ormanbekova, D. Crop wild relatives of Kazakhstani Tien Shan: Flora, vegetation, resources. Plant Divers. 2020, 42, 87–96. [Google Scholar] [CrossRef]
- Ledbetter, C.A. Apricots. In Temperate Fruit Crop Breeding: Germplasm to Genomics; Hancock, J.F., Ed.; Springer: Berlin, Germany, 2008; pp. 39–82. [Google Scholar]
- Baitenov, M.S. Flora of Kazakhstan; Generic Complex of Flora: Almaty, Kazakhstan, 2021; Volume 2, p. 280. (In Russian) [Google Scholar]
- Eastwood, A.; Lazkov, G.; Newton, A. The Red List of Trees of Central Asia; Fauna & Flora International: Cambridge, UK, 2009; p. 31. [Google Scholar]
- Lapeña, I.; Turdieva, M.; López Noriega, I.; Ayad, W.G. Conservation of Fruit Tree Diversity in Central Asia: Policy Options and Challenges; Bioversity International: Rome, Italy, 2014; p. 251. [Google Scholar]
- Vinceti, B.; Elias, M.; Azimov, R.; Turdieva, M.; Aaliev, S.; Bobokalonov, F.; Butkov, E.; Kaparova, E.; Mukhsimov, N.; Shamuradova, S.; et al. Home gardens of Central Asia: Reservoirs of diversity of fruit and nut tree species. PLoS ONE 2022, 17, e0271398. [Google Scholar] [CrossRef]
- Honkavaara, E.; Arbiol, R.; Markelin, L.; Martinez, L.; Cramer, M.; Bovet, S.; Chandelier, L.; Ilves, R.; Klonus, S.; Marshal, P.; et al. Digital airborne photogrammetry—A new tool for quantitative remote sensing? A state-of-the-art review on radiometric aspects of digital photogrammetric images. Remote Sens. 2009, 1, 577–605. [Google Scholar] [CrossRef]
- Fyleris, T.; Kriščiūnas, A.; Gružauskas, V.; Čalnerytė, D.; Barauskas, R. Urban Change Detection from Aerial Images Using Convolutional Neural Networks and Transfer Learning. ISPRS Int. J. Geo-Inf. 2022, 11, 246. [Google Scholar] [CrossRef]
- Linder, W. Digital Photogrammetry: A Practical Course; Springer: Berlin, Germany, 2009; p. 214. [Google Scholar] [CrossRef]
- Balenović, I.; Seletković, A.; Pernar, R.; Marjanović, H.; Vuletić, D.; Paladinić, E.; Kolić, J.; Benko, M. Digital Photogrammetry—State of the Art and Potential for Application in Forest Management in Croatia. S.-E. Eur. For. 2011, 2, 81–93. [Google Scholar] [CrossRef]
- Magnusson, M.; Fransson, J.E.S.; Olsson, H. Aerial photo-interpretation using Z/I DMC images for estimation of forest variables. Scand. J. For. Res. 2007, 22, 254–266. [Google Scholar] [CrossRef]
- Alsadik, B.; Ellsäßer, F.J.; Awawdeh, M.; Al-Rawabdeh, A.; Almahasneh, L.; Oude Elberink, S.; Abuhamoor, D.; Al Asmar, Y. Remote Sensing Technologies Using UAVs for Pest and Disease Monitoring: A Review Centered on Date Palm Trees. Remote Sens. 2024, 16, 4371. [Google Scholar] [CrossRef]
- Gobakken, T.; Bollandsås, O.M.; Næsset, E. Comparing biophysical forest characteristics estimated from photogrammetric matching of aerial images and airborne laser scanning data. Scand. J. For. Res. 2015, 30, 73–86. [Google Scholar] [CrossRef]
- Zeng, Y.; Hao, D.; Huete, A.; Dechant, B.; Berry, J.; Chen, J.M.; Joiner, J.; Frankenberg, C.; Bond-Lamberty, B.; Ryu, Y.; et al. Optical vegetation indices for monitoring terrestrial ecosystems globally. Nat. Rev. Earth Environ. 2022, 3, 477–493. [Google Scholar] [CrossRef]
- Uscanga, A. Tracking vegetation changes with time series of satellite images. Nat. Rev. Earth Environ. 2023, 4, 513. [Google Scholar] [CrossRef]
- Zhao, Q.; Qu, Y. The Retrieval of Ground NDVI (Normalized Difference Vegetation Index) Data Consistent with Remote-Sensing Observations. Remote Sens. 2024, 16, 1212. [Google Scholar] [CrossRef]
- Xue, J.; Su, B. Significant remote sensing vegetation indices: A review of developments and applications. J. Sens. 2017, 2017, 1353691. [Google Scholar] [CrossRef]
- Solymosi, K.; Kövér, G.; Romvári, R. The Development of Vegetation Indices: A Short Overview. Acta Agrar. Kaposvariensis 2019, 23, 75–90. [Google Scholar] [CrossRef]
- Nijland, W.; de Jong, R.; de Jong, S.M.; Wulder, M.A.; Bater, C.W.; Coops, N.C. Monitoring plant condition and phenology using infrared sensitive consumer grade digital cameras. Agric. For. Meteorol. 2014, 184, 98–106. [Google Scholar] [CrossRef]
- Huete, A.R.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Martinez, A.; Labib, S.M. Demystifying normalized difference vegetation index (NDVI) for greenness exposure assessments and policy interventions in urban greening. Environ. Res. 2023, 220, 115155. [Google Scholar] [CrossRef]
- Karnieli, A.; Agam, N.; Pinker, R.T.; Anderson, M.; Imhoff, M.L.; Gutman, G.G.; Panov, N.; Goldberg, A. Use of NDVI and land surface temperature for drought assessment: Merits and limitations. J. Clim. 2010, 23, 618–633. [Google Scholar] [CrossRef]
- Huang, J.; Wang, H.; Dai, Q.; Han, D. Analysis of NDVI Data for Crop Identification and Yield Estimation. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2014, 7, 4374–4384. [Google Scholar] [CrossRef]
- Reid, C.E.; Kubzansky, L.D.; Li, J.; Shmool, J.L.; Clougherty, J.E. It’s not easy assessing greenness: A comparison of NDVI datasets and neighborhood types and their associations with self-rated health in New York City. Health Place 2018, 54, 92–101. [Google Scholar] [CrossRef]
- Weier, J.; Herring, D. Measuring Vegetation (NDVI & EVI). NASA Earth Observatory; 2000. Available online: https://earthobservatory.nasa.gov/features/MeasuringVegetation (accessed on 1 December 2024).
- GISGeography. Available online: https://gisgeography.com/ndvi-normalized-difference-vegetation-index/ (accessed on 11 October 2024).
- Morawitz, D.F.; Blewett, T.M.; Cohen, A.; Alberti, M. Using NDVI to Assess Vegetative Land Cover Change in Central Puget Sound. Environ. Monit. Assess. 2006, 114, 85–106. [Google Scholar] [CrossRef]
- Jensen, J.R. Remote Sensing of the Environment: An Earth Resource Perspective, 2nd ed.; Pearson Prentice Hall: London, UK, 2006; p. 656. [Google Scholar]
- Brown, L.A.; Dash, J.; Ogutu, B.O.; Richardson, A.D. On the relationship between continuous measures of canopy greenness derived using near-surface remote sensing and satellite-derived vegetation products. Agric. For. Meteorol. 2017, 247, 280–292. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; Peña, J.M.; de Castro, A.I.; López-Granados, F. Multi-Temporal Mapping of the Vegetation Fraction in Early-Season Wheat Fields Using Images from UAV. Comput. Electron. Agric. 2014, 103, 104–113. [Google Scholar] [CrossRef]
- Dandois, J.P.; Ellis, E.C. High spatial resolution three-dimensional mapping of vegetation spectral dynamics using computer vision. Remote Sens. Environ. 2013, 136, 259–276. [Google Scholar] [CrossRef]
- Manapkanova, U.; Rymkhanova, N.; Reim, S.; Fritzsche, E.; Höfer, M.; Beshko, N.; Satekov, Y.; Kushnarenko, S.V. Genetic Diversity and Phenotypic Variation of Indigenous Wild Cherry Species in Kazakhstan and Uzbekistan. Plants 2025, 14, 1676. [Google Scholar] [CrossRef] [PubMed]
- Fiore, M.C.; Gugliuzza, G. Morphological and genetic evaluation of local germplasm: The case study of Sicilian apples. Acta Hortic. 2023, 1384, 121–128. [Google Scholar] [CrossRef]
- Corrado, G.; Forlani, M.; Rao, R.; Basile, B. Diversity and Relationships among Neglected Apricot (Prunus armeniaca L.) Landraces Using Morphological Traits and SSR Markers: Implications for Agro-Biodiversity Conservation. Plants 2021, 10, 1341. [Google Scholar] [CrossRef] [PubMed]
- Corrado, G.; La Mura, M.; Ambrosino, O.; Pugliano, G.; Varricchio, P.; Rao, R. Relationships of Campanian olive cultivars: Comparative analysis of molecular and phenotypic data. Genome 2009, 52, 692–700. [Google Scholar] [CrossRef]
- Manco, R.; Basile, B.; Capuozzo, C.; Scognamiglio, P.; Forlani, M.; Rao, R.; Corrado, G. Molecular and phenotypic diversity of traditional european plum (Prunus domestica L.) germplasm of southern Italy. Sustainability 2019, 11, 4112. [Google Scholar] [CrossRef]
- Keating, J.N.; Garwood, R.J.; Sansom, R.S. Phylogenetic congruence, conflict and consilience between molecular and morphological data. BMC Ecol. Evo. 2023, 23, 30. [Google Scholar] [CrossRef]
- Sommer, R.J. Phenotypic Plasticity: From Theory and Genetics to Current and Future Challenges. Genetics 2020, 215, 1–13. [Google Scholar] [CrossRef]
- van den Ende, C.; Puttick, M.N.; Urrutia, A.O.; Wills, M.A. Why should we compare morphological and molecular disparity? Methods Ecol. Evol. 2023, 14, 2390–2410. [Google Scholar] [CrossRef]
- Duminil, J.; Di Michele, M. Plant species delimitation: A comparison of morphological and molecular markers. Plant Biosyst. 2009, 143, 528–542. [Google Scholar] [CrossRef]
- Qin, Q.; Dong, Y.; He, J.; Chen, J.; Wu, D.; Zhang, S. Assessment of genetic diversity and construction of core germplasm in populations of Acorus tatarinowii based on SNP markers. J. Appl. Res. Med. Aromat. Plants 2025, 44, 100605. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Sun, H.; Ning, N.; Yang, L. Construction and evaluation of a primary core collection of apricot germplasm in China. Sci. Hortic. 2011, 128, 311–319. [Google Scholar] [CrossRef]
- Romadanova, N.V.; Aralbayeva, M.M.; Zemtsova, A.S.; Alexandrova, A.M.; Kazybayeva, S.Z.; Mikhailenko, N.V.; Kushnarenko, S.V.; Bettoni, J.C. In Vitro Collection for the Safe Storage of Grapevine Hybrids and Identification of the Presence of Plasmopara viticola Resistance Genes. Plants 2024, 13, 1089. [Google Scholar] [CrossRef] [PubMed]
- Stoeckel, S.; Grange, J.; Fernández-Manjarres, J.F.; Bilger, I.; Frascaria-Lacoste, N.; Mariette, S. Heterozygote excess in a self-incompatible and partially clonal forest tree species—Prunus avium L. Mol. Ecol. 2006, 8, 2109–2118. [Google Scholar] [CrossRef]
- Zhebentyayeva, T.N.; Reighard, G.L.; Gorina, V.M.; Abbott, A.G. Simple sequence repeat (SSR) analysis for assessment of genetic variability in apricot germplasm. Theor. Appl. Genet. 2003, 106, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Uzun, A.; Pinar, H.; Gürcan, K.; Turgunbaev, K.; Yıldız, E.; Ilgın, M.; Dolgikh, S. Genetic diversity and population structure of wild and cultivated apricots collected from Kyrgyzstan. Genet. Resour. Crop. Evol. 2024, 71, 4131–4140. [Google Scholar] [CrossRef]
- Ellstrand, N.C.; Rieseberg, L.H. When gene flow really matters: Gene flow in applied evolutionary biology. Evol. Appl. 2016, 9, 833–836. [Google Scholar] [CrossRef]
- Hu, X.; Zheng, P.; Ni, B.; Miao, X.; Zhao, Z.; Li, M. Population genetic diversity and structure analysis of wild apricot (Prunus armeniaca L.) revealed by SSR markers in the Tien-Shan Mountains of China. Pak. J. Bot. 2018, 5, 609–615. [Google Scholar]
- Bourguiba, H.; Krichen, L.; Trifi-Farah, N.; Khadari, B.; Mamouni, A.; Trabelsi, S.; D’Onofrio, C.; Egea-Caballero, J.; Ruiz, D.; Asma, B.M.; et al. Genetic structure of mediterranean apricots by SSR fingerprinting. Acta Hortic. 2011, 918, 309–314. [Google Scholar] [CrossRef]
- Bettoni, J.C.; Wang, M.-R.; Wang, Q.-C. In Vitro Regeneration, Micropropagation and Germplasm Conservation of Horticultural Plants. Horticulturae 2024, 10, 45. [Google Scholar] [CrossRef]
- Zhang, A.-L.; Wang, M.-R.; Li, Z.; Panis, B.; Bettoni, J.C.; Vollmer, R.; Xu, L.; Wang, Q.-C. Overcoming Challenges for Shoot Tip Cryopreservation of Root and Tuber Crops. Agronomy 2023, 13, 219. [Google Scholar] [CrossRef]
- Romadanova, N.V.; Mishustina, S.A.; Matakova, G.N.; Kushnarenko, S.V.; Rakhimbaev, I.R.; Reed, B.M. In vitro collectionmethods for Malus shoot cultures used for developing a cryogenic bank in Kazakhstan. Acta Hortic. 2016, 1113, 271–277. [Google Scholar] [CrossRef]
- Volk, G.M.; Bonnart, R.M.; Henk, A.D.; Chen, K.Y.; Bettoni, J.C.; Wang, Q.-C.; Kreckel, H.D.; Levinger, N.E. Fundamentals of plant cryopreservation: Dormant bud two-step cooling and shoot tip vitrification. Acta Hortic. 2025, 1421, 117–124. [Google Scholar] [CrossRef]
- Dobránszki, J.; Teixeira da Silva, J.A. Micropropagation of apple—A review. Biotechnol. Adv. 2010, 28, 462–488. [Google Scholar] [CrossRef] [PubMed]
- An, K.; Zhou, C.; Tong, B.; Liu, D.; Shan, X.; Zhang, X.; Bian, F. Population genetic differentiation and structure of rare plant Anemone shikokiana based on genotyping-by-sequencing (GBS). BMC Plant Biol. 2024, 24, 995. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Populations: Vol. 2. The Theory of Gene Frequencies, 1st ed.; The University of Chicago: Chicago, IL, USA, 1969; p. 520. [Google Scholar]
- Charlesworth, D.; Wright, S.I. Breeding systems and genome evolution. Curr. Opin. Genet. Dev. 2001, 11, 685–690. [Google Scholar] [CrossRef]
- Barrett, S.C.H. Mating strategies in flowering plants: The outcrossing–selfing paradigm and beyond. Phil. Trans. R. Soc. B Biol. Sci. 2003, 358, 991–1004. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W.; Sherman-Broyles, S.L. Factors influencing levels of genetic diversity in woody plant species. N. For. 1992, 6, 95–124. [Google Scholar] [CrossRef]
- Furches, M.S.; Small, R.L.; Furches, A. Genetic diversity in three endangered pitcher plant species (Sarracenia; Sarraceniaceae) is lower than widespread congeners. Am. J. Bot. 2013, 100, 2092–2101. [Google Scholar] [CrossRef]
- Sork, V.L. Gene flow and natural selection shape spatial patterns of genes in tree populations: Implications for evolutionary processes and applications. Evol. Appl. 2015, 9, 291–310. [Google Scholar] [CrossRef]
- Xiang-Rong, F.; Wagutu, G.K.; Wen, X.-Y.; Chen, S.-L.; Liu, Y.-L.; Chen, Y.-Y. Decreasing genetic connectivity in the endangered tree Magnolia patungensis in fragmented forests. Glob. Ecol. Conserv. 2020, 24, e01227. [Google Scholar] [CrossRef]
- Waqar, Z.; Moraes, R.C.S.; Benchimol, M.; Morante-Filho, J.C.; Mariano-Neto, E.; Gaiotto, F.A. Gene Flow and Genetic Structure Reveal Reduced Diversity between Generations of a Tropical Tree, Manilkara multifida Penn., in Atlantic Forest Fragments. Genes 2021, 12, 2025. [Google Scholar] [CrossRef]
- Bijlsma, R.; Loeschcke, V. Genetic erosion impedes adaptive responses to stressful environments. Evol. Appl. 2012, 5, 117–129. [Google Scholar] [CrossRef]
- Hoban, S.; Campbell, C.D.; da Silva, J.M.; Ekblom, R.; Funk, W.C.; Garner, B.A.; Godoy, J.A.; Kershaw, F.; MacDonald, A.J.; Mergeay, J.; et al. Genetic diversity is considered important but interpreted narrowly in country reports to the Convention on Biological Diversity: Current actions and indicators are insufficient. Biol. Conserv. 2021, 261, 109233. [Google Scholar] [CrossRef]
- Akzhunis, I.; Dinara, Z.; Nurzhaugan, D.; Aidyn, O.; Nazerke, T.; Gulmira, T. Unravelling the Chloroplast Genome of the Kazakh Apricot (Prunus armeniaca L.) Through MinION Long-Read Sequencing. Plants 2025, 14, 638. [Google Scholar] [CrossRef] [PubMed]
- Kuziev, R.K.; Sektimenko, V.E. Soils of Uzbekistan; Extremum Press Publishing House: Tashkent, Uzbekistan, 2009; p. 351. (In Russian) [Google Scholar]
- Kuziev, R.K.; Yuldashev, G.Y.U.; Akramov, I.A. Bonitization of Soils; The Way of Science Publishing House: Tashkent, Uzbekistan, 2004; p. 127. [Google Scholar]
- Rechlin. (n.d.). Almaty (Province) in Kazakhstan [Map]. Wikipedia. Available online: https://pt.m.wikipedia.org/wiki/Ficheiro:Almaty_%28province%29_in_Kazakhstan.svg (accessed on 19 June 2025).
- Matese, A.; Di Gennaro, S.F. Practical Applications of a Multisensor UAV Platform Based on Multispectral, Thermal and RGB High Resolution Images in Precision Viticulture. Agriculture 2018, 8, 116. [Google Scholar] [CrossRef]
- Małek, A. Assessment of the use of unmanned aerial vehicles for road pavement condition surveying. Roads Bridges/Drog. I Mosty 2023, 22, 331–345. [Google Scholar] [CrossRef]
- Baltsavias, E.; Gruen, A.; Eisenbeiss, H.; Zhang, L.; Waser, L. High-quality image matching and automated generation of 3D tree models. Int. J. Remote Sens. 2008, 29, 1243–1259. [Google Scholar] [CrossRef]
- Bohlin, J.; Wallerman, J.; Fransson, J. Forest variable estimation using photogrammetric matching of digital aerial images in combination with a high-resolution DEM. Scand. J. For. Res. 2012, 27, 692–699. [Google Scholar] [CrossRef]
- Darwin, N.; Ahmad, A. Fast Data Acquisition of Aerial Images Using Unmanned Aerial Vehicle System. IJ-ICT. 2014, 3, 162–170. [Google Scholar] [CrossRef]
- Järndstedt, J.; Pekkarinen, A.; Tuominen, S.; Ginzler, C.; Holopainen, M.; Viitala, R. Forest variable estimation using a high-resolution digital surface model. ISPRS J. Photogramm. 2012, 74, 78–84. [Google Scholar] [CrossRef]
- Vastaranta, M.; Wulder, M.A.; White, J.C.; Pekkarinen, A.; Tuominen, S.; Ginzler, C.; Kankare, V.; Holopainen, M.; Hyyppä, J.; Hyyppä, H. Airborne laser scanning and digital stereo imagery measures of forest structure: Comparative results and implications to forest mapping and inventory update. Can. J. Remote Sens. 2013, 39, 382–395. [Google Scholar] [CrossRef]
- Amani, S.; Shafizadeh-Moghadam, H. A review of machine learning models and influential factors for estimating evapotranspiration using remote sensing and ground-based data. Agric. Water Manag. 2023, 284, 108324. [Google Scholar] [CrossRef]
- Vani, V.; Venkata, M. Comparative study of NDVI and SAVI vegetation indices in Anantapur district semi-arid areas. Int. J. Civ. Eng. Technol. 2017, 8, 559–566. [Google Scholar]
- Cuénoud, P.; Savolainen, V.; Chatrou, L.W.; Powell, M.; Grayer, R.J.; Chase, M.W. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am. J. Bot. 2002, 89, 132–144. [Google Scholar] [CrossRef]
- Costion, C.; Ford, A.; Cross, H.; Crayn, D.; Harrington, M.; Lowe, A. Plant DNA barcodes can accurately estimate species richness in poorly known floras. PLoS ONE 2011, 6, e26841. [Google Scholar] [CrossRef]
- Gu, W.; Song, J.; Cao, Y.; Sun, Q.; Yao, H.; Wu, Q.; Chao, J.; Zhou, J.; Xue, W.; Duan, J.; et al. Application of the ITS2 Region for Barcoding Medicinal Plants of Selaginellaceae in Pteridophyta. PLoS ONE 2013, 8, e67818. [Google Scholar] [CrossRef]
- de Vere, N.; Rich, T.C.G.; Trinder, S.A.; Long, C. DNA barcoding for plants. Methods Mol. Biol. 2015, 1245, 101–118. [Google Scholar] [CrossRef]
- Berdimuratova, K.T.; Amirgazin, A.O.; Kuibagarov, M.A.; Lutsay, V.B.; Mukanov, K.K.; Shevtsov, A.B. Optimization of PCR Purification Using Silica-Coated Magnetic Beads. Eurasian J. Appl. Biotechnol. 2020, 1. [Google Scholar] [CrossRef]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar] [CrossRef]
- Hagen, L.S.; Chaïb, J.; Fady, B.; Decroocq, V.; Bouchet, J.P.; Lambert, P.; Audergon, J.M. Genomic and cDNA microsatellites from apricot (Prunus armeniaca L.). Mol. Ecol. Notes 2024, 4, 742–745. [Google Scholar] [CrossRef]
- Dirlewanger, E.; Cosson, P.; Tavaud, M.; Aranzana, M.; Poizat, C.; Zanetto, A.P.; Laigret, F. Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor. Appl. Genet. 2022, 105, 127–138. [Google Scholar] [CrossRef]
- Aranzana, M.J.; Garcia-Mas, J.; Carbo, J.; Arús, P. Development and variability analysis of microsatellite markers in peach. Plant Breed. 2008, 121, 87–92. [Google Scholar] [CrossRef]
- Testolin, R.; Marrazzo, T.; Cipriani, G.; Quarta, R.; Verde, I.; Dettori, M.T.; Pancaldi, M.; Sansavini, S. Microsatellite DNA in peach (Prunus persica L. Batsch) and its use in fingerprinting and testing the genetic origin of cultivars. Genome 2000, 43, 512–520. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P. GENALEX 6: Genetic analysis in excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Lemenkova, P. R Libraries {dendextend} and {magrittr} and Clustering Package scipy.cluster of Python For Modelling Diagrams of Dendrogram Trees. Carpathian J. Electr. Eng. 2020, 13, 5–12. [Google Scholar] [CrossRef]
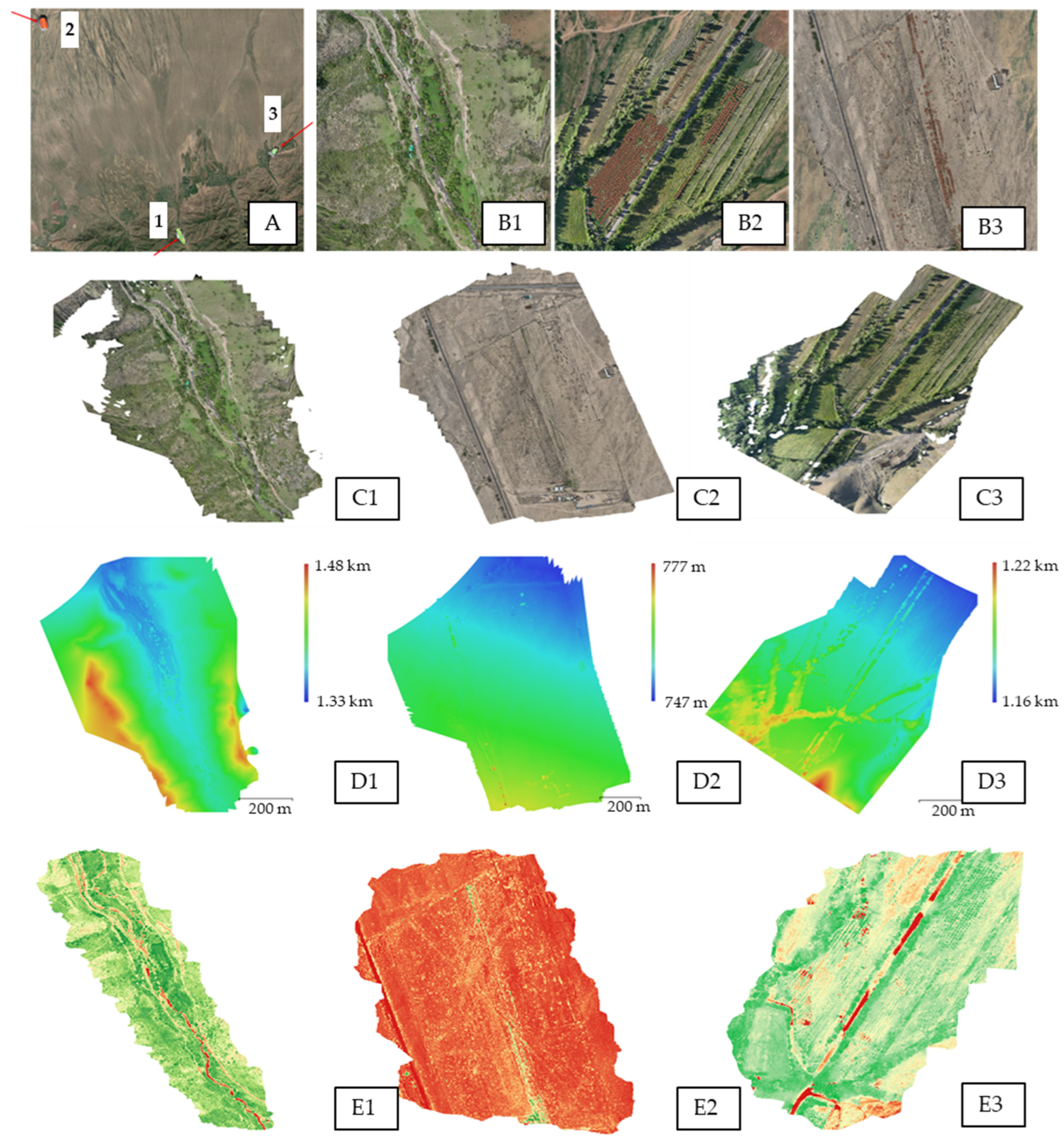

| Areas | Altitude (m) | Fight Duration | Number of Photos Taken | NDVI |
|---|---|---|---|---|
| 1 * | 80 | 19 min 23 s | 325 | 0.61 |
| 100 | 23 min 17 s | 474 | 0.63 | |
| 2 | 80 | 20 min 42 s | 382 | 0.33 |
| 100 | 26 min 05 s | 512 | 0.34 | |
| 3 | 80 | 24 min 30 s | 404 | 0.7 |
| 100 | 18 min 20 s | 296 | 0.68 |
| Population | Location | N | Na | Ne | I | Ho | He | uHe | F |
|---|---|---|---|---|---|---|---|---|---|
| Pop1 | Bolshoy Aksu, Site 1 | 3 | 3.769 | 3.226 | 1.214 | 0.615 | 0.667 | 0.800 | 0.104 |
| Pop2 | Bolshoy Aksu, Site 1 | 18 | 5.846 | 2.714 | 1.226 | 0.628 | 0.599 | 0.616 | −0.054 |
| Pop3 | Bolshoy Aksu, Site 1 | 1 | 1.846 | 1.846 | 0.587 | 0.846 | 0.423 | 0.846 | 1.000 |
| Pop4 | Bolshoy Aksu, Site 1 | 6 | 4.923 | 3.671 | 1.398 | 0.744 | 0.707 | 0.772 | −0.052 |
| Pop5 | Bolshoy Kyrgyzsay, Site 2 | 5 | 4.231 | 3.189 | 1.234 | 0.769 | 0.646 | 0.718 | −0.191 |
| Pop6 | Bolshoy Kyrgyzsay, Site 2 | 1 | 1.692 | 1.692 | 0.480 | 0.692 | 0.346 | 0.692 | 1.000 |
| Pop7 | Bolshoy Kyrgyzsay, Site 2 | 3 | 3.385 | 2.892 | 1.044 | 0.769 | 0.577 | 0.692 | −0.338 |
| Pop8 | Turgen | 6 | 4.231 | 2.998 | 1.215 | 0.782 | 0.650 | 0.709 | −0.206 |
| Pop9 | Turgen | 3 | 3.308 | 2.678 | 1.052 | 0.872 | 0.603 | 0.723 | −0.447 |
| Pop10 | Turgen | 4 | 4.231 | 3.424 | 1.285 | 0.904 | 0.680 | 0.777 | −0.341 |
| Pop11 | Bolshoy Kyrgyzsay, Site 2 | 4 | 4.077 | 3.198 | 1.223 | 0.731 | 0.656 | 0.750 | −0.131 |
| Mean | 4.91 | 3.776 | 2.866 | 1.087 | 0.759 | 0.596 | 0.736 | −0.303 |
| Locus | Fis | Fit | Fst | Nm |
|---|---|---|---|---|
| Locus1 | −0.185 | 0.191 | 0.317 | 0.537 |
| Locus2 | −0.387 | −0.079 | 0.222 | 0.876 |
| Locus3 | −0.237 | −0.054 | 0.148 | 1.435 |
| Locus4 | −0.275 | 0.040 | 0.247 | 0.762 |
| Locus5 | −0.365 | −0.194 | 0.125 | 1.745 |
| Locus6 | −0.166 | 0.117 | 0.243 | 0.780 |
| Locus7 | −0.309 | −0.044 | 0.202 | 0.987 |
| Locus8 | −0.294 | 0.033 | 0.253 | 0.740 |
| Locus9 | −0.294 | 0.009 | 0.235 | 0.816 |
| Locus10 | −0.129 | 0.109 | 0.211 | 0.936 |
| Locus11 | −0.237 | 0.132 | 0.298 | 0.589 |
| Locus12 | −0.269 | −0.052 | 0.171 | 1.212 |
| Locus13 | −0.413 | −0.147 | 0.188 | 1.079 |
| Mean | −0.274 | 0.005 | 0.220 | 0.961 |
| SE | 0.023 | 0.032 | 0.015 | 0.094 |
| Number of Accessions | GPS Coordinates, Elevations, m | Place of Collection, Year, Population |
|---|---|---|
| 23 | N43°22′.417′—N43°22′.832′ E077°35′.351′—E077°35′.868′, 980–1046 m | Turgen Gorge, Enbekshikazakh District, 2023–2024, population 1 |
| 43 | N43°17′.118′—N43°18′.456′ E079°37′.901′—E079°39′.901′, 1302–1691 m | Bolshoy Aksu Gorge, Uyghur District, 2023–2024, population 2/site 1 |
| 24 | N43°18′.072′—N43°18′.702′ E079°30′.658′—E079°32′.314′, 1580–1692 m | Bolshoy Kyrgyzsay Gorge, Uyghur District, 2023–2024, population 2/site 2 |
| 16 | N43°20′.784′—N43°20′.875′ E079°29′.827′—E079°29′.890′, 1234–1246 m | Foothills of Bolshoy Kyrgyzsay Gorge, Uyghur District, 2023–2024, population 2/site 3 |
| 5 | N43°31′.652′—N43°31′.674′ E079°27′.353′—E079°27′.849′, 862–864 m | Chundzha village, Karadala Forestry, 2024, population 2/site 4 |
| Name of Primers | Primer Sequence 5′-3′ | Primer Annealing Temperature | Reference |
|---|---|---|---|
| MatK_390-F | CGATCTATTCATTCAATATTTC | 53 °C | [165] |
| MatK_1326-R | TCTAGCACACGAAAGTCGAAGT | ||
| trnHF_05-F | CGCGCATGGTGGATTCACAATCC | 58 °C | [166] |
| psbA3f-R | GTTATGCATGAACGTAATGCTC | ||
| ITS-Bel3-F | GACGCTTCTCCAGACTACAAT | 60 °C | [167] |
| ITS-p5-R | CCTTATCACTTAGAGGAAGGAG | ||
| rbcLa-F | ATGTCACCACAAACAGAGACTAAAGC | 62 °C | [168] |
| rbcLr590 | AGTCCACCGCGTAGACATTCAT |
| Primer Code | Name | Fluorescent Label | Subsequence | Reference |
|---|---|---|---|---|
| Pr-1 | 240001 | FAM | cagtttgatttgtgtgcctctc | [171] |
| 240002 | gatccaccctttgcataaaatc | |||
| Pr-2 | 240003 | FAM | gtgcccacttacctgttttagg | [171] |
| 240004 | tcgacgatcagacttgctacag | |||
| Pr-3 | 240005 | VIC | ctgagtgatccatttgcagg | [172] |
| 240006 | agggcatctagacctcattgtt | |||
| Pr-4 | 240007 | NED | ttaagagtttgtgatgggaacc | [172] |
| 240008 | aagcataatttagcataaccaagc | |||
| Pr-5 | 240009 | FAM | tcctgcgtagaagaaggtagc | [172] |
| 240010 | cgacataaagtccaaatggc | |||
| Pr-6 | 240011 | PET | aattgtacttgccaatgctatga | [172] |
| 240012 | ctgccttctgctcacacc | |||
| Pr-7 | 240013 | FAM | tatattgttggcttcttgcatg | [172] |
| 240014 | tgaaagtgaaacaatggaagc | |||
| Pr-8 | 240015 | NED | atgaggacgtgtctgaatgg | [172] |
| 240016 | agccaaacccctcttatacg | |||
| Pr-9 | 240017 | VIC | aattaactccaacagctcca | [173] |
| 240018 | atggttgcttaattcaatgg | |||
| Pr-10 | 240019 | FAM | caattagctagagagaattattg | [173] |
| 240020 | gacaagaagcaagtagtttg | |||
| Pr-11 | 240021 | PET | tgaatattgttcctcaattc | [173] |
| 240022 | ctctaggcaagagatgaga | |||
| Pr-12 | 240023 | VIC | tcagcaaactagaaacaaa | [173] |
| 240024 | ccttgcaatctggttgatgtt | |||
| Pr-13 | 240025 | PET | tcggtttttaaaattccaaaa | [173] |
| 240026 | gttacccttatttgcacccaaca | |||
| Pr-15 | 240029 | PET | agggaaagtttctgctgcac | [174] |
| 240030 | gctgaagacgacgatgatga |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romadanova, N.V.; Altayeva, N.A.; Zemtsova, A.S.; Artimovich, N.A.; Shevtsov, A.B.; Kakimzhanova, A.; Nurtaza, A.; Tolegen, A.B.; Kushnarenko, S.V.; Bettoni, J.C. Geobotanical Study, DNA Barcoding, and Simple Sequence Repeat (SSR) Marker Analysis to Determine the Population Structure and Genetic Diversity of Rare and Endangered Prunus armeniaca L. Plants 2025, 14, 2333. https://doi.org/10.3390/plants14152333
Romadanova NV, Altayeva NA, Zemtsova AS, Artimovich NA, Shevtsov AB, Kakimzhanova A, Nurtaza A, Tolegen AB, Kushnarenko SV, Bettoni JC. Geobotanical Study, DNA Barcoding, and Simple Sequence Repeat (SSR) Marker Analysis to Determine the Population Structure and Genetic Diversity of Rare and Endangered Prunus armeniaca L. Plants. 2025; 14(15):2333. https://doi.org/10.3390/plants14152333
Chicago/Turabian StyleRomadanova, Natalya V., Nazira A. Altayeva, Alina S. Zemtsova, Natalya A. Artimovich, Alexandr B. Shevtsov, Almagul Kakimzhanova, Aidana Nurtaza, Arman B. Tolegen, Svetlana V. Kushnarenko, and Jean Carlos Bettoni. 2025. "Geobotanical Study, DNA Barcoding, and Simple Sequence Repeat (SSR) Marker Analysis to Determine the Population Structure and Genetic Diversity of Rare and Endangered Prunus armeniaca L." Plants 14, no. 15: 2333. https://doi.org/10.3390/plants14152333
APA StyleRomadanova, N. V., Altayeva, N. A., Zemtsova, A. S., Artimovich, N. A., Shevtsov, A. B., Kakimzhanova, A., Nurtaza, A., Tolegen, A. B., Kushnarenko, S. V., & Bettoni, J. C. (2025). Geobotanical Study, DNA Barcoding, and Simple Sequence Repeat (SSR) Marker Analysis to Determine the Population Structure and Genetic Diversity of Rare and Endangered Prunus armeniaca L. Plants, 14(15), 2333. https://doi.org/10.3390/plants14152333






