ConvTransNet-S: A CNN-Transformer Hybrid Disease Recognition Model for Complex Field Environments
Abstract
1. Introduction
- 1.
- We constructed a multi-crop dataset consisting of 10,441 field-condition images with complex backgrounds, encompassing 12 crop categories and 37 disease types, all of which were collected in real-world field environments.
- 2.
- We propose a hybrid network model named ConvTransNet-S, which achieves a dynamic equilibrium between local feature representations and global dependency modeling.
- 3.
- The model introduces the three key components: LPU, LMHSA, and IRFFN. This design effectively addresses the limitations of conventional CNNs in modeling long-range dependencies and mitigates the deficiencies of transformers in capturing local details, while significantly enhancing the stability of gradient propagation across network layers through hierarchical residual connections.
2. Materials and Methods
2.1. Experimental Data
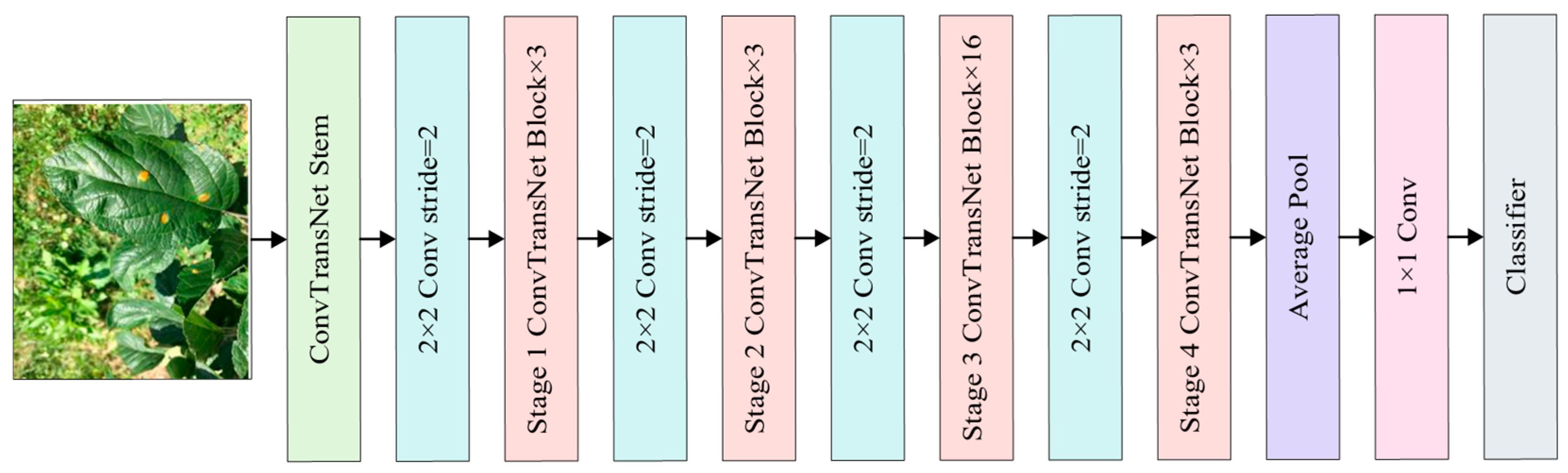
2.2. Construction of Leaf Disease Identification Model
| Output Size | Layer Name | ConvTransNet-S |
|---|---|---|
| Stem | , stride 2
| |
| Patch Embedding | , 64, stride 2 | |
| Stage 1 | LPU LMHSA IRFFN | |
| Patch Embedding | 128, stride 2 | |
| Stage 2 | LPU LMHSA IRFFN | |
| Patch Embedding | 256, stride 2 | |
| Stage 3 | LPU LMHSA IRFFN | |
| Patch Embedding | 512, stride 2 | |
| Stage 4 | LPU LMHSA IRFFN | |
| Projection | , 1280 | |
| Classifier | Fully connected layer, 1000 |
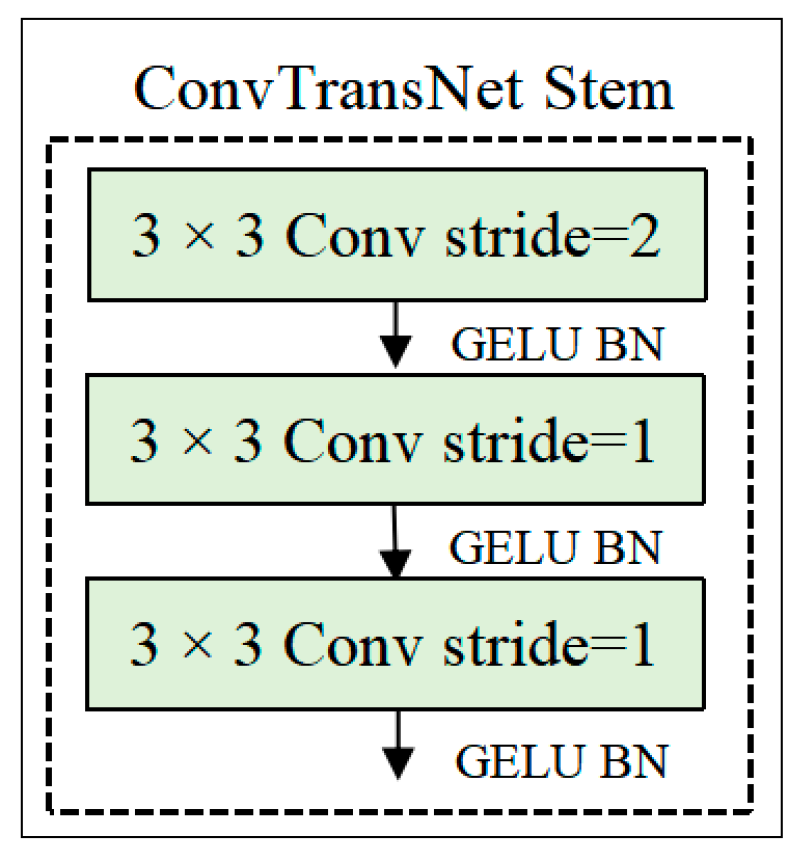
2.2.1. Stem Module
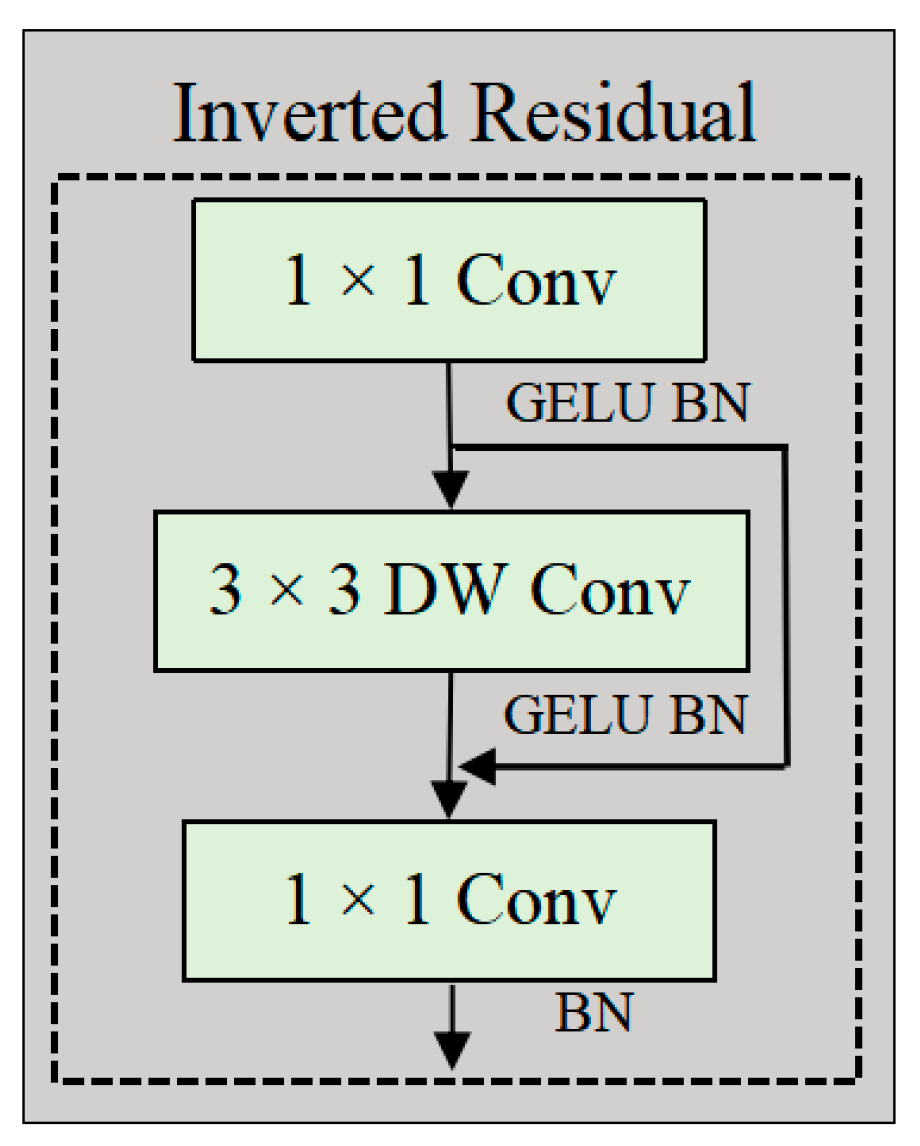
2.2.2. ConvTransNet Block
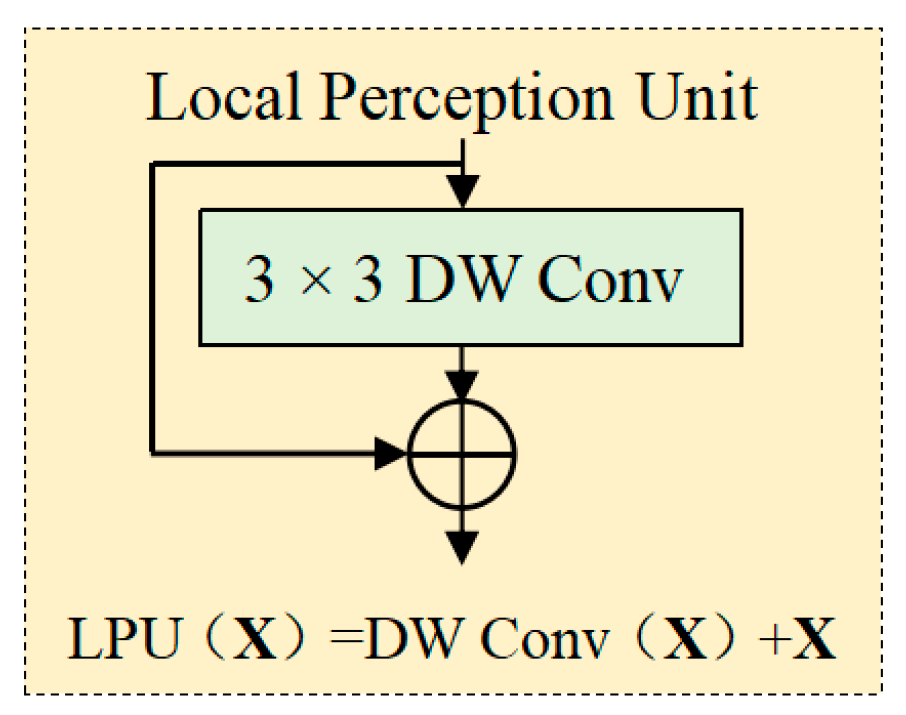
2.2.3. Local Perception Unit
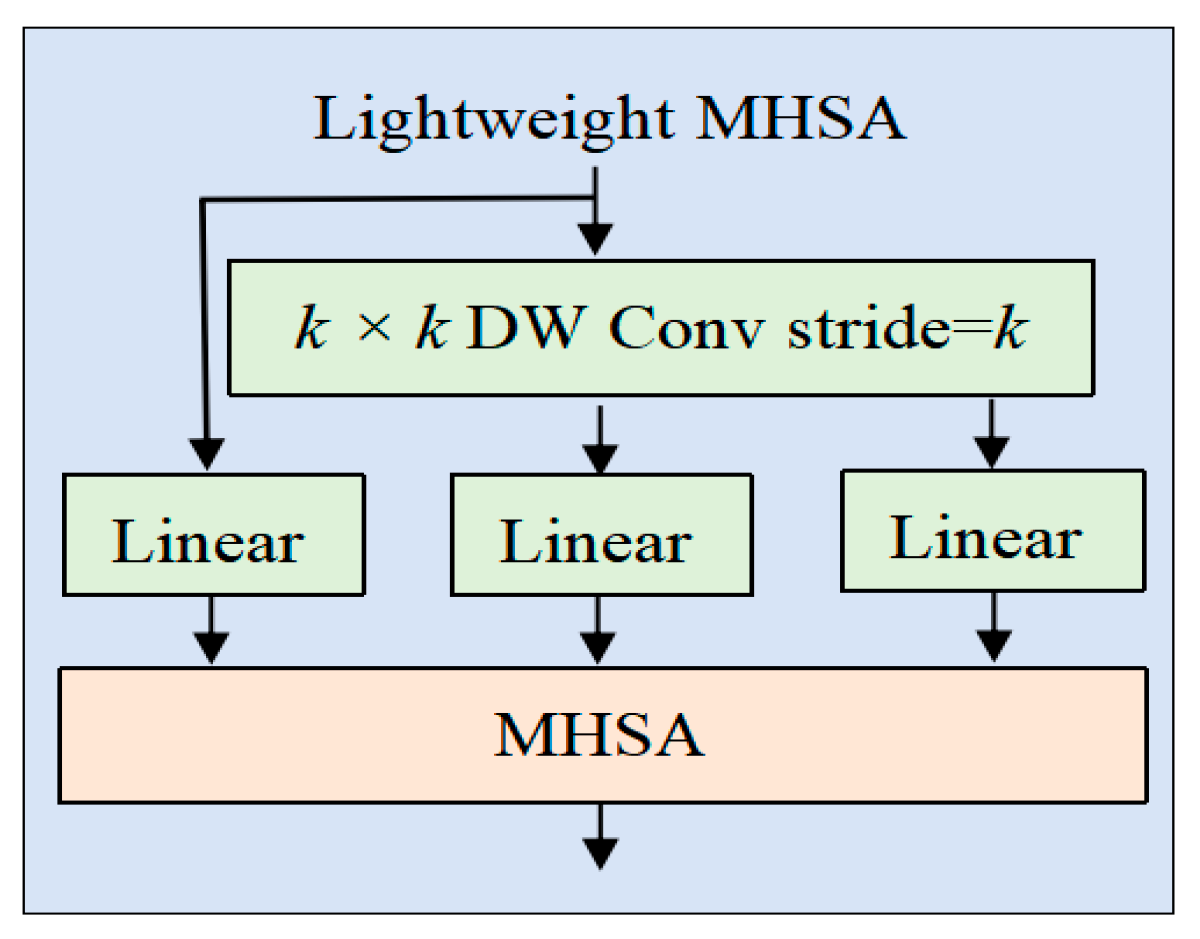
2.2.4. Lightweight Multi-Head Self-Attention
2.2.5. Inverted Residual Feed-Forward Network
2.3. Experimental Environment
2.4. Model Evaluation
3. Experimental Results and Analysis
3.1. Performance Comparison of Different Classification Models
3.2. Application of the ConvTransNet-S Model in Field Environments
3.3. Ablation Studies
3.4. Fine-Grained Performance Evaluation on Sub-Class Datasets
4. Discussion
5. Conclusions
- 1.
- On the PlantVillage dataset, ConvTransNet-S achieved an accuracy of 98.85% with only 25.14M parameters and 3.762 GFLOPs. This model outperforms EfficientNetV2, ViT and Swin-Transformer when evaluated under identical training protocols.
- 2.
- In robustness evaluations conducted under complex field conditions, ConvTransNet-S demonstrated significant superiority with a recognition accuracy of 88.53%, achieving improvements of 14.22% and 0.34% over EfficientNetV2 and Swin Transformer, respectively, highlighting its precision-separation capability for pathological features amidst cluttered backgrounds.
- 3.
- Ablation studies have confirmed the effectiveness of LPU in maintaining translation invariance and capturing subtle disease features. LMHSA reduces the computational overhead by 46.8% compared to the standard self-attention mechanism. Furthermore, the phased design strategy enhances hierarchical features without sacrificing model scalability. The findings demonstrate that ConvTransNet-S, by harmonizing local-global feature interactions and balancing accuracy with computational efficiency, provides a robust, efficient, and scalable solution for plant disease recognition in practical agricultural scenarios.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, J.; Wang, X. Plant diseases and pests detection based on deep learning: A review. Plant Methods 2021, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Srinidhi, V.V.; Sahay, A.; Deeba, K. Plant pathology disease detection in apple leaves using deep convolutional neural networks: Apple leaves disease detection using EfficientNet and DenseNet. In Proceedings of the 2021 5th International Conference on Computing Methodologies and Communication (ICCMC), Erode, India, 8–10 April 2021. [Google Scholar]
- Zhou, J.; Li, J.; Wang, C.; Wu, H.; Zhao, C.; Wang, Q. A Vegetable Disease Recognition Model for Complex Background Based on Region Proposal and Progressive Learning. Comput. Electron. Agric. 2021, 184, 106101. [Google Scholar] [CrossRef]
- Rashid, J.; Khan, I.; Ali, G.; Almotiri, S.H.; AlGhamdi, M.A.; Masood, K. Multilevel deep learning model for potato leaf disease recognition. Electronics 2021, 10, 2064. [Google Scholar] [CrossRef]
- Jiang, Z.; Dong, Z.; Jiang, W. Recognition of rice leaf diseases and wheat leaf diseases based on multitask deep transfer learning. Comput. Electron. Agric. 2021, 186, 106184. [Google Scholar] [CrossRef]
- Kumar, A.; Vani, M. Imagebased tomato leaf disease detection. In Proceedings of the 2019 10th International Conference on Computing Communication and Networking Technologies (ICCCNT), Kanpur, India, 6–8 July 2019. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.M.; Zhang, X.Y.; Ren, S.Q.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Picon, A.; Alvarez-Gila, A.; Seitz, M. Deep convolutional neural networks for mobile capture device-based crop disease classification in the wild. Comput. Electron. Agric. 2019, 161, 280–290. [Google Scholar] [CrossRef]
- Jiang, H.H.; Yang, X.H.; Ding, R.R. Identification of apple leaf diseases based on improved ResNet18. Trans. Chin. Soc. Agric. Mach. 2023, 54, 295–303. (In Chinese) [Google Scholar]
- Zhao, S.; Liu, J. Crop pest recognition in real agricultural environment using convolutional neural networks by a parallel attention mechanism. Front. Plant Sci. 2022, 13, 839572. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Rashid, R.; Aslam, W.; Aziz, R.; Aldehim, G. A Modified MobileNetv3 Coupled with Inverted Residual and Channel Attention Mechanisms for Detection of Tomato Leaf Diseases. IEEE Access 2025, 13, 52683–52696. [Google Scholar] [CrossRef]
- Yao, J.; Tran, S.N.; Garg, S. Deep Learning for Plant Identification and Disease Classification from Leaf Images: Multi-prediction Approaches. ACM Comput. Surv. 2024, 56, 153. [Google Scholar] [CrossRef]
- Sun, J.; Cao, W.; Fu, X. Few-shot learning for plant disease recognition: A review. Agron. J. 2024, 116, 1204–1216. [Google Scholar] [CrossRef]
- Mu, J.; Feng, Q.; Yang, J. Few-shot disease recognition algorithm based on supervised contrastive learning. Front. Plant Sci. 2024, 15, 1341831. [Google Scholar] [CrossRef] [PubMed]
- Sajitha, P.; Andrushia, A.D.; Anand, N. A review on machine learning and deep learning image-based plant disease classification for industrial farming systems. J. Ind. Inf. Integr. 2024, 38, 100572. [Google Scholar] [CrossRef]
- Ahmad, I.; Hamid, M.; Yousaf, S. Optimizing pretrained convolutional neural networks for tomato leaf disease detection. Complexity 2020, 2020, 8810499. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Proceedings of the Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Thai, H.-T.; Le, K.-H.; Nguyen, N.L.-T. FormerLeaf: An efficient vision Transformer for Cassava Leaf Disease detection. Comput. Electron. Agric. 2023, 204, 107518. [Google Scholar] [CrossRef]
- Zhang, Q.; Sun, B.; Cheng, Y. Residual Self-Calibration and Self-Attention Aggregation Network for Crop Disease Recognition. Int. J. Environ. Res. Public Health 2021, 18, 8700. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Xie, L.; Huang, Q. Inception convolutional vision Transformers for plant disease identification. Internet Things 2023, 21, 100650. [Google Scholar] [CrossRef]
- Li, G.; Wang, Y.; Zhao, Q. PMVT: A lightweight vision Transformer for plant disease identification on mobile devices. Front. Plant Sci. 2023, 14, 1256773. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Li, M. Crop leaf disease recognition based on Self-Attention convolutional neural network. Comput. Electron. Agric. 2020, 172, 105341. [Google Scholar] [CrossRef]
- Thakur, P.S.; Chaturvedi, S.; Khanna, P. Vision Transformer meets convolutional neural network for plant disease classification. Ecol. Inform. 2023, 77, 102245. [Google Scholar] [CrossRef]
- Lee, C.P.; Lim, K.M.; Song, Y. Plant-CNN-ViT: Plant Classification with Ensemble of Convolutional Neural Networks and Vision Transformer. Plants 2023, 12, 2642. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jiang, W.; Zeng, J. FOTCA: Hybrid Transformer-CNN architecture using AFNO for accurate plant leaf disease image recognition. Front. Plant Sci. 2023, 14, 1231903. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, J.; Naithani, S.; Singh, U.; Raj, H. Plant disease detection using convolutional neural network. In Proceedings of the 2023 International Conference on Smart Devices (ICSD), Dehradun, India, 2–3 December 2023; Available online: https://ieeexplore.ieee.org/document/10751329 (accessed on 3 May 2024).
- Guo, Y.; Lan, Y.; Chen, X. CST: Convolutional Swin Transformer for detecting the degree and types of plant diseases. Comput. Electron. Agric. 2022, 202, 107407. [Google Scholar] [CrossRef]
- Guo, J.; Han, K.; Wu, H.; Tang, Y.; Chen, X.; Wang, Y.; Xu, C. CMT: Convolutional Neural Networks Meet Vision Transformers. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022. [Google Scholar]
- Lespinats, S.; Colange, B.; Dutykh, D. Nonlinear Dimensionality Reduction Techniques: A Data Structure Preservation Approach; Springer International Publishing: Cham, Switzerland, 2022. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A. An image is worth 16x16 words: Transformers for image recognition at scale. In Proceedings of the 9th International Conference on Learning Representations (ICLR), Virtual Event, 3–7 May 2021. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y. Swin Transformer: Hierarchical vision Transformer using shifted windows. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 11–17 October 2021. [Google Scholar]
- Peng, Z.L.; Huang, W.; Gu, S.Z. Conformer: Local features coupling global representations for visual recognition. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 11–17 October 2021. [Google Scholar]
- Chen, C.F.; Fan, Q.F.; Panda, R. CrossViT: Cross-attention multi-scale vision Transformer for image classification. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 11–17 October 2021. [Google Scholar]
- Chen, Y.; Dai, X.; Chen, D. Mobile-Former: Bridging MobileNet and Transformer. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022. [Google Scholar]
- Singh, A.K.; Rao, A.; Chattopadhyay, P.; Maurya, R.; Singh, L. Effective plant disease diagnosis using Vision Transformer trained with leafy-generative adversarial network-generated images. Expert Syst. Appl. 2024, 254, 124387. [Google Scholar] [CrossRef]
- Rezaei, M.; Diepeveen, D.; Laga, H. Plant disease recognition in a low data scenario using few-shot learning. Comput. Electron. Agric. 2024, 219, 108812. [Google Scholar] [CrossRef]
- Zhang, Y.B.; Zhu, Y.; Liu, J.L.; Yu, W.; Jiang, C. An interpretability optimization method for deep learning networks based on Grad-CAM. IEEE Internet Things J. 2024, 21, 3961–3970. [Google Scholar] [CrossRef]


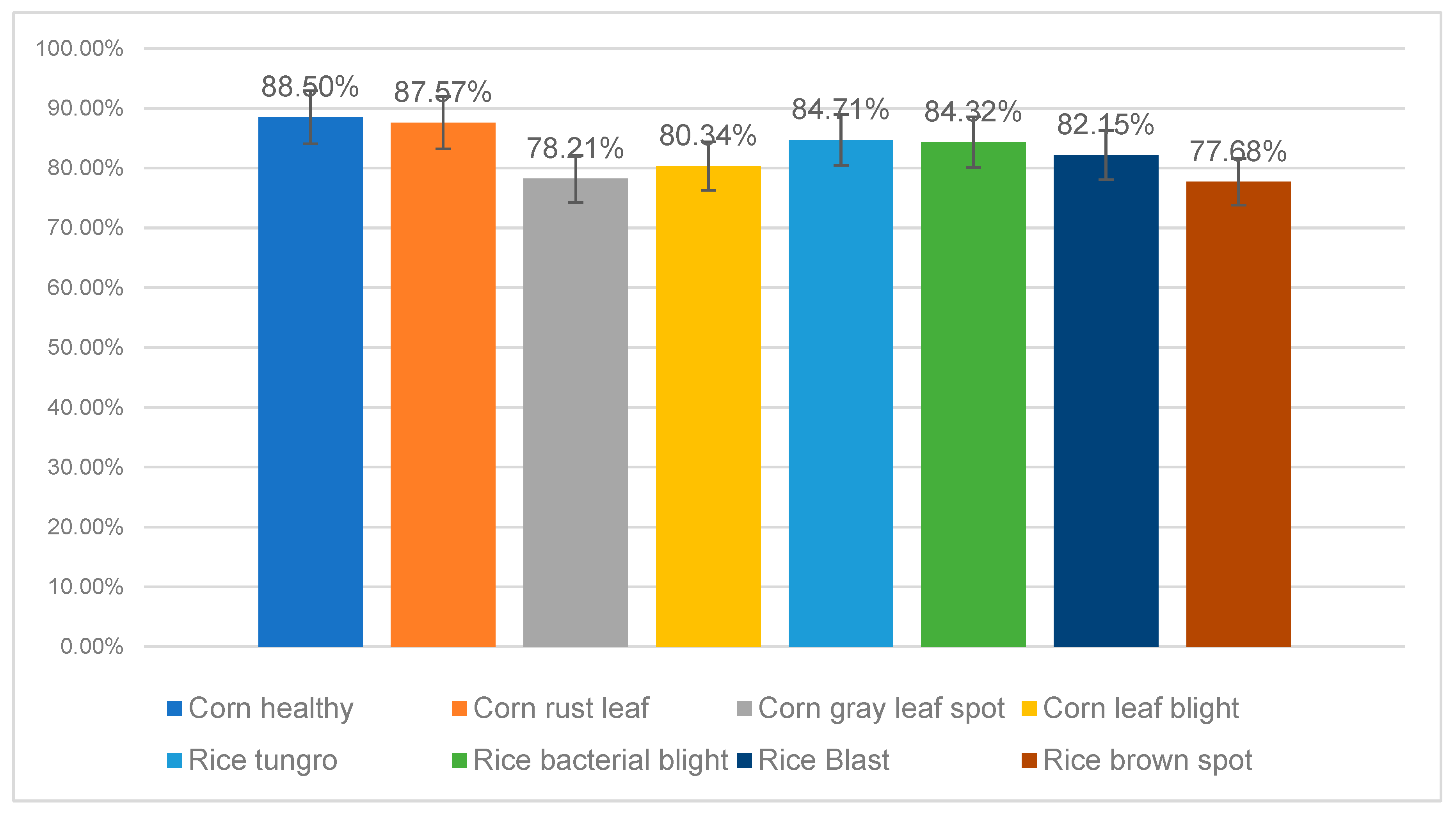

| Type | Label Name | Number of Sample Images | Type | Label Name | Number of Sample Images |
|---|---|---|---|---|---|
| Apple | Apple healthy | 516 | Rice | Rice bacterial blight | 1584 |
| Apple black rot | 91 | Rice blast | 1440 | ||
| Apple rust | 622 | Rice brown spot | 1595 | ||
| Apple scab | 592 | Rice tungro | 1308 | ||
| Corn | Corn gray leaf spot | 62 | Grape | Grape black rot | 9 |
| Corn healthy | 28 | Grape esca (black measles) | 11 | ||
| Corn leaf blight | 178 | Grape healthy | 20 | ||
| Corn rust leaf | 106 | Grape leaf blight (isariopsis leaf spot) | 12 | ||
| Cotton | Cotton areolate mildew | 30 | Tomato | Tomato target spot | 11 |
| Cotton bacterial blight | 452 | Tomato bacterial spot | 76 | ||
| Cotton cercospora leaf spot | 30 | ||||
| Cotton curl virus | 337 | Tomato early blight | 78 | ||
| Cotton healthy | 107 | Tomato late blight | 87 | ||
| Cotton target spot | 68 | Tomato mold leaf | 76 | ||
| Cotton verticillium wilt | 32 | Tomato mosaic virus | 49 | ||
| Wheat | Wheat healthy | 102 | Tomato healthy | 70 | |
| Wheat septoria | 97 | Tomato septoria spot | 119 | ||
| Wheat stripe rust | 196 | Tomato yellow virus | 106 | ||
| Potato | Potato healthy | 49 | Tomato spider mites Two-spotted spider mite | 11 | |
| Potato early blight | 40 | ||||
| Potato late blight | 44 |
| Model | Acc (%) | Inference Time (ms) | GFLOPs | Parameters (M) | Model Volume (MB) |
|---|---|---|---|---|---|
| EfficientNetV2 | 95.47 | 16.06 | 0.07859 | 0.121 | 29.58 |
| Transformer | 98.65 | 8.42 | 33.73 | 85.68 | 322.53 |
| Vision Transformer | 98.54 | 8.94 | 56.01 | 86.4 | 327.41 |
| Swin Transformer | 98.80 | 16.81 | 8.74 | 27.53 | 105.09 |
| CrossViT | 92.57 | 20.29 | 10.16 | 26.15 | 100.33 |
| ConFormer | 83.99 | 14.10 | 9.80 | 22.91 | 87.38 |
| MobileFormer | 96.91 | 22.59 | 329.89 | 6.46 | 24.63 |
| Model | Acc (%) | Inference Time (ms) | GFLOPs | Parameters (M) |
|---|---|---|---|---|
| ConvTransNet-B | 94.49 | 22.47 | 8.689 | 45.72 |
| ConvTransNet-T | 89.79 | 10.05 | 0.596 | 9.49 |
| ConvTransNet-XS | 99.12 | 10.94 | 1.434 | 15.24 |
| ConvTransNet-S | 98.85 | 7.56 | 3.762 | 25.14 |
| Model | Acc (%) | Inference Time (ms) |
|---|---|---|
| EfficientNetV2 | 74.31 | 16.18 |
| Vision Transformer | 85.78 | 8.17 |
| Transformer | 86.98 | 8.42 |
| Swin Transformer | 88.19 | 15.30 |
| Con Former | 86.73 | 23.50 |
| CrossVit | 86.88 | 12.49 |
| Mobile Former | 88.54 | 21.60 |
| ConvTransNet-B | 82.11 | 8.91 |
| ConvTransNet-T | 76.91 | 8.32 |
| ConvTransNet-XS | 88.06 | 8.21 |
| ConvTransNet-S | 88.53 | 8.31 |
| Model | Acc (%) | Inference Time (ms) | GFLOPs | Parameters (M) |
|---|---|---|---|---|
| ResNet50 | 86.08 | 6.19 | 0.02359 | 89.97 |
| ViT-S | 85.33 | 8.56 | 3.52 | 22.02 |
| ConvTransNet-S | 88.53 | 8.31 | 3.762 | 25.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, S.; Wang, G.; Li, H.; Liu, Y.; Shi, L.; Yang, S. ConvTransNet-S: A CNN-Transformer Hybrid Disease Recognition Model for Complex Field Environments. Plants 2025, 14, 2252. https://doi.org/10.3390/plants14152252
Jia S, Wang G, Li H, Liu Y, Shi L, Yang S. ConvTransNet-S: A CNN-Transformer Hybrid Disease Recognition Model for Complex Field Environments. Plants. 2025; 14(15):2252. https://doi.org/10.3390/plants14152252
Chicago/Turabian StyleJia, Shangyun, Guanping Wang, Hongling Li, Yan Liu, Linrong Shi, and Sen Yang. 2025. "ConvTransNet-S: A CNN-Transformer Hybrid Disease Recognition Model for Complex Field Environments" Plants 14, no. 15: 2252. https://doi.org/10.3390/plants14152252
APA StyleJia, S., Wang, G., Li, H., Liu, Y., Shi, L., & Yang, S. (2025). ConvTransNet-S: A CNN-Transformer Hybrid Disease Recognition Model for Complex Field Environments. Plants, 14(15), 2252. https://doi.org/10.3390/plants14152252







