Major Genes for Powdery Mildew Resistance in Research and Breeding of Barley: A Few Brief Narratives and Recommendations
Abstract
1. Introduction
2. Major Genes and Crop Resistance
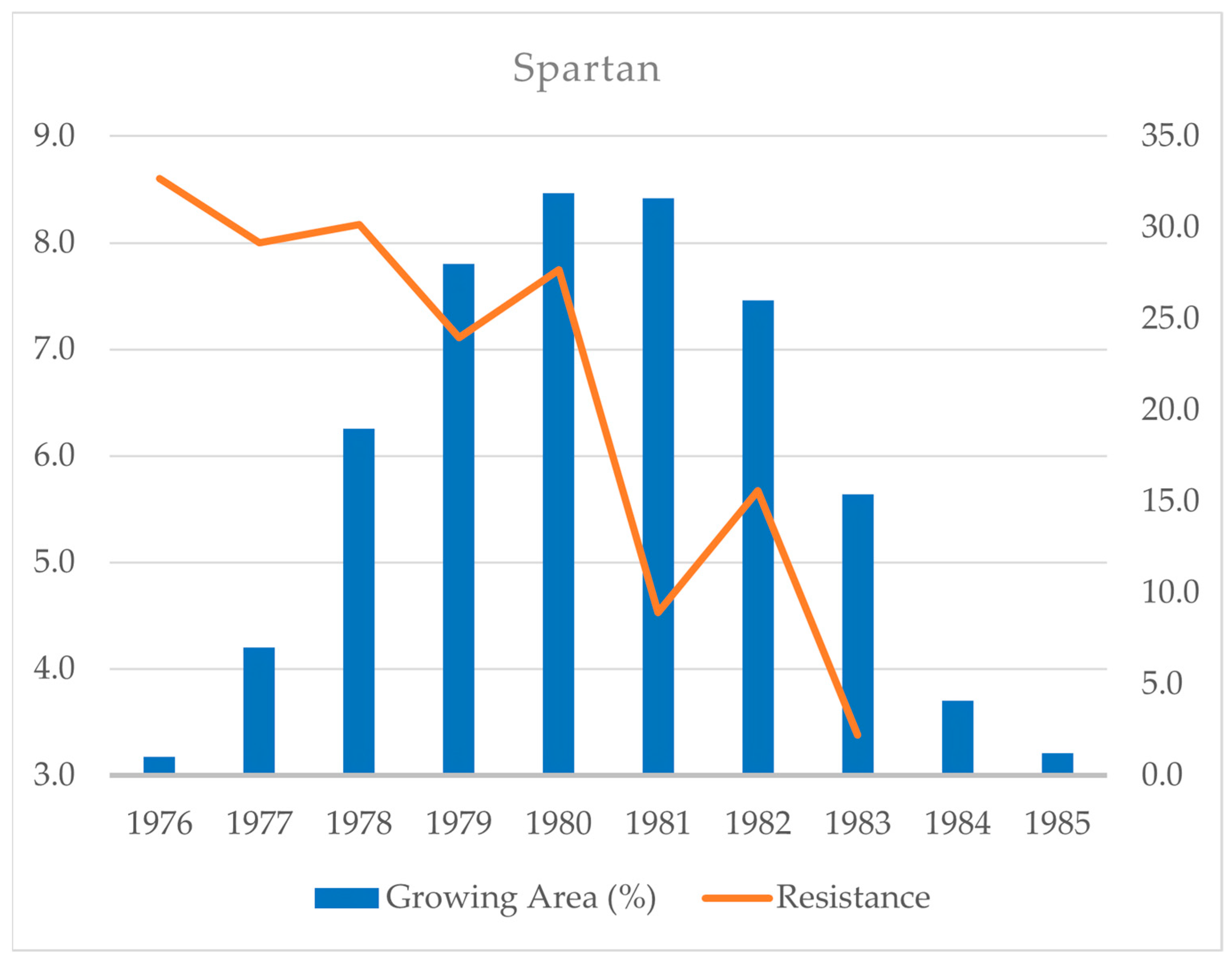
2.1. Non-Durable Major Resistance Genes
2.2. Durable Major Resistance Genes
2.3. Pathogen Variation
2.4. Postulation of Major Resistance Genes
2.5. Other Uses of Major Resistance Genes
3. Conclusions
4. Future Directions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bourras, S.; Praz, C.R.; Spanu, P.D.; Keller, B. Cereal powdery mildew effectors: A complex toolbox for an obligate pathogen. Curr. Opin. Microbiol. 2018, 46, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, J.H. Erysiphe graminis, powdery mildew of cereals and grasses. In Advances in Plant Pathology; Academic Press: London, UK, 1988; Volume 6, pp. 137–157. [Google Scholar]
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Ann. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef] [PubMed]
- Kusch, S.; Qian, J.Z.; Loos, A.; Kümmel, F.; Spanu, P.D.; Panstruga, R. Long-term and rapid evolution in powdery mildew fungi. Mol. Ecol. 2024, 33, 10. [Google Scholar] [CrossRef]
- Brückner, F. Powdery mildew (Erysiphe graminis DC.) on barley. Survey of the occurrence of physiological races on the territory of Czechoslovakia in 1960−1961. Rostl. Vyrob. 1963, 9, 1–8. [Google Scholar]
- Brückner, F. Powdery mildew (Erysiphe graminis DC.) on barley. V. The resistance of barley varieties to physiological races detected in Czechoslovakia and the possibility to use it in breeding for resistance. Rostl. Vyrob. 1964, 10, 395–408. [Google Scholar]
- Brückner, F. A complementary effect of different alleles for mildew resistance in barley. Z. Pflanzenzücht. 1967, 58, 122–127. [Google Scholar]
- Brückner, F. The breeding of the malting barley cultivar of new morphotype Forum. Genet. Šlecht. 1993, 29, 199–203. [Google Scholar]
- Dreiseitl, A. Analysis of growing Czechoslovak spring barley varieties resistant to powdery mildew. Rostl. Vyrob. 1993, 39, 337–344. [Google Scholar]
- Dreiseitl, A. Powdery mildew resistance in winter barley cultivars. Plant Breed. 2007, 126, 268–273. [Google Scholar] [CrossRef]
- Dreiseitl, A. A novel way to identify specific powdery mildew resistance genes in hybrid barley cultivars. Sci. Rep. 2020, 10, 18930. [Google Scholar] [CrossRef]
- Dreiseitl, A. Rare virulences and great pathotype diversity of a Central European Blumeria hordei population. J. Fungi 2023, 9, 1045. [Google Scholar] [CrossRef] [PubMed]
- Dreiseitl, A.; Jørgensen, J.H. Powdery mildew resistance in Czech and Slovak barley cultivars. Plant Breed. 2000, 119, 203–209. [Google Scholar] [CrossRef]
- Nover, I.; Brückner, F.; Wiberg, A.; Wolfe, M.S. Races of Erysiphe graminis DC. f. sp. hordei Marchal in Europe. Z. Pfl.-Krank. Pfl. Schutz 1968, 75, 350–353. [Google Scholar]
- Brown, J.K.M.; Hovmoller, M.S. Epidemiology—Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 2002, 297, 537–541. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, R.B.; Brown, J.K.M. Movement of barley powdery mildew within field plots. Plant Pathol. 1998, 47, 394–400. [Google Scholar] [CrossRef]
- Dreiseitl, A. Adaptation of Blumeria graminis f.sp. hordei to barley resistance genes in the Czech Republic in 1971–2000. Plant Soil Environ. 2003, 49, 241–248. [Google Scholar] [CrossRef]
- Jørgensen, J.H. Genetics of powdery mildew resistance in barley. Crit. Rev. Plant Sci. 1994, 13, 97–119. [Google Scholar] [CrossRef]
- Dreiseitl, A. Specific resistance of barley to powdery mildew, its use and beyond: A concise critical review. Genes 2020, 11, 971. [Google Scholar] [CrossRef]
- Dreiseitl, A. Heterogeneity of powdery mildew resistance revealed in accessions of the ICARDA wild barley collection. Front. Plant Sci. 2017, 8, 202. [Google Scholar] [CrossRef]
- Fischbeck, G.; Schwarzbach, E.; Sobel, Z.; Wahl, I. Mildew resistance in Israeli populations of 2-rowed wild barley (Hordeum spontaneum). Z. Pflanzenzücht. 1976, 76, 163–166. [Google Scholar]
- Moseman, J.G.; Baenzinger, P.S.; Kilpatrick, R.A. Genes conditioning resistance of Hordeum spontaneum to Erysiphe graminis f. sp. hordei. Crop Sci. 1981, 21, 229–232. [Google Scholar] [CrossRef]
- Flor, H.H. Current status of the gene-for-gene concept. Ann. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Dreiseitl, A. Mlo-mediated broad-spectrum and durable resistance against powdery mildews and its current and future applications. Plants 2024, 13, 138. [Google Scholar] [CrossRef]
- Xu, Y.H.; Jia, Q.J.; Zhou, G.F.; Zhang, X.Q.; Angessa, T.; Broughton, S.; Yan, G.; Zhang, W.Y.; Li, C.D. Characterization of the sdw1 semi-dwarf gene in barley. BMC Plant Biol. 2017, 17, 11. [Google Scholar] [CrossRef]
- Dreiseitl, A. Differences in powdery mildew epidemics in spring and winter barley based on 30-year variety trials. Ann. Appl. Biol. 2011, 159, 49–57. [Google Scholar] [CrossRef]
- Brown, J.K.M.; Jørgensen, J.H. A catalogue of mildew resistance genes in European barley varieties. In Integrated Control of Cereal Mildews: Virulence and Their Change, Proceedings of the Second European Workshop on Integrated Control of Cereal Mildews, Risø, Denmark, 23–25 January 1990; Jørgensen, J.H., Ed.; Risø National Laboratory: Roskilde, Denmark, 1991; pp. 263–286. [Google Scholar]
- Dreiseitl, A. Genes for resistance to powdery mildew in European barley cultivars registered in the Czech Republic from 2011 to 2015. Plant Breed. 2017, 136, 351–356. [Google Scholar] [CrossRef]
- Wolfe, M.S.; Brändle, U.; Koller, B.; Limpert, E.; McDermott, J.M.; Müller, K.; Schaffner, D. Barley mildew in Europe: Population biology and host resistance. Euphytica 1992, 63, 125–139. [Google Scholar] [CrossRef]
- Abebe, T.D.; Abate, A.; Leon, J. Genetic diversity within landraces of barley (Hordeum vulgare L.) and its implications on germplasm collection and utilization. Genet. Resour. Crop Evol. 2023, 70, 1985–1998. [Google Scholar] [CrossRef]
- Czembor, J.H. Resistance to powdery mildew in selections from Moroccan barley landraces. Euphytica 2002, 125, 397–409. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, H.J. Selections from barley landrace collected in Libya as new sources of effective resistance to powdery mildew (Blumeria graminis f.sp. hordei). Rostl. Vyrob. 2002, 48, 217–223. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, E. Sources of resistance to powdery mildew in wild barley (Hordeum vulgare subsp. spontaneum) collected in Jordan, Lebanon, and Libya. Agronomy 2023, 13, 2462. [Google Scholar] [CrossRef]
- Czembor, J.H.; Johnston, M.R. Resistance to powdery mildew in selections from Tunisian landraces of barley. Plant Breed. 1999, 118, 503–509. [Google Scholar] [CrossRef]
- Piechota, U.; Czembor, P.C.; Slowacki, P.; Czembor, J.H. Identifying a novel powdery mildew resistance gene in a barley landrace from Morocco. J. Appl. Genet. 2019, 60, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Freisleben, R.; Lein, A. Über die Auffindung einer mehltauresistenten Mutante nach Röentgenbestrahlung einer anfälligen reinen Linie von Sommergerste. Naturwissenschaften 1942, 30, 608. [Google Scholar] [CrossRef]
- Jørgensen, J.H. Discovery, characterisation and exploitation of Mlo powdery mildew resistance in barley. Euphytica 1992, 63, 141–152. [Google Scholar] [CrossRef]
- Favret, E.A. Different categories of mutations for disease reaction in the host organism. In Mutation Breeding for Disease Resistance; International Atomic Energy Agency: Vienna, Austria, 1971; pp. 107–116. [Google Scholar]
- Schwarzbach, E. Recessive total resistance of barley to mildew (Erysiphe graminis D.C. f. sp. hordei Marchal) as a mutation induced by ethylmethansulfonate. Genet. Šlechteni. 1967, 3, 159–162. [Google Scholar]
- Kusch, S.; Frantzeskakis, L.; Lassen, B.D.; Kümmel, F.; Pesch, L.; Barsoum, M.; Walden, K.D.; Panstruga, R. A fungal plant pathogen overcomes mlo-mediated broad-spectrum disease resistance by rapid gene loss. New Phytol. 2024, 244, 262–279. [Google Scholar] [CrossRef]
- Schwarzbach, E. Response to selection for virulence against the ml-o based mildew resistance in barley, not fitting the gene-for-gene hypothesis. Barley Gen. Newsl. 1979, 9, 85–88. [Google Scholar]
- Schwarzbach, E. Heat induced susceptibility of mlo-barley to powdery mildew. (Blumeria graminis D.C. f.sp. hordei Marchal). Czech J. Genet. Plant Breed. 2001, 37, 82–87. [Google Scholar]
- Yaeno, T.; Wahara, M.; Nagano, M.; Wanezaki, H.; Toda, H.; Inoue, H.; Eishima, A.; Nishiguchi, M.; Hisano, H.; Kobayashi, K. RACE1, a Japanese Blumeria graminis f. sp. hordei isolate, is capable of overcoming partially mlo-mediated penetration resistance in barley in an allele-specific manner. PLoS ONE 2021, 16, e0256574. [Google Scholar] [CrossRef]
- Kusch, S.; Panstruga, R. mlo-based resistance: An apparently universal “weapon” to defeat powdery mildew disease. Mol. Plant-Microbe Interact. 2017, 30, 179–189. [Google Scholar] [CrossRef]
- Bui, T.P.; Le, H.; Ta, D.T.; Nguyen, C.X.; Le, N.T.; Tran, T.T.; Nguyen, P.V.; Stacey, G.; Stacey, M.G.; Pham, N.B. Enhancing powdery mildew resistance in soybean by targeted mutation of MLO genes using the CRISPR/Cas9 system. BMC Plant Biol. 2023, 23, 533. [Google Scholar] [CrossRef]
- Giuseppe, A.; Raffaella, E.M. The first genome-wide mildew locus O genes characterization in the Lamiaceae plant family. Int. J. Molec. Sci. 2023, 24, 13627. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.B.; Wang, Y.Q.; Zhang, X.Y.; Zhang, C.H. Cloning and expression analysis of mildew locus O (MLO) genes related to powdery mildew resistance in Vitis pseudoreticulata, and functional characterisation of VpMLO13. New Zealand J. Crop Hortic. Sci. 2023. Early Access. [Google Scholar] [CrossRef]
- Liu, J.; Wu, Y.P.; Zhang, X.; Gill, R.A.; Hu, M.; Bai, Z.T.; Zhao, C.J.; Zhang, Y.; Liu, Y.Y.; Hu, Q. Functional and evolutionary study of MLO gene family in the regulation of Sclerotinia stem rot resistance in Brassica napus L. Biotechnol. Biofuels Bioprod. 2023, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Deng, Y.; Ning, Y.; He, Z.; Wang, G.L. Exploiting broad-spectrum disease resistance in crops: From molecular dissection to breeding. Ann. Rev. Plant Biol. 2020, 71, 575–603. [Google Scholar] [CrossRef] [PubMed]
- Sirangelo, T.M. NLR- and mlo-based resistance mechanisms against powdery mildew in Cannabis sativa. Plants 2024, 13, 105. [Google Scholar] [CrossRef]
- Tek, M.I.; Calis, O.; Fidan, H.; Shah, M.D.; Celik, S.; Wani, S.H. CRISPR/Cas9 based mlo-mediated resistance against Podosphaera xanthii in cucumber (Cucumis sativus L.). Front. Plant Sci. 2022, 13, 1081506. [Google Scholar] [CrossRef]
- Traore, S.M.; Han, S.; Binagwa, P.; Xu, W.; Chen, X.G.; Liu, F.Z.; He, G.H. Genome-wide identification of mlo genes in the cultivated peanut (Arachis hypogaea L.). Euphytica 2021, 217, 61. [Google Scholar] [CrossRef]
- Wang, J.; Wu, X.H.; Wang, Y.; Wu, X.Y.; Wang, B.G.; Lu, Z.F.; Li, G.J. Genome-wide characterization and expression analysis of the MLO gene family sheds light on powdery mildew resistance in Lagenaria siceraria. Heliyon 2023, 9, e14624. [Google Scholar] [CrossRef]
- Xu, J.P.; Naing, A.H.; Kang, H.H.; Lee, S.Y.; Li, W.L.; Chung, M.Y.; Kim, C.K. CRISPR/Cas9-mediated editing of PhMLO1 confers powdery mildew resistance in petunia. Plant Biotech. Rep. 2023, 17, 767–775. [Google Scholar] [CrossRef]
- Hiura, U.; Heta, H. Studies on the disease resistance in barley. III. Further studies on the physiologic races of Erysiphe graminis hordei in Japan. Berichte Ohara Inst. landwirtschaft. Biol. 1955, 10, 135–156. [Google Scholar]
- Jørgensen, J.H.; Jensen, H.P. Powdery mildew resistance gene Ml-a8 (Reg1h8) in northwest European spring barley varieties. Barley Genet. Newsl. 1983, 13, 51–52. [Google Scholar]
- Czembor, J.H.; Czembor, E. Sources of resistance to powdery mildew in barley landraces from Turkey. Agriculture 2021, 11, 1017. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, H.J. Powdery mildew resistance in selections from Moroccan barley landraces. Phytoparasitica 2000, 28, 65–78. [Google Scholar] [CrossRef]
- Jensen, H.P.; Christensen, E.; Jørgensen, J.H. Powdery mildew resistance genes in 127 Northwest European spring barley varieties. Plant Breed. 1992, 108, 210–228. [Google Scholar] [CrossRef]
- Moseman, J.G. Genes for specific resistance-powdery mildew of barley. Phytopathology 1971, 61, 617–620. [Google Scholar] [CrossRef]
- Dreiseitl, A. Resistance of ‘Roxana’ to powdery mildew and its presence in some European spring barley cultivars. Plant Breed. 2011, 130, 419–422. [Google Scholar] [CrossRef]
- McVey, D.V.; Roelfs, A.P. Postulation of genes for stem rust resistance in the entries of the Fourth international winter wheat performance nursery. Crop Sci. 1975, 15, 335–337. [Google Scholar] [CrossRef]
- Dreiseitl, A.; Steffenson, B.J. Postulation of leaf rust resistance genes in Czech and Slovak barley cultivars and breeding lines. Plant Breed. 2000, 119, 211–214. [Google Scholar] [CrossRef]
- Kaur, H.; Kaur, J.; Bala, R.; Sharma, A.; Kumari, J.; Mavi, G.S.; Kaur, S. Postulation of leaf rust resistance genes in Indian and exotic wheat germplasm using near-isogenic lines (NILs) and molecular markers. Crop Prot. 2023, 174, 106431. [Google Scholar] [CrossRef]
- Mebrate, S.A.; Dehne, H.W.; Pillen, K.; Oerke, E.C. Postulation of seedling leaf rust resistance genes in selected Ethiopian and German bread wheat cultivars. Crop Sci. 2008, 48, 507–516. [Google Scholar] [CrossRef]
- Randhawa, M.; Bansal, U.; Lillemo, M.; Miah, H.; Bariana, H. Postulation of rust resistance genes in Nordic spring wheat genotypes and identification of widely effective sources of resistance against the Australian rust flora. J. Appl. Genet. 2016, 57, 453–465. [Google Scholar] [CrossRef]
- Singh, D.; Park, R.F.; McIntosh, R.A. Postulation of leaf (brown) rust resistance genes in 70 wheat cultivars grown in the United Kingdom. Euphytica 2001, 120, 205–218. [Google Scholar] [CrossRef]
- Xu, X.D.; Feng, J.; Lin, R.M.; Hussain, K.; Xu, S.C.; Lin, F. Postulation of stripe rust resistance genes in 44 Chinese wheat cultivars. Int. J. Agric. Biol. 2011, 13, 665–670. [Google Scholar]
- Yang, H.L.; Diao, W.D.; Yan, X.C.; Gebrewahid, T.W.; Li, Z.F.; Yao, Z.J. Identification of genes for leaf rust resistance in seedlings of wheat cultivars from the Yellow-Huai Basin in China and slow rusting observations in field trials. Czech J. Genet. Plant Breed. 2023, 59, 219–234. [Google Scholar] [CrossRef]
- Liu, S.B.; Wang, H.G.; Zhang, X.Y.; Li, X.F.; Li, D.Y.; Duan, X.Y.; Zhou, Y.L. Molecular cytogenetic identification of a wheat-Thinopyron intermedium (Host) Barkworth & DR Dewey partial amphiploid resistant to powdery mildew. J. Integr. Plant Biol. 2005, 47, 726–733. [Google Scholar] [CrossRef]
- Hysing, S.C.; Merker, A.; Liljeroth, E.; Koebner, R.M.D.; Zeller, F.J.; Hsam, S.L.K. Powdery mildew resistance in 155 Nordic bread wheat cultivars and landraces. Hereditas 2007, 144, 102–119. [Google Scholar] [CrossRef]
- Yang, G.T.; Tong, C.Y.; Li, H.W.; Li, B.; Li, Z.S.; Zheng, Q. Cytogenetic identification and molecular marker development of a novel wheat-Thinopyrum ponticum translocation line with powdery mildew resistance. Theor. Appl. Genet. 2022, 135, 2041–2057. [Google Scholar] [CrossRef]
- Silvar, C.; Flath, K.; Kopahnke, D.; Gracia, M.P.; Lasa, J.M.; Casas, A.M.; Igartua, E.; Ordon, F. Analysis of powdery mildew resistance in the Spanish barley core collection. Plant Breed. 2011, 130, 195–202. [Google Scholar] [CrossRef]
- Surlan-Momirovic, G.; Flath, K.; Silvar, C.; Brankovic, G.; Kopahnke, D.; Knezevic, D.; Schliephake, E.; Ordon, F.; Perovic, D. Exploring the Serbian GenBank barley (Hordeum vulgare L. subsp vulgare) collection for powdery mildew resistance. Genet. Resour. Crop Evol. 2016, 63, 275–287. [Google Scholar] [CrossRef]
- Brown, J.K.M. The choice of molecular marker methods for population genetic studies of plant pathogens. New Phytol. 1996, 133, 183–195. [Google Scholar] [CrossRef]
- Czembor, P.C.; Czembor, J.H. Identification of RAPD marker for the Mlat powdery mildew resistance gene in barley. Mikol. Fitopatol. 2005, 39, 66–73. [Google Scholar]
- Hinze, K.; Thompson, R.D.; Ritter, E.; Salamini, F.; Schulze-Lefert, P. Restriction fragment length polymorphism-mediated targeting of the ML-O resistance locus in barley (Hordeum vulgare). Proc. Nat. Acad. Sci. USA 1991, 88, 3691–3695. [Google Scholar] [CrossRef]
- Schuller, C.; Backes, G.; Fischbeck, G. RFLP markers to identity the alleles on the Mla locus confering powdery mildew resistance in barley. Theor. Appl. Genet. 1992, 84, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Piechota, U.; Slowacki, P.; Czembor, P.C. Identification of a novel recessive gene for resistance to powdery mildew (Blumeria graminis f. sp. hordei) in barley (Hordeum vulgare). Plant Breed. 2020, 139, 730–742. [Google Scholar] [CrossRef]
- af Sätra, J.S.; Troggio, M.; Odilbekov, F.; Sehic, J.; Mattisson, H.; Hjalmarsson, I.; Ingvarsson, P.K.; Garkava-Gustavsson, L. Genetic status of the Swedish central collection of heirloom apple cultivars. Sci. Hortic. 2020, 272, 109599. [Google Scholar] [CrossRef]
- Girma, G.; Korie, S.; Dumet, D.; Franco, J. Improvement of accession distinctiveness as an added value to the global worth of the yam (Dioscorea ssp) genebank. Int. J. Conserv. Sci. 2012, 3, 199–206. [Google Scholar]
- Hempel, P.; Hohe, A.; Trankner, C. Molecular reconstruction of an old pedigree of diploid and triploid Hydrangea macrophylla genotypes. Front. Plant Sci. 2018, 9, 429. [Google Scholar] [CrossRef]
- Jreisat, C.S.; Laten, H.M. Ribosomal RNA internal transcribed regions identify possible misidentification or mislabeling among Trifolium (Clover) specimens from germplasm collections. Crop Sci. 2017, 57, 322–326. [Google Scholar] [CrossRef]
- Shan, F.; Clarke, H.C.; Plummer, J.A.; Yan, G.; Siddique, K.H.M. Geographical patterns of genetic variation in the world collections of wild annual Cicer characterized by amplified fragment length polymorphisms. Theor. Appl. Genet. 2005, 110, 381–391. [Google Scholar] [CrossRef] [PubMed]
- van de Wouw, M.; van Treuren, R.; van Hintum, T. Authenticity of old cultivars in genebank collections: A case study on Lettuce. Crop Sci. 2011, 51, 736–746. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, Y.Z.; Liu, J.; Xu, C.; Zou, X.H.; Chen, X.; Liu, Y.L.; Wu, P.; Yang, X.Y.; Zhou, S.L. DNA barcoding of Oryza: Conventional, specific, and super barcodes. Plant Mol. Biol. 2021, 105, 215–228. [Google Scholar] [CrossRef]
- Mascher, M.; Wicker, T.; Jenkins, J.; Plott, C.; Lux, T.; Koh, C.S.; Ens, J.; Gundlach, H.; Boston, L.B.; Tulpová, Z.; et al. Long-read sequence assembly: A technical evaluation in barley. Plant Cell 2021, 33, 1888–1906. [Google Scholar] [CrossRef] [PubMed]
- Pont, L.; Compte, I.; Sanz-Nebot, V.; Barbosa, J.; Benavente, F. Analysis of hordeins in barley grain and malt by capillary electrophoresis-mass spectrometry. Food Anal. Methods 2020, 13, 325–336. [Google Scholar] [CrossRef]
- Dreiseitl, A.; Zavřelová, M. Non-authenticity of spring barley genotypes revealed in gene bank accessions. Plants 2022, 11, 3059. [Google Scholar] [CrossRef]
- Dreiseitl, A.; Zavřelová, M. Identification of barley powdery mildew resistances in gene bank accessions and the use of gene diversity for verifying seed purity and authenticity. PLoS ONE 2018, 13, e0208719. [Google Scholar] [CrossRef]
- Nover, I.; Lehmann, C.O. Resistenzeigenschaften im Gersten- und Weizensortiment Gatersleben. 17. Prüfung von Sommergersten auf ihr Verhalten gegen Mehltau (Erysiphe graminis DC. f. sp. hordei Marchal). Kulturpflanze 1973, 21, 275–294. [Google Scholar] [CrossRef]
- Silvar, C.; Casas, A.M.; Igartua, E.; Ponce-Molina, L.J.; Gracia, M.P.; Schweizer, G.; Herz, M.; Flath, K.; Waugh, R.; Kopahnke, D.; et al. Resistance to powdery mildew in Spanish barley landraces is controlled by different sets of quantitative trait loci. Theor. Appl. Genet. 2011, 123, 1019–1028. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, E.; Suchecki, R.; Watson-Haigh, N.S. Genome-wide association study for powdery mildew and rusts adult plant resistance in European spring barley from Polish gene bank. Agronomy 2022, 12, 7. [Google Scholar] [CrossRef]
- Pickering, R.A.; Rennie, W.F.; Cromey, M.G. Disease resistant material available from the wide hybridization programme at DSIR. Barley Newsl. 1987, 31, 248–259. [Google Scholar]
- Pickering, R.A.; Hill, A.M.; Michel, M.; Timmerman-Vaughan, G.M. The transfer of a powdery mildew resistance gene from Hordeum bulbosum L. to barley (H. vulgare L.) chromosome 2 (2I). Theor. Appl. Genet. 1995, 91, 1288–1292. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Kasha, K.J. Transfer of a dominant gene for powdery mildew resistance and DNA from Hordeum bulbosum into cultivated barley (Hordeum vulgare). Theor. Appl. Genet. 1992, 84, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Hoseinzadeh, P.; Ruge-Wehling, B.; Schweizer, P.; Stein, N.; Pidon, H. High resolution mapping of a Hordeum bulbosum-derived powdery mildew resistance locus in barley using distinct homologous introgression lines. Front. Plant Sci. 2020, 11, 225. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dreiseitl, A. Major Genes for Powdery Mildew Resistance in Research and Breeding of Barley: A Few Brief Narratives and Recommendations. Plants 2025, 14, 2091. https://doi.org/10.3390/plants14142091
Dreiseitl A. Major Genes for Powdery Mildew Resistance in Research and Breeding of Barley: A Few Brief Narratives and Recommendations. Plants. 2025; 14(14):2091. https://doi.org/10.3390/plants14142091
Chicago/Turabian StyleDreiseitl, Antonín. 2025. "Major Genes for Powdery Mildew Resistance in Research and Breeding of Barley: A Few Brief Narratives and Recommendations" Plants 14, no. 14: 2091. https://doi.org/10.3390/plants14142091
APA StyleDreiseitl, A. (2025). Major Genes for Powdery Mildew Resistance in Research and Breeding of Barley: A Few Brief Narratives and Recommendations. Plants, 14(14), 2091. https://doi.org/10.3390/plants14142091









