A Non-Specific Phytohormone Regulatory Network in Saccharina japonica Coordinates Growth and Environmental Adaptation
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Metabolite Sample Extraction
2.3. UPLC Conditions and LC-MS/MS Analysis
2.4. Total RNA Extraction
2.5. RNA-Seq
2.6. Identification of Differentially Expressed Genes (DEGs)
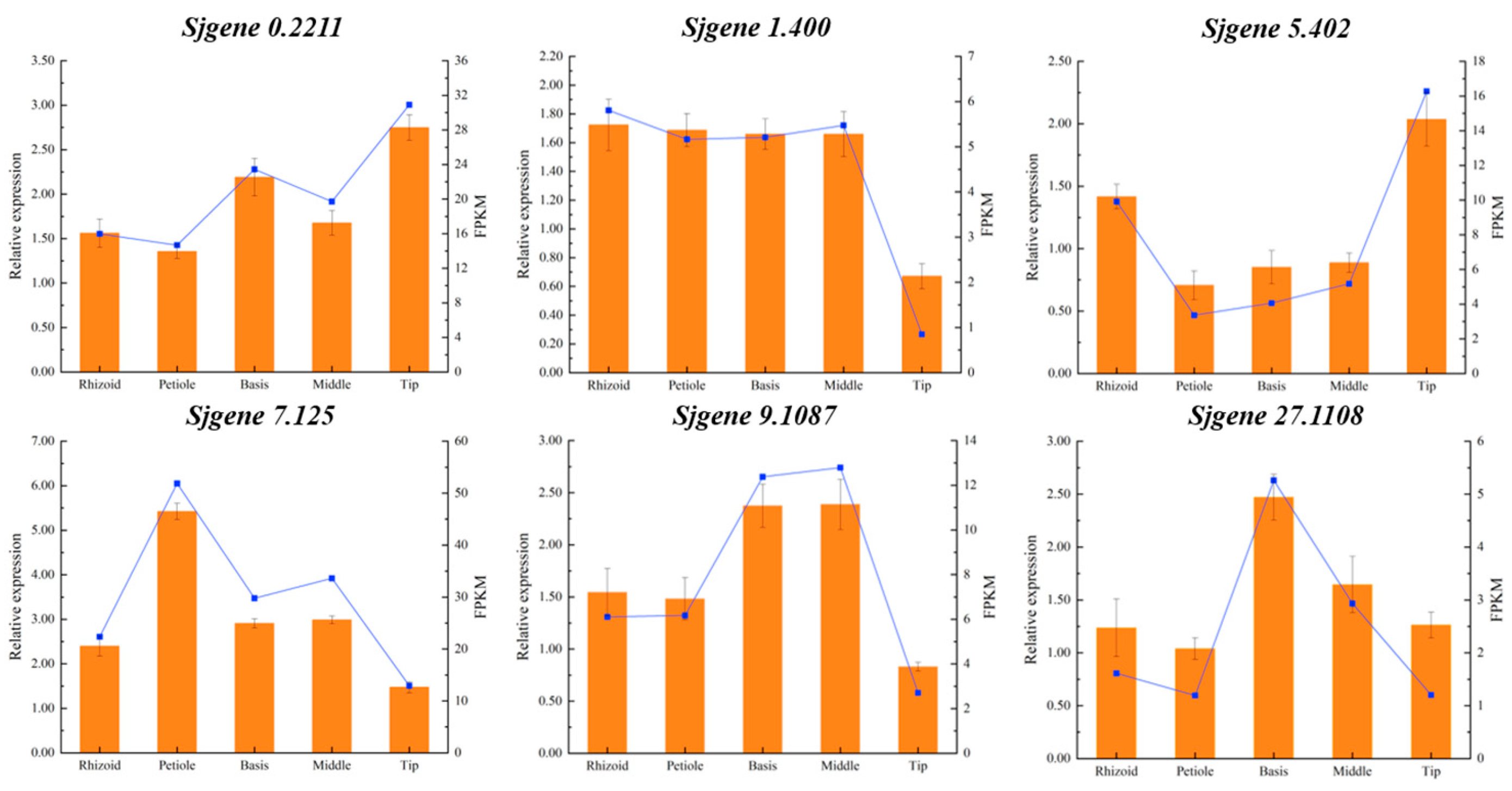
2.7. Real-Time Fluorescent Quantitative Analysis
2.8. Data Analysis
3. Results
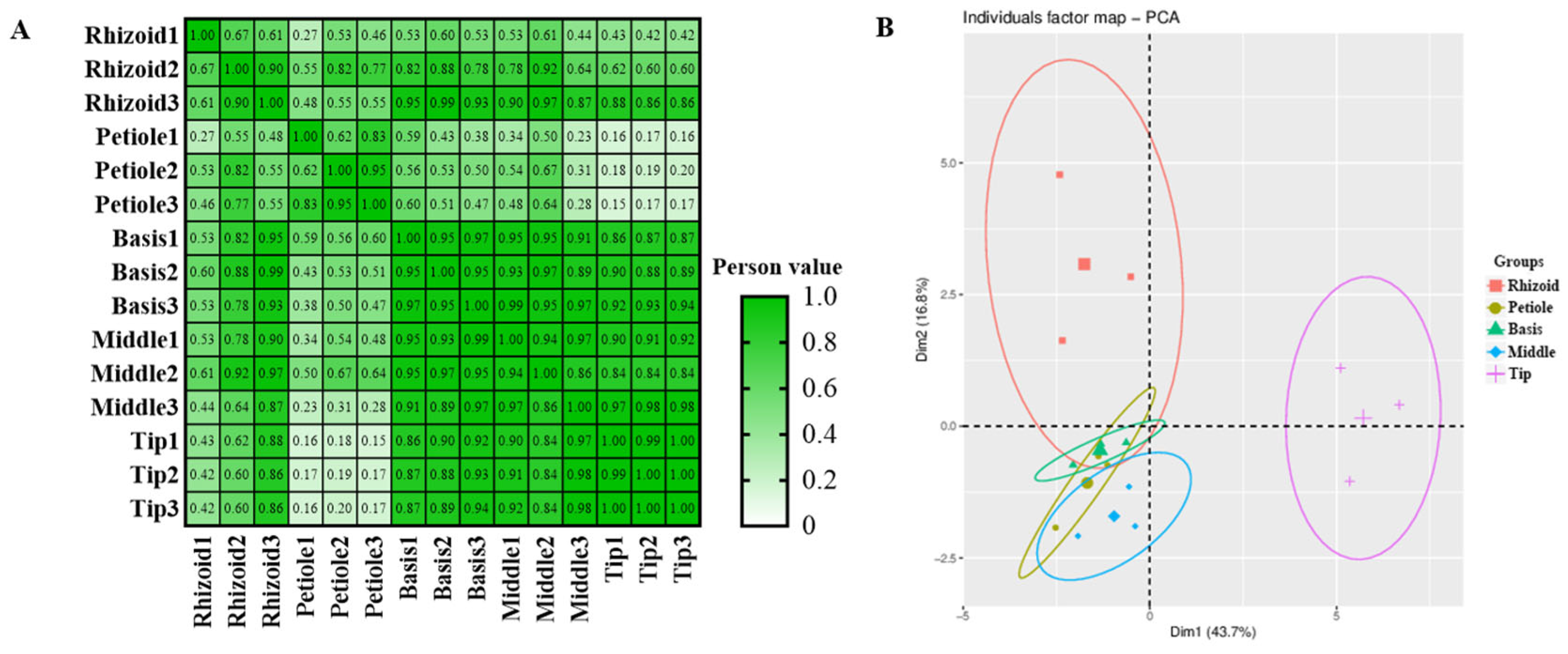
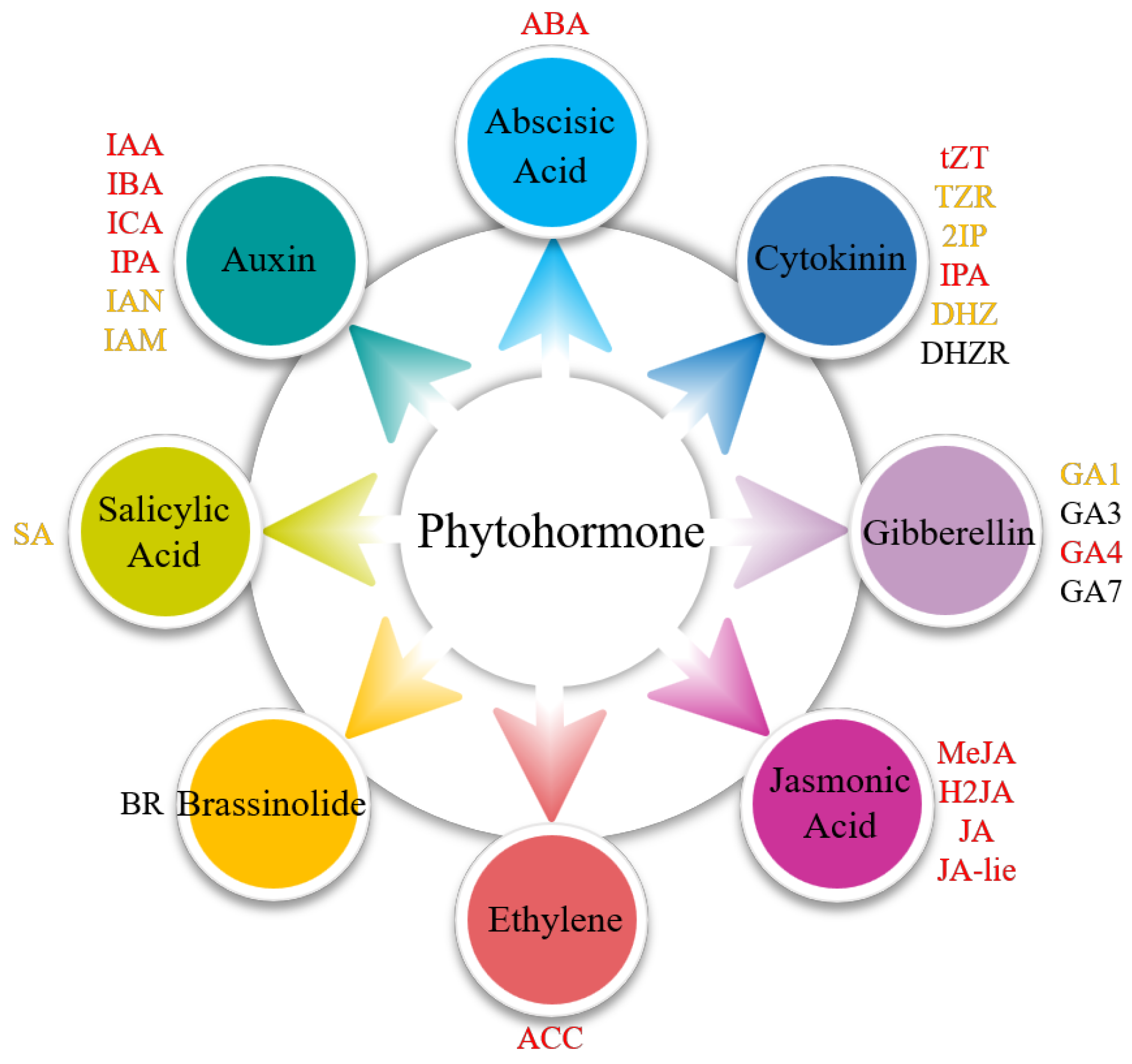
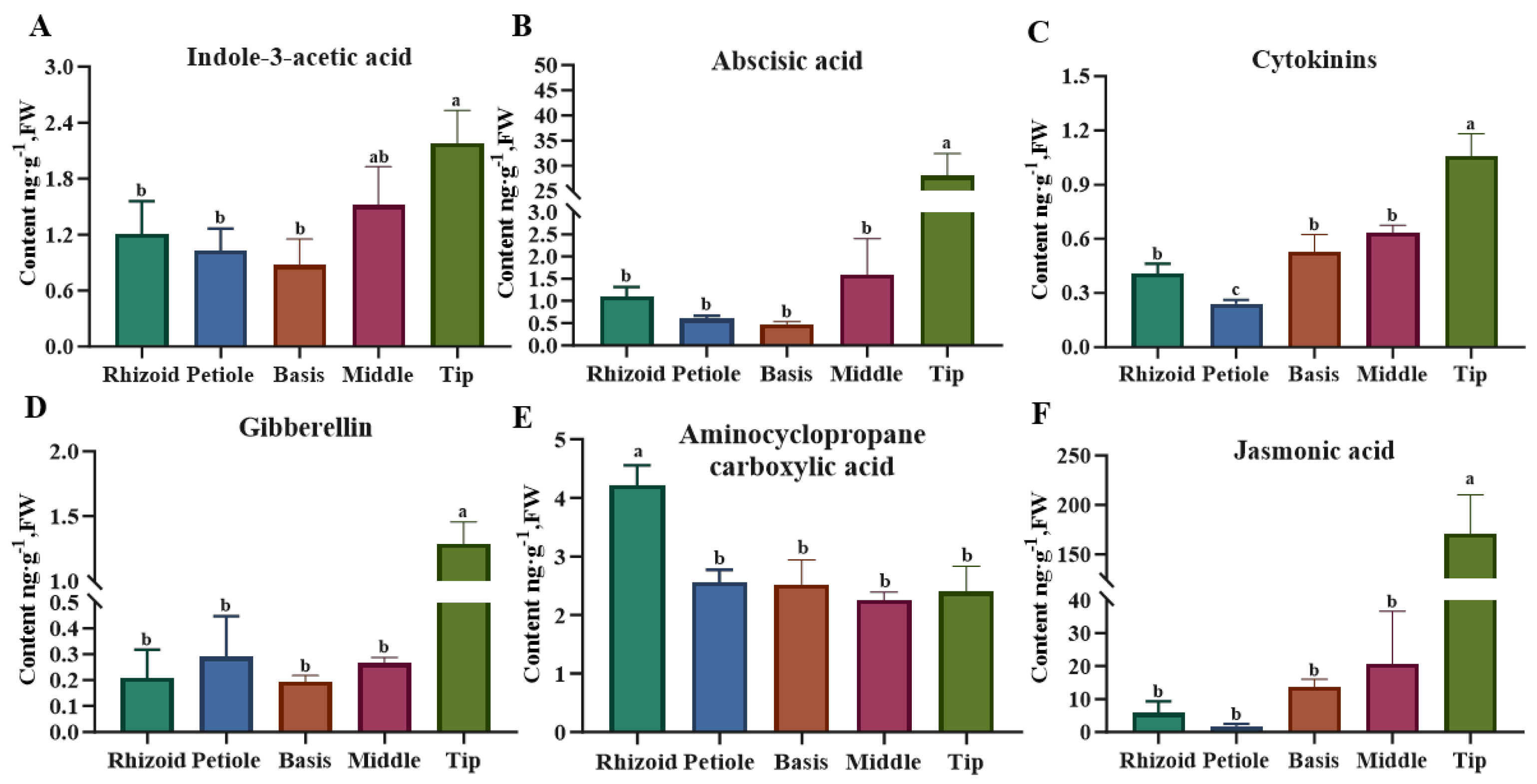
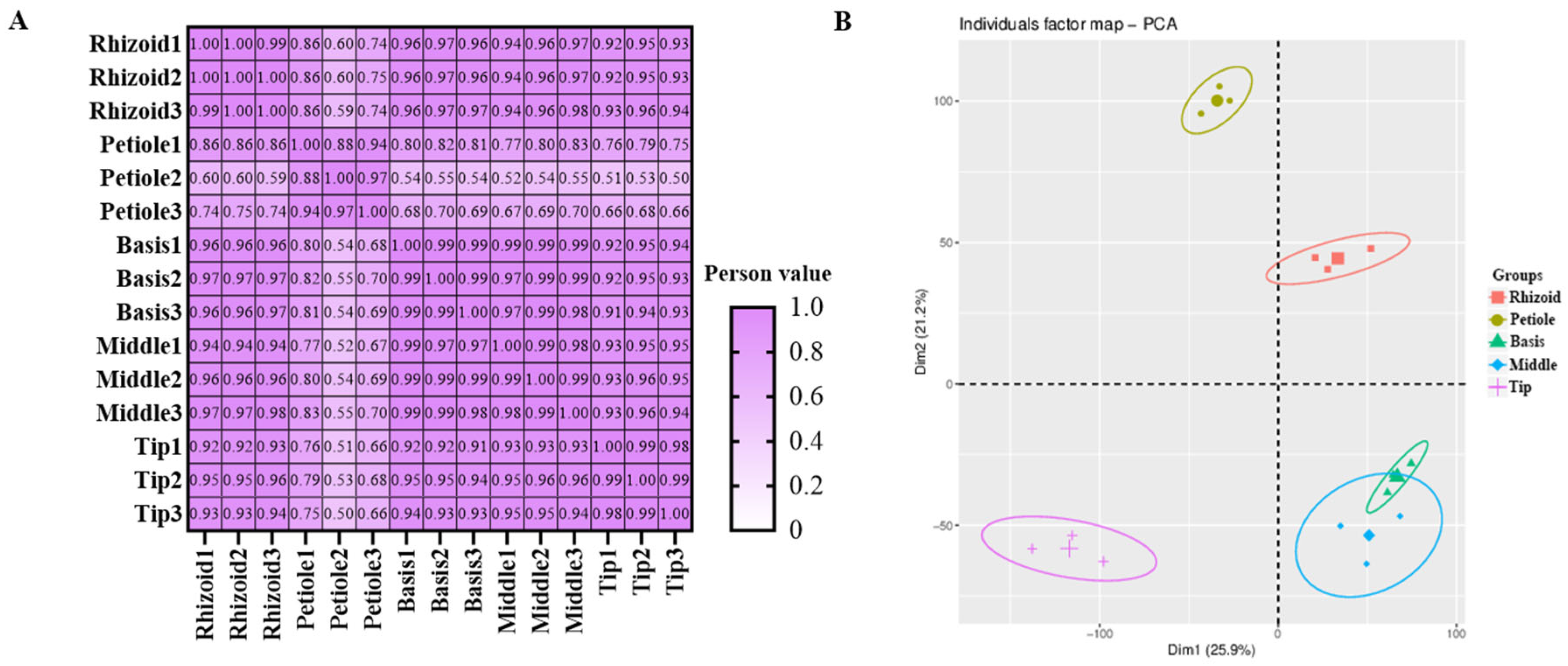
3.1. Phytohormone Content Assays in Different Parts of S. japonica
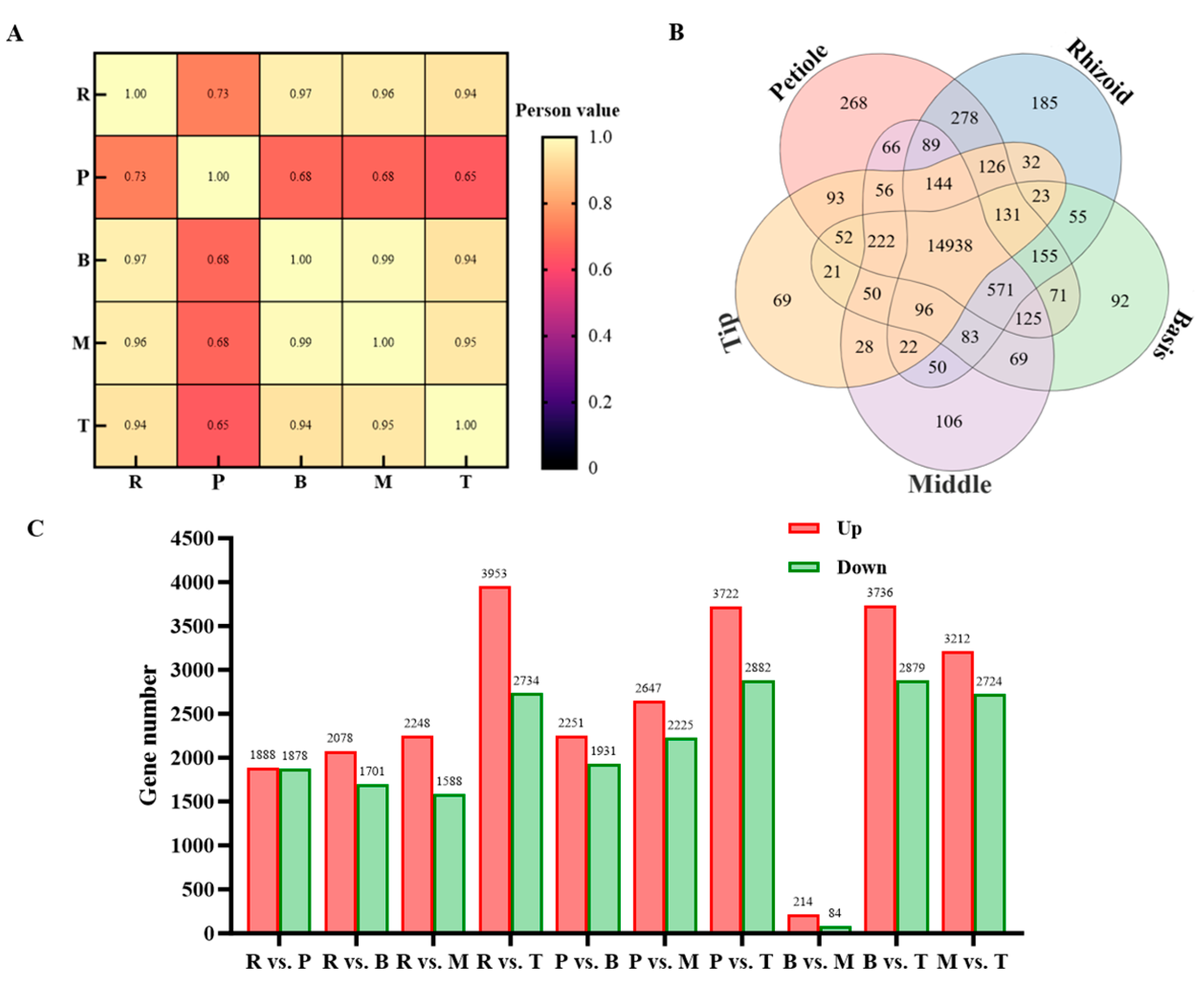
3.2. Quality Check of Transcriptome Sequencing Data
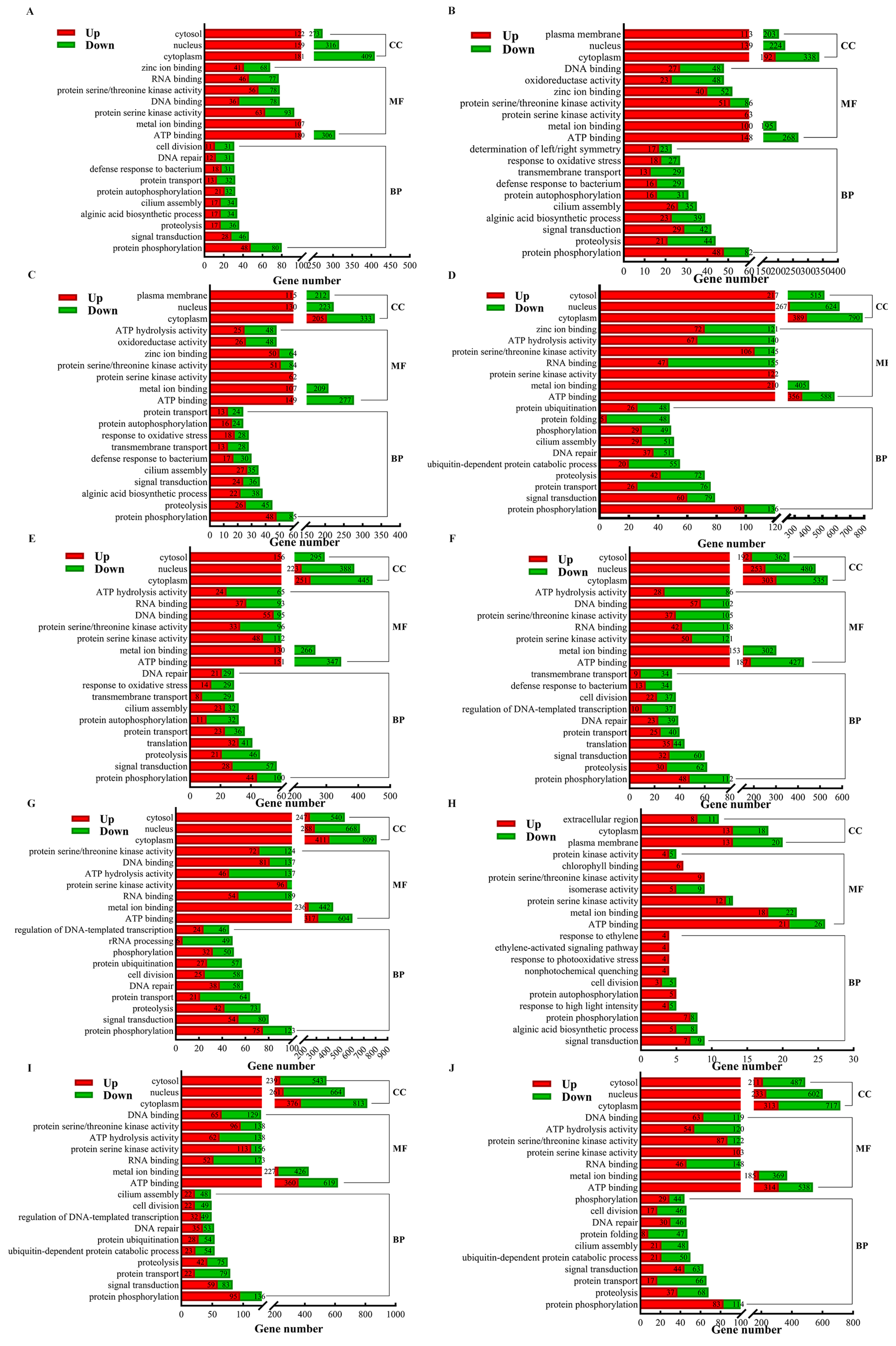
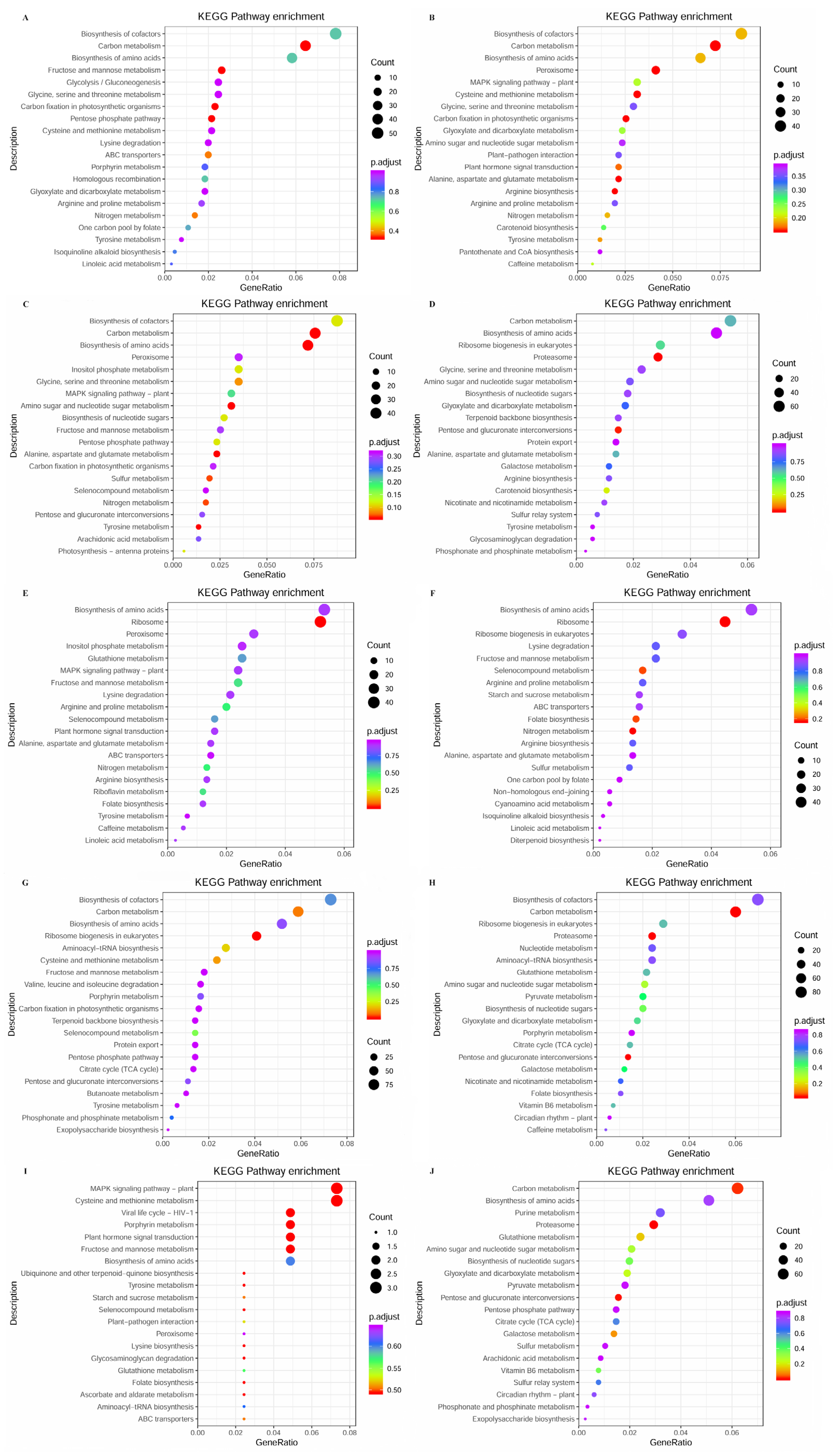
3.3. Transcriptome Analysis of S. japonica Parts
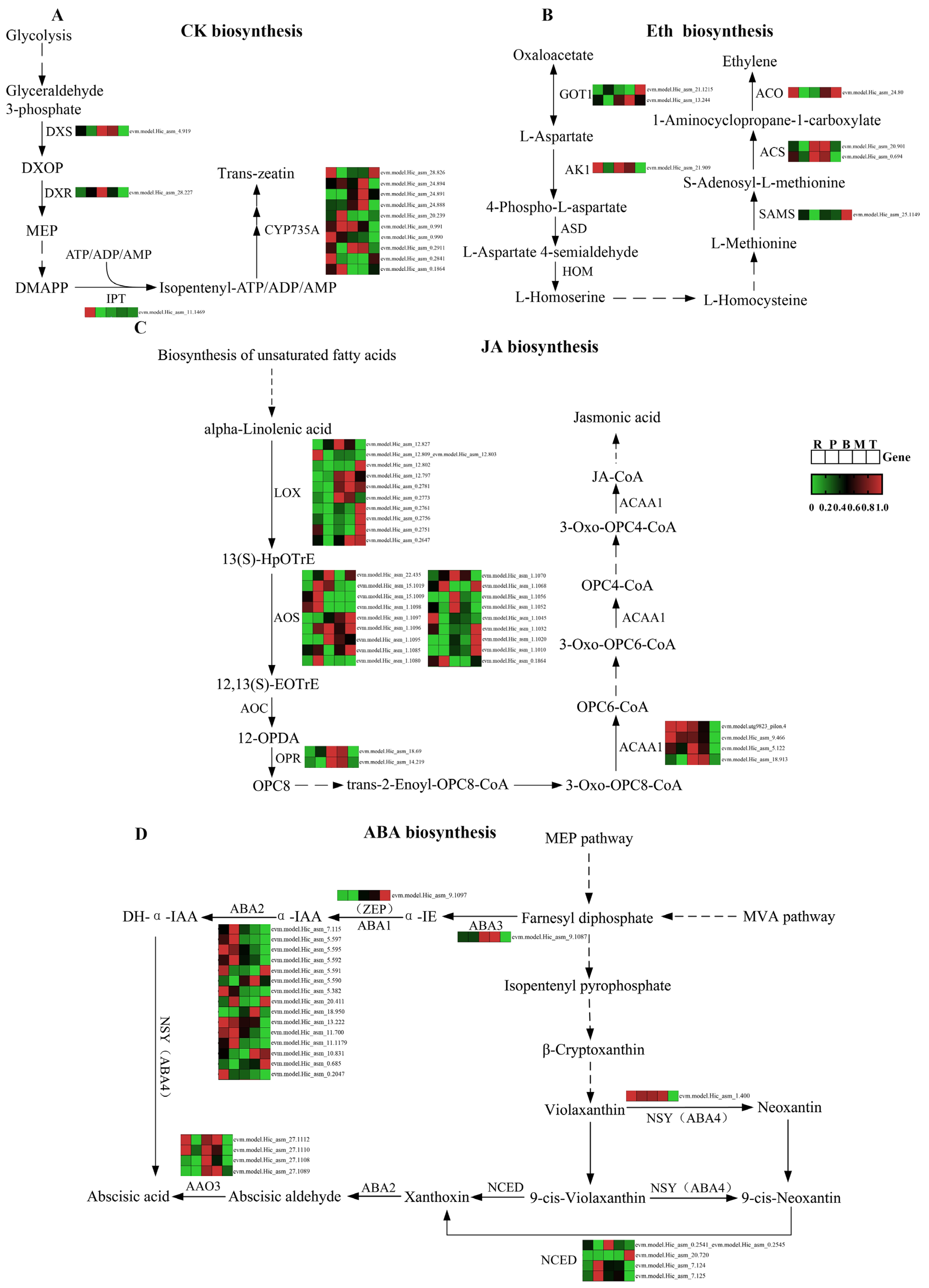
3.4. Expression Analysis of Phytohormone Synthesis-Related Genes in Different Parts of S. japonica
4. Discussion
4.1. Comparison of Phytohormone Content of S. japonica with Other Algae
4.2. Relationship Between Tissue Specificity and Phytohormone Content in S. japonica
4.3. Tissue Specificity of Phytohormone Biosynthetic Pathway in S. japonica
4.4. Differences in Transcriptional Regulation of DEGs in Different Parts of S. japonica
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Xu, D.; Wang, D.; Li, B.; Fan, X.; Zhang, X.; Ye, N.; Wang, Y.; Mou, S.; Zhuang, Z. Effects of CO2 and seawater acidification on the early stages of Saccharina japonica development. Environ. Sci. Technol. 2015, 49, 3548–3556. [Google Scholar] [CrossRef] [PubMed]
- Meng, X. The Progress of the Methods to Identify and Classify Phytoplankton. Biol. Chem. Eng. 2019, 5, 102–104. [Google Scholar]
- Li, X.; Zhong, C.; Jin, Z.; Lin, Q.; Shan, T.; Pang, S. Genetic structure, major chemical component contents and agronomic characters of the principal cultivars of the kelp Saccharina japonica in the largest kelp production province in China. J. Appl. Phycol. 2023, 35, 763–772. [Google Scholar] [CrossRef]
- Wiesemeier, T.; Jahn, K.; Pohnert, G. No evidence for the induction of brown algal chemical defense by the phytohormones jasmonic acid and methyl jasmonate. J. Chem. Ecol. 2008, 34, 1523–1531. [Google Scholar] [CrossRef][Green Version]
- Gao, G.; Beardall, J.; Jin, P.; Gao, L.; Xie, S.; Gao, K. A review of existing and potential blue carbon contributions to climate change mitigation in the Anthropocene. J. Appl. Ecol. 2022, 59, 1686–1699. [Google Scholar] [CrossRef]
- Gao, G.; Burgess, J.; Wu, M.; Wang, S.; Gao, K. Using macroalgae as biofuel: Current opportunities and challenges. Bot. Mar. 2020, 63, 355–370. [Google Scholar] [CrossRef]
- Liu, T. Re-Sequencing and Comparative Genomics of Saccharina japonica Genome. Ph.D. Thesis, Ocean University of China, Qingdao, China, 2020. [Google Scholar]
- Wang, L.; Zou, Y.; Kaw, H.; Wang, G.; Sun, H.; Cai, L.; Li, C.; Meng, L.; Li, D. Recent developments and emerging trends of mass spectrometric methods in plant hormone analysis: A review. Plant Methods 2020, 16, 54. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.; Hsu, P.; Takahashi, Y.; Munemasa, S.; Schroeder, J. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Anees, M.; Gao, L.; Gong, C.; Umer, M.; Yuan, P.; Zhu, H.; Lu, X.; He, N.; Kaseb, M.; Yang, D.; et al. Aux/IAA gene Cla004102, is involved in synergistic regulation of various endogenous hormones, regulating flesh firmness in watermelon. Sci. Hort. 2023, 310, 111719. [Google Scholar] [CrossRef]
- Puja, O.; Renu, B.; Shagun, B.; Ravinderjit, K.; Shivam, J.; Anjali, K.; Parihar, R. The common molecular players in plant hormone crosstalk and signaling. Curr. Protein Pept. Sci. 2015, 16, 369–388. [Google Scholar]
- Li, J.; Li, C. Seventy-year major research progress in plant hormones by Chinese scholars. Chin. Sci. Life Sci. 2019, 49, 1227–1281. [Google Scholar]
- Qiu, Q.; Tian, X.; Wu, G.; Wu, J.; Fan, X.; Yuan, D. Comparative analysis of the transcriptome during single-seed formation of Castanea henryi: Regulation of starch metabolism and endogenous hormones. BMC Plant Biol. 2023, 23, 90. [Google Scholar] [CrossRef] [PubMed]
- Quinta-Nunes, F.; Brandão, P.; Barreto, C.M.; Glick, B.R.; Nascimento, F.X. Plant growth promotion, phytohormone production and genomics of the rhizosphere-associated microalga, Micractinium rhizosphaerae sp. nov. Plants 2023, 12, 651. [Google Scholar] [CrossRef] [PubMed]
- Malek, M.; Ghaderi, F.F.; Torabi, B.; Sadeghipour, H.R. Dynamics of seed dormancy and germination at high temperature stress is affected by priming and phytohormones in rapeseed (Brassica napus L.). J. Plant Physiol. 2021, 269, 153614. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, C.; Yan, X.; Zhang, J.; Xu, J. Simultaneous analysis of ten phytohormones in Sargassum horneri by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. J. Sep. Sci. 2016, 39, 1804–1813. [Google Scholar] [CrossRef]
- Górka, B.; Wieczorek, P.P. Simultaneous determination of nine phytohormones in seaweed and algae extracts by HPLC-PDA. J. Chromatogr. B. 2017, 1057, 32–39. [Google Scholar] [CrossRef]
- Liu, X. Research on Phytohormones of Several Macroalgaes. Master’s Thesis, Ningbo University, Ningbo, China, 2012. [Google Scholar]
- Salama, E.S.; Kabra, A.N.; Ji, M.K.; Kim, J.R.; Min, B.; Jeon, B.H. Enhancement of microalgae growth and fatty acid content under the influence of phytohormones. Bioresour. Technol. 2014, 172, 97–103. [Google Scholar] [CrossRef]
- Piotrowska, A.; Czerpak, R. Cellular response of light/dark-grown green alga Chlorella vulgaris Beijerinck (Chlorophyceae)to exogenous adenine-and phenylurea-type cytokinins. ActaPhysiol Plant 2009, 31, 573–585. [Google Scholar] [CrossRef]
- Kiseleva, A.A.; Tarachovskaya, E.R.; Shishova, M.F. Biosynthesis of phytohormones in algae. Russ. J. Plant Physiol. 2012, 59, 595–610. [Google Scholar] [CrossRef]
- Inka, B.; Christian, W.; Kai, B.; Cornelia, M.B.; Bela, H.B.; Anja, E.; Peter, F.; Dieter, H.; Sabine, J.; Rolf, K.; et al. The genus Laminaria sensulato: Recent insights and developments. Eur. J. Phycol. 2008, 43, 1–86. [Google Scholar]
- Cock, J.M.; Coelho, S.M.; Brownlee, C.; Taylor, A.R. The Ectocarpus genome sequence: Insights into brown algal biology and the evolutionary diversity of the eukaryotes. New Phytol. 2010, 188, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Wu, C. Advances in research on plant hormones in algae. Stud. Mar. Sin. 1998, 167–176. Available online: https://kns.cnki.net/kcms2/article/abstract?v=ywQ10qv17SHI4RxohSmpInxApG0g024Vgz4QtXmt3JbEVdPIchHceAzOXzSNK_PuGx-o-WkagBXjvmju1kkBWQDsHW-X_sfAomCXgFy-qjnVgxp-j3sxycdQi1qYDVAVCm9agCJx4q8NM_4ASXWL3NW0oxiiGLWjsX-jAmK2ZFlrE_F9KJl1ShJbRSgr8QIy&uniplatform=NZKPT&language=CHS (accessed on 8 June 2025).
- Wang, Z.; Sun, H.; Liu, T.; Zhai, Y.; Xing, L.; Miao, J.; Leng, G. Variations of endogenous hormones in Rongfu Laminaria saccharia at different development stage. Prog. Fish. Sci. 2011, 32, 94–98. [Google Scholar]
- Cui, J.; Dai, Y.; Lai, Y.; Tan, Y.; Liu, T. Effects of Abscisic Acid on the Physiological and Biochemical Responses of Saccharina japonica Under High-Temperature Stress. Int. J. Mol. Sci. 2024, 25, 11581. [Google Scholar] [CrossRef]
- Garbary, D.J.; Kim, K.Y. Anatomical Differentiation and Photosynthetic Adaptation in Brown Algae. Algae 2005, 20, 233–238. [Google Scholar] [CrossRef]
- Niklas, K.J. Plant Evolution: An Introduction to the History of Life; University of Chicago Press: Chicago, IL, USA, 2016; Chapter 6. [Google Scholar]
- Cock, J.M.; Sterck, L.; Rouzé, P.; Scornet, D.; Allen, A.E.; Amoutzias, G.; Anthouard, V.; Artiguenave, F.; Aury, J.M.; Badger, J.H.; et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 2010, 465, 617–621. [Google Scholar] [CrossRef]
- Albaseer, S.S.; Rao, R.N.; Swamy, Y.V.; Mukkanti, K. An overview of sample preparation and extraction of synthetic pyrethroids from water, sediment and soil. J. Chromatogr. A 2010, 1217, 5537–5554. [Google Scholar] [CrossRef]
- Balcke, G.U.; Handrick, V.; Bergau, N.; Fichtner, M.; Henning, A.; Stellmach, H.; Tissier, A.; Hause, B.; Frolov, A. An UPLC-MS/MS method for highly sensitive high-throughput analysis of phytohormones in plant tissues. Plant Methods 2012, 8, 47. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Sakakibara, H. Cytokinin Biosynthesis and Regulation. Vitam. Horm. 2005, 72, 271–72287. [Google Scholar] [PubMed]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.; Li, Q.B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Peeters, A.; Koiwai, H.; Oritani, T.; Marion, P.A.; Zeevaart, J.; Koornneef, M.; Kamiya, Y.; Koshiba, T. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. USA 2000, 97, 12908–12913. [Google Scholar] [CrossRef]
- Li, T.; Wang, C.; Miao, J. Identification and quantification of indole-3-acetic acid in the kelp Laminaria japonica Areschoug and its effect on growth of marine microalgae. J. Appl. Phycol. 2007, 19, 479–484. [Google Scholar] [CrossRef]
- Li, J. The auxin concentration in sixteen Chinese marine algae. Chin. J. Ocean. Limnol. 2006, 24, 329–332. [Google Scholar]
- Liu, X.; Zhao, P.; Xu, J.; Luo, Q.; Wang, X.; Chen, H.; Yan, X. LC-MS simultaneous determination of nine phytohormones in macroalgaes. Chin. J. Pharm. Anal. 2012, 32, 1747–1752. [Google Scholar]
- Stirk, W.A.; Bálint, P.; Tarkowská, D.; Novák, O.; Strnad, M.; Ördög, V.; Staden, J. Hormone profiles in microalgae: Gibberellins and brassinosteroids. Plant Physiol. Biochem. 2013, 70, 348–353. [Google Scholar] [CrossRef]
- Tarakhovskaya, E.R.; Maslov, Y.I.; Shishova, M.F. Phytohormones in algae. Russ. J. Plant Physiol. 2007, 54, 163–170. [Google Scholar] [CrossRef]
- Cai, X.; Shao, M.; Sun, X.; Xu, N. Detection of multiple phytohormones by GC-MS technique in Gracilaria lemaneiformis and the response to nitrogen stresses. Oceanol. Limnol. Sin. 2007, 42, 753–758. [Google Scholar]
- Collén, J.; Porcel, B.; Carré, W.; Ball, S.G.; Chaparro, C.; Tonon, T.; Barbeyron, T.; Michel, G.; Noel, B.; Valentin, K. Genome structureand metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc. Natl. Acad. Sci. USA 2013, 110, 5247–5252. [Google Scholar] [CrossRef]
- Yokota, T.; Kim, S.K.; Fukui, Y.; Takahashi, N.; Takeuchi, Y.; Takematsu, T. Brassinosteroids and sterols from a green alga, Hydrodictyon reticulatum: Configuration at C-24. Phytochemistry 1987, 26, 503–506. [Google Scholar] [CrossRef]
- Bajguz, A. Isolation and characterization of brassinosteroids from algal cultures of Chlorella vulgaris Beijerinck (Trebouxiophyceae). J. Plant Physiol. 2009, 166, 1946–1949. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yao, J.; Zhang, J.; Duan, D. Status of genetic studies and breeding of Saccharina japonica in China. J. Ocean. Limnol. 2020, 38, 1064–1079. [Google Scholar] [CrossRef]
- Raven, J.A.; Hurd, C.L. Ecophysiology of photosynthesis in macroalgae. Photosynth. Res. 2012, 113, 105–125. [Google Scholar] [CrossRef] [PubMed]
- Norm, K. Plant AUometry: The Scaling of Form and Process. Écoscience 1995, 2, 416–417. [Google Scholar]
- Gomes, G.L.B.; Scortecci, K.C. Auxin and its role in plant development: Structure, signalling, regulation and response mechanisms. Plant Biol. 2021, 23, 894–904. [Google Scholar] [CrossRef]
- Jan, P.; Jirí, F. Auxin transport routes in plant development. Development 2009, 136, 2675–2688. [Google Scholar]
- Ljung, K.; Hull, A.K.; Celenza, J.; Yamada, M.; Estelle, M.; Normanly, J.; Sandberg, G. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 2005, 17, 1090–1104. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Fan, X.; Han, L. Reviews of research on auxins in algae. Stud. Mar. Sin. 2000, 65–73. Available online: https://kns.cnki.net/kcms2/article/abstract?v=ywQ10qv17SH63cjFar1FGqCKpfDYVeGuuFXqtq66uHGXFkDR_WKLNt1hvWJh2Nm-NVgqGCiugo6DJ8WDRbhcBDE4BavZja4Ue1TfIasrQvO7TqOB0H1os7sYdfmzhmy-by321GKKMfGt_SyLQlmvuYS5ys_3ttpBX34QwOsEYF_x2hZX_wzA8cM5yyyo8C2v&uniplatform=NZKPT&language=CHS (accessed on 8 June 2025).
- Zhao, J.; Deng, X.; Qian, J.; Liu, T.; Ju, M.; Li, J.; Yang, Q.; Zhu, X.; Li, W.; Liu, C.J.; et al. Arabidopsis ABCG14 forms a homodimeric transporter for multiple cytokinins and mediates long-distance transport of isopentenyladenine-type cytokinins. Plant Commun. 2023, 4, 100468. [Google Scholar] [CrossRef]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Schmülling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Davière, J.M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef] [PubMed]
- Laure, C.D.; Patrick, B.; Yuri, K.; Mitsunori, S.; Françoise, D.V.; Sylvie, F.M. N availability modulates the role of NPF3.1, a gibberellin transporter, in GA-mediated phenotypes in Arabidopsis. Planta 2016, 244, 1315–1328. [Google Scholar]
- Xiong, L.; Zhu, J. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003, 133, 29–36. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.; Bressan, R.A.; Song, C.; Zhu, J.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Ying, W.; Liao, L.; Wei, H.; Gao, Y.; Liu, X.; Sun, L. Structural basis for abscisic acid efflux mediated by ABCG25 in Arabidopsis thaliana. Nat. Plants 2023, 9, 1697–1708. [Google Scholar] [CrossRef]
- Rehman, M.; Saeed, M.S.; Fan, X.; Salam, A.; Munir, R.; Yasin, M.U.; Khan, A.R.; Muhammad, S.; Ali, B.; Ali, I.; et al. The Multifaceted Role of Jasmonic Acid in Plant Stress Mitigation: An Overview. Plants 2023, 12, 3982. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Xin, X.F. Regulation and integration of plant jasmonate signaling: A comparative view of monocot and dicot. J. Genet. Genomics 2022, 49, 704–714. [Google Scholar] [CrossRef] [PubMed]
- An, N.; Huang, X.; Yang, Z.; Zhang, M.; Ma, M.; Yu, F.; Jing, L.; Du, B.; Wang, Y.F.; Zhang, X.; et al. Cryo-EM structure and molecular mechanism of the jasmonic acid transporter ABCG16. Nat. Plants 2024, 10, 2052–2061. [Google Scholar] [CrossRef] [PubMed]
- Rosado, D.; Ackermann, A.; Spassibojko, O.; Rossi, M.; Pedmale, U.V. WRKY transcription factors and ethylene signaling modify root growth during the shade-avoidance response. Plant Physiol. 2022, 188, 1294–1311. [Google Scholar] [CrossRef]
- Larsen, P.B. Mechanisms of ethylene biosynthesis and response in plants. Essays Biochem. 2015, 58, 61–70. [Google Scholar]
- Souza, G.M.; Trewavas, T.; de Menezes Daloso, D. Systems plant physiology: An integrated view of plants life. Prog. Biophys. Mol. Biol. 2019, 146, 1–2. [Google Scholar] [CrossRef]
- Santner, A.; Estelle, M. Recent advances and emerging trends in plant hormone signalling. Nature 2009, 459, 1071–1078. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Lu, Y.; Xu, J. Phytohormones in microalgae: A new opportunity for microalgal biotechnology? Trends Plant Sci. 2015, 20, 273–282. [Google Scholar] [CrossRef]
- Takei, K.; Yamaya, T.; Sakakibara, H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J. Biol. Chem. 2004, 279, 41866–41872. [Google Scholar] [CrossRef]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perroud, P.F.; Lindquist, E.A.; Kamisugi, Y.; et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 2008, 319, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Laudert, D.; Weiler, E.W. Allene oxide synthase: A major control point in Arabidopsis thaliana octadecanoid signalling. Plant J. 1998, 15, 675–684. [Google Scholar] [CrossRef]
- Laule, O.; Fürholz, A.; Chang, H.S.; Zhu, T.; Wang, X.; Heifetz, P.B.; Gruissem, W.; Lange, M. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 6866–6871. [Google Scholar] [CrossRef]
- Gamalero, E.; Lingua, G.; Glick, B.R. Ethylene, ACC, and the Plant Growth-Promoting Enzyme ACC Deaminase. Biology 2023, 12, 1043. [Google Scholar] [CrossRef]
- Glauser, G.; Grata, E.; Dubugnon, L.; Rudaz, S.; Farmer, E.E.; Wolfender, J.L. Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J. Biol. Chem. 2008, 283, 16400–16407. [Google Scholar] [CrossRef]
- Angelovici, R.; Mittler, R. Redox regulation of plant stress and development. Plant Cell Environ. 2024, 47, 2713–2715. [Google Scholar] [CrossRef]
- Smith, S.R.; Glé, C.; Abbriano, R.M.; Traller, J.C.; Davis, A.; Trentacoste, E.; Vernet, M.; Allen, A.E.; Hildebrand, M. Transcript level coordination of carbon pathways during silicon starvation-induced lipid accumulation in the diatom Thalassiosira pseudonana. New Phytol. 2016, 210, 890–904. [Google Scholar] [CrossRef]
- Aubry, E.; Dinant, S.; Vilaine, F.; Bellini, C.; Le, H.R. Lateral Transport of Organic and Inorganic Solutes. Plants 2019, 8, 20. [Google Scholar] [CrossRef]
- Pate, J.S.; Atkins, C.A.; Hamel, K.; McNeil, D.L.; Layzell, D.B. Transport of organic solutes in Phloem and xylem of a nodulated legume. Plant Physiol. 1979, 63, 1082–1088. [Google Scholar] [CrossRef]
- Shah, K.; Zhu, X.; Zhang, T.; Chen, J.; Chen, J.; Qin, Y. The poetry of nitrogen and carbon metabolic shifts: The role of C/N in pitaya phase change. Plant Sci. 2024, 348, 112240. [Google Scholar] [CrossRef]
- Szal, B.; Podgórska, A. The role of mitochondria in leaf nitrogen metabolism. Plant Cell Environ. 2012, 35, 1756–1768. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, F.M.; van Koningsbruggen, S.; Navascués, J.; Lamond, A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007, 8, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Shaw, P.; Brown, J. Nucleoli: Composition, function, and dynamics. Plant Physiol. 2012, 158, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Eberhard, S.; Finazzi, G.; Wollman, F.A. The dynamics of photosynthesis. Annu. Rev. Genet. 2008, 42, 463–515. [Google Scholar] [CrossRef]
- Charrier, B.; Coelho, S.M.; Le, B.A.; Tonon, T.; Michel, G.; Potin, P.; Kloareg, B.; Boyen, C.; Peters, A.F.; Cock, J.M. Development and physiology of the brown alga Ectocarpus siliculosus: Two centuries of research. New Phytol. 2008, 177, 319–332. [Google Scholar] [CrossRef]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef]
- Cappadocia, L.; Lima, C.D. Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism. Chem. Rev. 2018, 118, 889–918. [Google Scholar] [CrossRef]
- Brezo, M.; Arenas, F.; Rubal, M.; Burgués, S.; Esteban, R.; García-Plazaola, I.; Figueroa, F.L.; Pereira, R.; Saldaña, L.; Sousa-Pinto, I.; et al. Physical factors driving intertidal macroalgae distribution: Physiological stress of a dominant fucoid at its southern limit. Oecologia 2012, 170, 341–353. [Google Scholar]
- Hildebrandt, T.M.; Nunes, N.A.; Araújo, W.L.; Braun, H.P. Amino Acid Catabolism in Plants. Mol. Plant 2015, 8, 1563–1579. [Google Scholar] [CrossRef]
- Kensuke, K.; Hiromitsu, T.; Ali, F.; Masami, Y.H. The Roles of Functional Amino Acids in Plant Growth and Development. Plant Cell Physiol. 2023, 64, 1482–1493. [Google Scholar]

| Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| SjEF1α | GTGATGGAGGAGAACCC | TTGATGACACCCACAGC |
| Sjgene 0.2211 | ACAAGGACGGGAACCACATC | TCACGAACTGATGCGTGTGA |
| Sjgene 1.400 | AAGGGAGCTGAAGTGGAACG | GTCCACGTCAAGTGTGTTGC |
| Sjgene 5.402 | CCGCGAGTACGACAAGATGA | AGAACGACACCTGCAGACAG |
| Sjgene 7.125 | GGCGAATGCGTGTTCATACC | GCTTCGCGTTCATGGTCTTC |
| Sjgene 9.1087 | TGTACTTCACGCACTCCACC | AGCCATCTCGTGCTCGTAAG |
| Sjgene 27.1108 | CAACATGTACAAGGAGGGCG | CTTGGTCGGAATGACCGAAAG |
| Phytohormone | Test Substances | Parts | ||||
|---|---|---|---|---|---|---|
| Rhizoid | Petiole | Basis | Middle | Tip | ||
| Auxin | 3-Indoleacetamide | 0.064 ± 0.009 a | NA | 0.040 ± 0.014 a | NA | 0.028 ± 0.013 a |
| 3-Indolecarboxylic Acid | 0.456 ± 0.084 a | 0.329 ± 0.071 ab | 0.324 ± 0.061 ab | 0.258 ± 0.019 b | 0.336 ± 0.105 ab | |
| Indole-3-Acetic Acid | 1.206 ± 0.288 b | 1.028 ± 0.192 b | 0.872 ± 0.229 b | 1.516 ± 0.337 ab | 2.180 ± 0.288 a | |
| 3-Indolepropionic Acid | 5.368 ± 0.504 a | 5.688 ± 0.419 a | 5.686 ± 0.396 a | 5.971 ± 0.297 a | 5.317 ± 0.836 a | |
| 3-Indolebutyric Acid | 1.881 ± 0.157 a | 1.978 ± 0.082 a | 1.849 ± 0.183 a | 1.889 ± 0.197 a | 1.518 ± 0.291 a | |
| 3-Indoleacetonitrile | NA | NA | NA | NA | 0.185 ± 0.008 | |
| Cytokinin | Trans-zeatin | 0.116 ± 0.006 a | 0.087 ± 0.008 b | 0.092 ± 0.018 ab | 0.087 ± 0.011 b | 0.102 ± 0.031 ab |
| Dihydrozeatin | NA | NA | 0.052 ± 0.010 a | 0.017 ± 0.005 b | 0.021 ± 0.012 b | |
| Trans-zeatin-riboside | NA | NA | NA | NA | NA | |
| N6-(delta2-Isopentenyl) Adenine | 0.045 ± 0.016 a | 0.029 ± 0.009 a | 0.065 ± 0.025 a | 0.033 ± 0.007 a | NA | |
| N6-(delta2-Isopentenyl) Adenosine | 0.246 ± 0.060 cd | 0.125 ± 0.018 d | 0.280 ± 0.050 c | 0.495 ± 0.022 b | 0.897 ± 0.053 a | |
| Gibberellins | Gibberellin A1 | NA | 0.008 ± 0.002 c | 0.013 ± 0.007 bc | 0.029 ± 0.008 b | 1.154 ± 0.112 a |
| Gibberellic A3 | NA | NA | NA | NA | NA | |
| Gibberellin A4 | 0.208 ± 0.089 a | 0.282 ± 0.128 a | 0.185 ± 0.017 a | 0.236 ± 0.025 a | 0.131 ± 0.056 a | |
| Gibberellin A7 | NA | NA | NA | NA | NA | |
| Abscisic Acid | Abscisic Acid | 1.101 ± 0.174 b | 0.602 ± 0.051 c | 0.476 ± 0.046 d | 1.595 ± 0.662 ab | 28.013 ± 3.562 a |
| Ethylene | Aminocyclopropane Carboxylic Acid | 4.215 ± 0.281 a | 2.560 ± 0.171 b | 2.522 ± 0.343 b | 2.255 ± 0.110 b | 2.401 ± 0.352 b |
| Jasmonic Acid | Methyl Jasmonate | 3.073 ± 1.188 b | 8.224 ± 2.475 ab | 5.436 ± 2.338 ab | 3.475 ± 0.726 b | 9.657 ± 0.580 a |
| Jasmonic Acid | 5.761 ± 2.891 b | 1.865 ± 0.521 b | 13.643 ± 1.936 b | 20.552 ± 13.119 b | 170.665 ± 32.253 a | |
| Dihydrojasmonic Acid | 8.345 ± 2.764 b | 2.082 ± 0.973 b | 11.206 ± 1.420 b | 12.591 ± 2.042 b | 128.375 ± 35.707 a | |
| Jasmonic Acid-Isoleucine | 0.129 ± 0.016 ab | 0.170 ± 0.026 a | 0.159 ± 0.027 ab | 0.192 ± 0.028 a | 0.104 ± 0.020 b | |
| Brassinosteroids | Brassinolide | NA | NA | NA | NA | NA |
| Salicylic Acid | Salicylic Acid | 9.517 ± 1.486 | NA | NA | NA | NA |
| Sample | Raw Reads | Clean Reads | Clean Reads Rate (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|
| Rhizoid1 | 103,336,502 | 93,647,616 | 90.62 | 94.65 | 54.86 |
| Rhizoid2 | 99,422,620 | 90,020,886 | 90.54 | 94.72 | 54.84 |
| Rhizoid3 | 92,234,828 | 81,998,948 | 88.9 | 94.45 | 55.03 |
| Petiole1 | 98,000,366 | 88,596,096 | 90.4 | 94.53 | 54.97 |
| Petiole2 | 93,811,592 | 83,146,322 | 88.63 | 94.54 | 55.29 |
| Petiole3 | 91,132,630 | 82,764,530 | 90.82 | 94.2 | 54.83 |
| Basis1 | 101,323,062 | 91,874,834 | 90.68 | 94.13 | 55.32 |
| Basis2 | 90,507,826 | 81,967,646 | 90.56 | 94.56 | 54.78 |
| Basis3 | 103,537,970 | 93,138,382 | 89.96 | 94.27 | 54.87 |
| Middle1 | 104,956,876 | 92,055,950 | 87.71 | 94.17 | 55.03 |
| Middle2 | 103,128,310 | 93,312,852 | 90.48 | 94.25 | 55.16 |
| Middle3 | 90,793,274 | 81,220,748 | 89.46 | 93.94 | 55.06 |
| Tip1 | 95,509,248 | 86,173,916 | 90.23 | 94.52 | 55.69 |
| Tip2 | 96,078,054 | 87,428,612 | 91 | 94.55 | 56.01 |
| Tip3 | 91,853,966 | 82,491,570 | 89.81 | 94.5 | 55.88 |
| ID | Metabolic Pathway | Gene Number | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R vs. P | R vs. B | R vs. M | R vs. T | P vs. B | P vs. M | P vs. T | B vs. M | B vs. T | M vs. T | ||
| ko00380 | Tryptophan metabolism | 4 (1) | 3 (2) | 2 (1) | 12 (7) | 7 (6) | 7 (7) | 15 (10) | 0 | 14 (7) | 11 (4) |
| ko00904 | Diterpenoid biosynthesis | 0 | 1 (1) | 1 (1) | 0 | 1 (1) | 2 (2) | 0 | 0 | 0 | 0 |
| ko00906 | Carotenoid biosynthesis | 5 (3) | 7 (3) | 4 (3) | 13 (8) | 5 (1) | 5 (3) | 10 (6) | 0 | 8 (7) | 8 (6) |
| ko00270 | Cysteine and methionine metabolism | 14 (5) | 16 (7) | 12 (9) | 21 (14) | 11 (7) | 15 (12) | 30 (20) | 3 (2) | 21 (13) | 19 (10) |
| ko00592 | Alpha-Linolenic acid metabolism | 5 (3) | 5 (3) | 6 (4) | 6 (3) | 3 (2) | 5 (3) | 7 (4) | 0 | 3 (1) | 4 (1) |
| ko00240 | Pyrimidine metabolism | 8 (0) | 7 (4) | 6 (6) | 16 (10) | 11 (9) | 11 (10) | 16 (10) | 0 | 19 (8) | 19 (8) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, J.; Zhu, J.; Dai, Y.; Yuan, J.; Lin, W.; Liu, T. A Non-Specific Phytohormone Regulatory Network in Saccharina japonica Coordinates Growth and Environmental Adaptation. Plants 2025, 14, 1821. https://doi.org/10.3390/plants14121821
Cui J, Zhu J, Dai Y, Yuan J, Lin W, Liu T. A Non-Specific Phytohormone Regulatory Network in Saccharina japonica Coordinates Growth and Environmental Adaptation. Plants. 2025; 14(12):1821. https://doi.org/10.3390/plants14121821
Chicago/Turabian StyleCui, Jiexin, Jinli Zhu, Yinru Dai, Jincheng Yuan, Wen Lin, and Tao Liu. 2025. "A Non-Specific Phytohormone Regulatory Network in Saccharina japonica Coordinates Growth and Environmental Adaptation" Plants 14, no. 12: 1821. https://doi.org/10.3390/plants14121821
APA StyleCui, J., Zhu, J., Dai, Y., Yuan, J., Lin, W., & Liu, T. (2025). A Non-Specific Phytohormone Regulatory Network in Saccharina japonica Coordinates Growth and Environmental Adaptation. Plants, 14(12), 1821. https://doi.org/10.3390/plants14121821






