In Silico Analysis of miRNA-mRNA Binding Sites in Arabidopsis thaliana as a Model for Drought-Tolerant Plants
Abstract
1. Introduction
2. Results
2.1. Predicted miRNA–mRNA Interactions Targeting Key ROS Detoxification Genes Involved in Drought Stress Response in A. thaliana
2.2. Characteristics of miRNA Binding Sites in mRNAs of Drought-Responsive Hormonal Signaling Genes in A. thaliana
2.3. Predicted miRNA Target Interactions Regulating Drought-Responsive Transcription Factor Genes in A. thaliana
3. Discussion
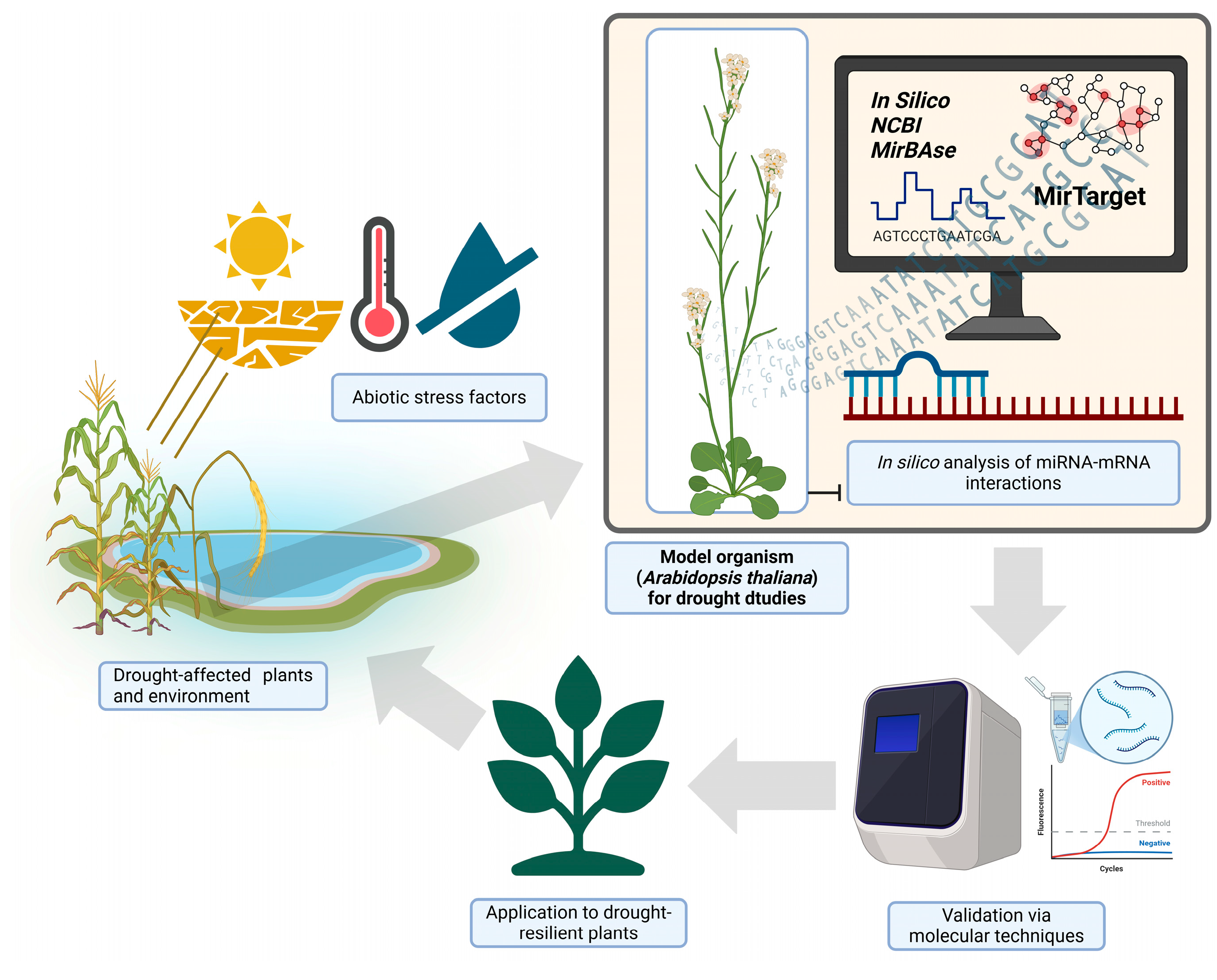
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| BS | Binding site |
| CDS | Coding sequence |
| miRNA | mRNA-inhibitory RNAs |
| mRNA | Messenger RNA |
| ROS | Reactive oxygen species |
| UTR | Untranslated region |
References
- Zhakypbek, Y.; Belkozhayev, A.M.; Kerimkulova, A.; Kossalbayev, B.D.; Murat, T.; Tursbekov, S.; Turysbekova, G.; Tursunova, A.; Tastambek, K.T.; Allakhverdiev, S.I. MicroRNAs in Plant Genetic Regulation of Drought Tolerance and Their Function in Enhancing Stress Adaptation. Plants 2025, 14, 410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, J.; Li, R.; Ge, Y.; Li, Y.; Li, R. Plants’ Response to Abiotic Stress: Mechanisms and Strategies. Int. J. Mol. Sci. 2023, 24, 10915. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lu, M.; Wang, Y.; Wang, Y.; Liu, Z.; Chen, S. Response Mechanism of Plants to Drought Stress. Horticulturae 2021, 7, 50. [Google Scholar] [CrossRef]
- Parums, D.V. A Review of the Increasing Global Impact of Climate Change on Human Health and Approaches to Medical Preparedness. Med. Sci. Monit. 2024, 30, e945763. [Google Scholar] [CrossRef]
- Tang, H.; Du, L.; Xia, C.; Luo, J. Bridging Gaps and Seeding Futures: A Synthesis of Soil Salinization and the Role of Plant-Soil Interactions under Climate Change. iScience 2024, 27, 110804. [Google Scholar] [CrossRef]
- Tarolli, P.; Luo, J.; Park, E.; Barcaccia, G.; Masin, R. Soil Salinization in Agriculture: Mitigation and Adaptation Strategies Combining Nature-Based Solutions and Bioengineering. iScience 2024, 27, 108830. [Google Scholar] [CrossRef]
- Lattuada, M.; Albrecht, C.; Wilke, T. Differential Impact of Anthropogenic Pressures on Caspian Sea Ecoregions. Mar. Pollut. Bull. 2019, 142, 274–281. [Google Scholar] [CrossRef]
- Yazdanpanah Dero, Q.; Yari, E.; Charrahy, Z. Global Warming, Environmental Security and Its Geo-Economic Dimensions Case Study: Caspian Sea Level Changes on the Balance of Transit Channels. J. Environ. Health Sci. Eng. 2020, 18, 541–557. [Google Scholar] [CrossRef]
- Sheffield, J.; Wood, E.F. Projected Changes in Drought Occurrence under Future Global Warming from Multi-Model, Multi-Scenario, IPCC AR4 Simulations. Clim. Dyn. 2008, 31, 79–105. [Google Scholar] [CrossRef]
- Haghpanah, M.; Hashemipetroudi, S.; Arzani, A.; Araniti, F. Drought Tolerance in Plants: Physiological and Molecular Responses. Plants 2024, 13, 2962. [Google Scholar] [CrossRef]
- Kossalbayev, B.D.; Yilmaz, G.; Sadvakasova, A.K.; Zayadan, B.K.; Belkozhayev, A.M.; Kamshybayeva, G.K.; Sainova, G.A.; Bozieva, A.M.; Alharby, H.F.; Tomo, T.; et al. Biotechnological production of hydrogen: Design features of photobioreactors and improvement of conditions for cultivating cyanobacteria. Int. J. Hydrogen Energy 2024, 49, 413–432. [Google Scholar] [CrossRef]
- Monroe, J.G.; Powell, T.; Price, N.; Mullen, J.L.; Howard, A.; Evans, K.; Lovell, J.T.; McKay, J.K. Drought Adaptation in Arabidopsis thaliana by Extensive Genetic Loss-of-Function. elife 2018, 7, e41038. [Google Scholar] [CrossRef]
- Thonglim, A.; Bortolami, G.; Delzon, S.; Larter, M.; Offringa, R.; Keurentjes, J.J.B.; Smets, E.; Balazadeh, S.; Lens, F. Drought Response in Arabidopsis Displays Synergistic Coordination between Stems and Leaves. J. Exp. Bot. 2023, 74, 1004–1021. [Google Scholar] [CrossRef]
- Ferjani, A.; Tsukagoshi, H.; Vassileva, V. Editorial: Model Organisms in Plant Science: Arabidopsis thaliana. Front. Plant Sci. 2023, 14, 1279230. [Google Scholar] [CrossRef]
- Kaur, S.; Seem, K.; Kumar, S.; Kaundal, R.; Mohapatra, T. Comparative Genome-Wide Analysis of MicroRNAs and Their Target Genes in Roots of Contrasting Indica Rice Cultivars under Reproductive-Stage Drought. Genes 2023, 14, 1390. [Google Scholar] [CrossRef] [PubMed]
- Rakhmetullina, A.; Pyrkova, A.; Aisina, D.; Ivashchenko, A. In Silico Prediction of Human Genes as Potential Targets for Rice miRNAs. Comput. Biol. Chem. 2020, 87, 107305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yang, J.; Zhang, N.; Wu, J.; Si, H. Roles of microRNAs in Abiotic Stress Response and Characteristics Regulation of Plant. Front. Plant Sci. 2022, 13, 919243. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Li, W.; Li, F.; Ren, M.; Wang, W. microRNAs: Key Regulators in Plant Responses to Abiotic and Biotic Stresses via Endogenous and Cross-Kingdom Mechanisms. Int. J. Mol. Sci. 2024, 25, 1154. [Google Scholar] [CrossRef]
- Samynathan, R.; Venkidasamy, B.; Shanmugam, A.; Ramalingam, S.; Thiruvengadam, M. Functional Role of microRNA in the Regulation of Biotic and Abiotic Stress in Agronomic Plants. Front. Genet. 2023, 14, 1272446. [Google Scholar] [CrossRef]
- Singroha, G.; Sharma, P.; Sunkur, R. Current Status of microRNA-Mediated Regulation of Drought Stress Responses in Cereals. Physiol. Plant. 2021, 172, 1808–1821. [Google Scholar] [CrossRef]
- Lei, Z.; Zhang, X.; Wang, M.; Mao, J.; Hu, X.; Lin, Y.; Xiong, X.; Qin, Y. Silencing of miR169a Improves Drought Stress by Enhancing Vascular Architecture, ROS Scavenging, and Photosynthesis of Solanum tuberosum L. Front. Plant Sci. 2025, 16, 1553135. [Google Scholar]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional Induction of Two Cu/Zn Superoxide Dismutase Genes in Arabidopsis Is Mediated by Downregulation of miR398 and Important for Oxidative Stress Tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [PubMed]
- Islam, W.; Waheed, A.; Naveed, H.; Zeng, F. MicroRNAs Mediated Plant Responses to Salt Stress. Cells 2022, 11, 2806. [Google Scholar] [CrossRef] [PubMed]
- Dash, P.K.; Gupta, P.; Pradhan, S.K.; Shasany, A.K.; Rai, R. Analysis of Homologous Regions of Small RNAs MIR397 and MIR408 Reveals the Conservation of Microsynteny among Rice Crop-Wild Relatives. Cells 2022, 11, 3461. [Google Scholar] [CrossRef]
- Santhoshi, Y.; Anjana, A.B.; Zala, H.; Bosamia, T.; Tiwari, K.; Prajapati, K.; Patel, P.; Soni, N.; Patel, N.; Solanki, S.; et al. Comprehensive Analysis of the NHX Gene Family and Its Regulation under Salt and Drought Stress in Quinoa (Chenopodium quinoa Willd.). Genes 2025, 16, 70. [Google Scholar] [CrossRef]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide Dismutases: Dual Roles in Controlling ROS Damage and Regulating ROS Signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef]
- Steinwand, B.J.; Kieber, J.J. The Role of Receptor-Like Kinases in Regulating Cell Wall Function. Plant Physiol. 2010, 153, 479–484. [Google Scholar] [CrossRef]
- Guo, H.; Ye, H.; Li, L.; Yin, Y. A Family of Receptor-Like Kinases Are Regulated by BES1 and Involved in Plant Growth in Arabidopsis thaliana. Plant Signal Behav. 2009, 4, 784–786. [Google Scholar] [CrossRef]
- Liu, L.; Yahaya, B.S.; Li, J.; Wu, F. Enigmatic Role of Auxin Response Factors in Plant Growth and Stress Tolerance. Front. Plant Sci. 2024, 15, 1398818. [Google Scholar] [CrossRef]
- Vaseva, I.I.; Mishev, K.; Depaepe, T.; Vassileva, V.; Van Der Straeten, D. The Diverse Salt-Stress Response of Arabidopsis ctr1-1 and ein2-1 Ethylene Signaling Mutants Is Linked to Altered Root Auxin Homeostasis. Plants 2021, 10, 452. [Google Scholar] [CrossRef]
- Gao, L.; Lv, Q.; Wang, L.; Han, S.; Wang, J.; Chen, Y.; Zhu, W.; Zhang, X.; Bao, F.; Hu, Y.; et al. Abscisic Acid-Mediated Autoregulation of the MYB41-BRAHMA Module Enhances Drought Tolerance in Arabidopsis. Plant Physiol. 2024, 196, 1608–1626. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lai, Z.; Shi, J.; Xiao, Y.; Chen, Z.; Xu, X. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 Transcription Factors in Plant Responses to Abscisic Acid and Abiotic Stress. BMC Plant Biol. 2010, 10, 281. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Song, Q.; Zuo, Z.-F.; Liu, L. MicroRNA398: A master regulator of plant development and stress responses. Int. J. Mol. Sci. 2022, 23, 10803. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Li, Z.; Xiong, L. A Plant MicroRNA Regulates the Adaptation of Roots to Drought Stress. FEBS Lett. 2012, 586, 1742–1747. [Google Scholar] [CrossRef]
- Sorin, C.; Declerck, M.; Christ, A.; Blein, T.; Ma, L.; Lelandais-Brière, C.; Njo, M.F.; Beeckman, T.; Crespi, M.; Hartmann, C. A miR169 Isoform Regulates Specific NF-YA Targets and Root Architecture in Arabidopsis. New Phytol. 2014, 202, 1197–1211. [Google Scholar] [CrossRef]
- Ji, J.; Zeng, Y.; Zhang, S.; Chen, F.; Hou, X.; Li, Q. The miR169b/NFYA1 module from the halophyte Halostachys caspica endows salt and drought tolerance in Arabidopsis through multi-pathways. Front. Plant Sci. 2023, 13, 1026421. [Google Scholar] [CrossRef]
- Haberman, N.; Digby, H.; Faraway, R.; Cheung, R.; Chakrabarti, A.M.; Jobbins, A.M.; Parr, C.; Yasuzawa, K.; Kasukawa, T.; Yip, C.W.; et al. Widespread 3′UTR capped RNAs derive from G-rich regions in proximity to AGO2 binding sites. BMC Biol. 2024, 22, 254. [Google Scholar] [CrossRef]
- Toledo-Stuardo, K.; Ribeiro, C.H.; Campos, I.; Tello, S.; Latorre, Y.; Altamirano, C.; Dubois-Camacho, K.; Molina, M.C. Impact of MICA 3′UTR allelic variability on miRNA binding prediction, a bioinformatic approach. Front. Genet. 2023, 14, 1273296. [Google Scholar] [CrossRef]
- Liu, J.N.; Ma, X.; Yan, L.; Liang, Q.; Fang, H.; Wang, C.; Dong, Y.; Chai, Z.; Zhou, R.; Bao, Y.; et al. MicroRNA and degradome profiling uncover defense response of Fraxinus velutina Torr. to salt stress. Front. Plant Sci. 2022, 13, 847853. [Google Scholar] [CrossRef]
- Joshi, R.K.; Bharat, S.S.; Mishra, R. Engineering drought tolerance in plants through CRISPR/Cas genome editing. 3 Biotech 2020, 10, 400. [Google Scholar] [CrossRef]
- Yang, Y.; Cao, G.; Tang, L. Selection and validation of reference genes for qRT-PCR normalization in dayflower (Commelina communis) based on transcriptome profiling. BMC Plant Biol. 2024, 24, 1131. [Google Scholar] [CrossRef]
- Ivashchenko, A.T.; Pyrkova, A.Y.; Niyazova, R.Y.; Alybayeva, A.; Baskakov, K. Prediction of miRNA Binding Sites in mRNA. Bioinformation 2016, 12, 237–240. [Google Scholar] [CrossRef]
- Ivashchenko, A.; Berillo, O.; Pyrkova, A.; Niyazova, R.; Atambayeva, S. MiR-3960 binding sites with mRNA of human genes. Bioinformation. 2014, 10, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Kool, E.T. Hydrogen Bonding, Base Stacking, and Steric Effects in DNA Replication. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 1–22. [Google Scholar] [CrossRef]
- Leontis, N.B.; Stombaugh, J.; Westhof, E. The Non-Watson-Crick Base Pairs and Their Associated Isostericity Matrices. Nucleic Acids Res. 2002, 30, 3497–3531. [Google Scholar] [CrossRef]
- Belkozhayev, A.; Niyazova, R.; Wilson, C.; Jainakbayev, N.; Pyrkova, A.; Ashirbekov, Y.; Akimniyazova, A.; Sharipov, K.; Ivashchenko, A. Bioinformatics Analysis of the Interaction of miRNAs and piRNAs with Human mRNA Genes Having Di- and Trinucleotide Repeats. Genes 2022, 13, 800. [Google Scholar] [CrossRef]
- Ivashchenko, A.; Pyrkova, A.; Niyazova, R. A Method for Clustering of miRNA Sequences Using Fragmented Programming. Bioinformation 2016, 12, 15–18. [Google Scholar] [CrossRef]
- Belkozhayev, A.; Niyazova, R.; Kamal, M.A.; Ivashchenko, A.; Sharipov, K.; Wilson, C.M. Differential microRNA Expression in the SH-SY5Y Human Cell Model as Potential Biomarkers for Huntington’s Disease. Front. Cell Neurosci. 2024, 18, 1399742. [Google Scholar] [CrossRef]
- Bari, A.; Orazova, S.; Ivashchenko, A. miR156- and miR171-Binding Sites in the Protein-Coding Sequences of Several Plant Genes. Biomed. Res. Int. 2013, 2013, 307145. [Google Scholar] [CrossRef]
- Ivashchenko, A.; Berillo, O.; Pyrkova, A.; Niyazova, R. Binding Sites of miR-1273 Family on the mRNA of Target Genes. Biomed. Res. Int. 2014, 2014, 620530. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
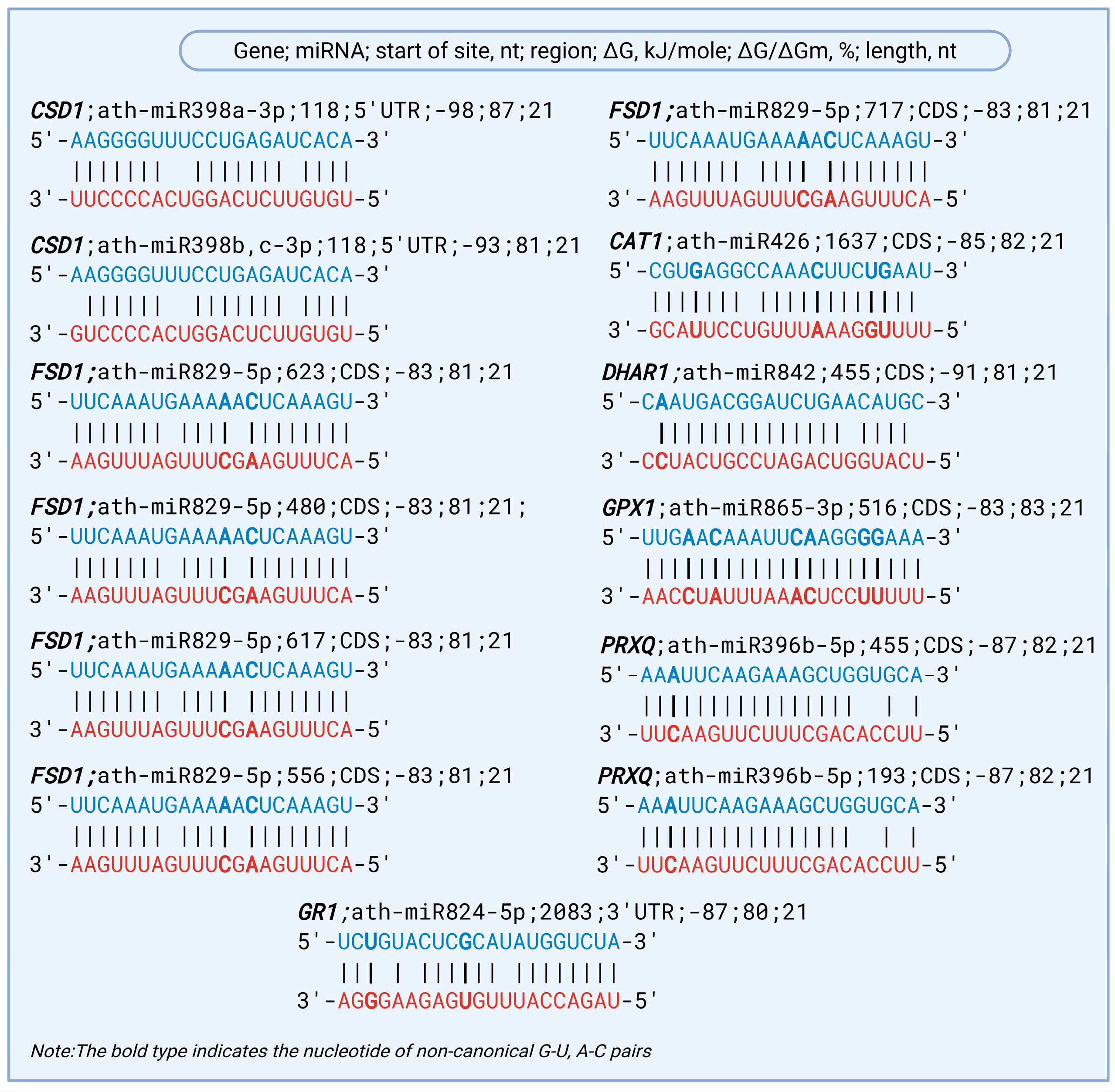
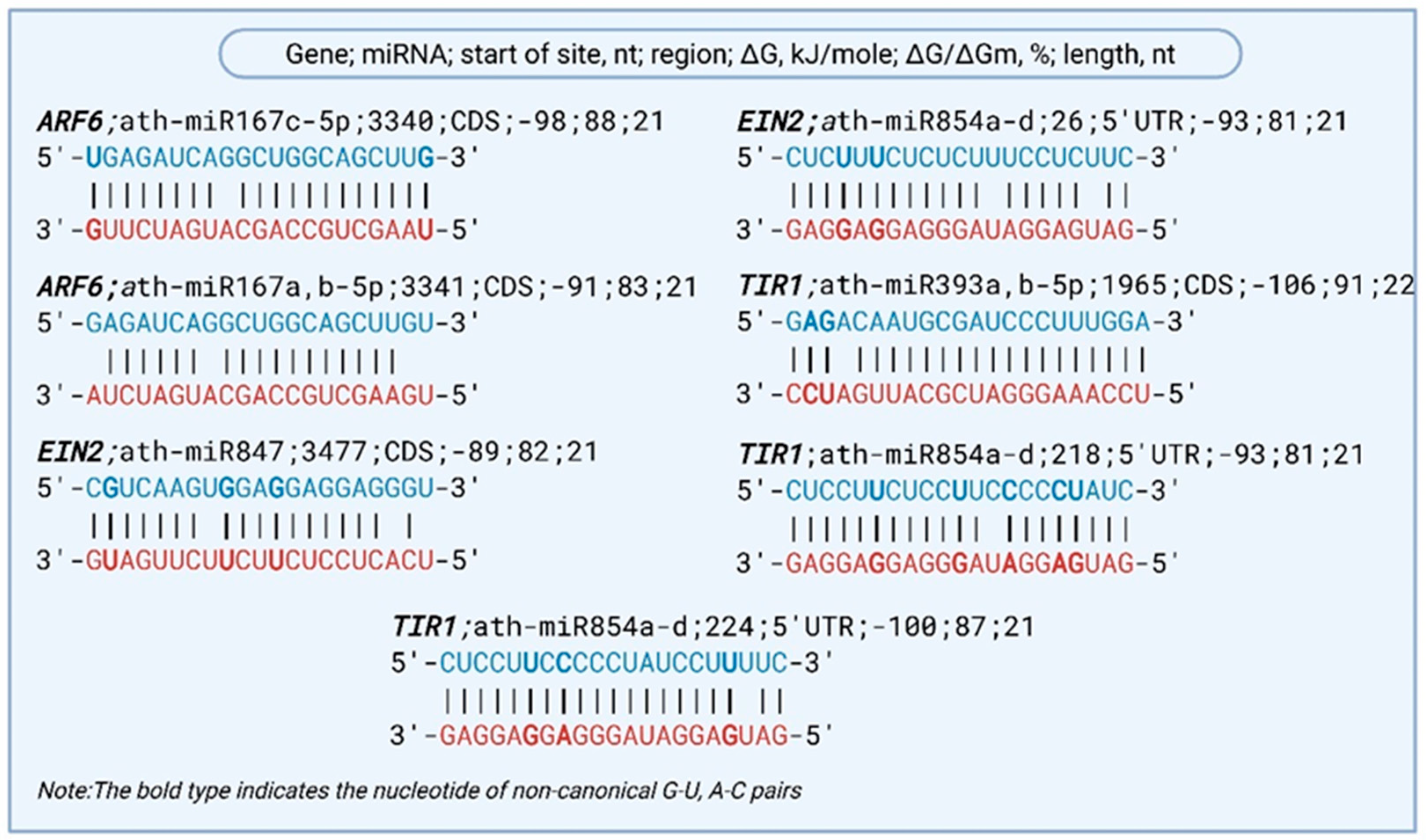


| Gene | NCBI Reference Sequence | miRNA | Start of Site, nt | Region | ΔG, kJ/mole | ΔG/ΔGm, % | Protein/Function |
|---|---|---|---|---|---|---|---|
| CSD1 | NM_100757.4 | ath-miR398a-3p | 118 | 5′UTR | −98 | 87 | Cu/Zn superoxide dismutase—detoxifies superoxide radicals |
| NM_100757.4 | ath-miR398b,c-3p | 118 | 5′UTR | −93 | 81 | ||
| FSD1 | NM_179109.3 | ath-miR829-5p | 623 | CDS | −83 | 81 | Fe superoxide dismutase—detoxifies superoxide radicals |
| NM_179110.2 | ath-miR829-5p | 480 | CDS | −83 | 81 | ||
| NM_118642.2 | ath-miR829-5p | 617 | CDS | −83 | 81 | ||
| NM_001203905.1 | ath-miR829-5p | 556 | CDS | −83 | 81 | ||
| NM_001036633.2 | ath-miR829-5p | 717 | CDS | −83 | 81 | ||
| CAT1 | NM_101914.4 | ath-miR426 | 1637 | CDS | −85 | 82 | Catalase 1—breaks down hydrogen peroxide |
| DHAR1 | NM_101814.5 | ath-miR842 | 455 | CDS | −91 | 81 | Dehydroascorbate reductase 1—regenerates ascorbate |
| GPX1 | NM_128065.5 | ath-miR865-3p | 516 | CDS | −83 | 83 | Glutathione peroxidase 1—reduces hydrogen peroxide and lipid peroxides |
| PRXQ | NM_001203050.1 | ath-miR396b-5p | 455 | CDS | −87 | 82 | Peroxiredoxin Q—reduces peroxides in chloroplasts |
| NM_001338777.1 | ath-miR396b-5p | 193 | CDS | −87 | 82 | ||
| GR1 | NM_113322.5 | ath-miR824-5p | 2083 | 3′UTR | −87 | 80 | Glutathione reductase 1—regenerates GSH |
| Gene | NCBI Reference Sequence | miRNA | Start of Site, nt | Region | ΔG, kJ/mole | ΔG/ΔGm, % | Protein/Function |
|---|---|---|---|---|---|---|---|
| ABI2 | NM_001125976.2 | ath-miR781a | 164 | 5′UTR | −87 | 85 | Protein phosphatase 2C—negative regulator of ABA signaling |
| ARF2 | NM_001203662.1 | ath-miR866-3p | 308 | 5′UTR | −85 | 85 | Auxin response factor 2—represses auxin-responsive gene expression |
| ARF6 | NM_102771.4 | ath-miR167c-5p | 3340 | CDS | −98 | 88 | Auxin response factor 6—regulates auxin-responsive gene expression |
| NM_102771.4 | ath-miR167a,b-5p | 3341 | CDS | −91 | 83 | ||
| CKX1 | NM_001336920.1 | ath-miR407 | 939 | CDS | −81 | 83 | Cytokinin oxidase/dehydrogenase 1—degrades cytokinins |
| NM_001336920.1 | ath-miR870-3p | 2050 | 3′UTR | −85 | 82 | ||
| NM_001336920.1 | ath-miR390b-3p | 2070 | 3′UTR | −87 | 80 | ||
| EIN2 | NM_120406.5 | ath-miR847 | 3477 | CDS | −89 | 82 | Ethylene-insensitive 2—central regulator of ethylene signaling |
| NM_120406.5 | ath-miR854a-d | 26 | 5′UTR | −93 | 81 | ||
| EIN3 | NM_112968.4 | ath-miR172e-3p | 1589 | CDS | −89 | 82 | Ethylene-insensitive 3—activates ethylene-responsive transcription |
| NM_112968.4 | ath-miR2111b-3p | 759 | CDS | −89 | 81 | ||
| HK2 | NM_122966.3 | ath-miR835-5p | 1075 | CDS | −87 | 85 | Histidine kinase 2—cytokinin receptor in signaling |
| NM_122966.3 | ath-miR858a | 3525 | CDS | −87 | 80 | ||
| JAZ1 | NM_001332386.1 | ath-miR847 | 355 | 5′UTR | −87 | 80 | Jasmonate ZIM-domain protein 1—represses jasmonate-responsive transcription |
| PYL4 | NM_129387.3 | ath-miR1886.3 | 879 | CDS | −81 | 83 | ABA receptor—mediates abscisic acid stress responses |
| RCAR1 | NM_100018.5 | ath-miR840-3p | 126 | 5′UTR | −85 | 80 | ABA receptor—inhibits PP2Cs in response to abscisic acid |
| RCAR3 | NM_124695.4 | ath-miR866-3p | 180 | 5′UTR | −81 | 81 | ABA receptor—inhibits PP2Cs upon ABA perception |
| NM_124695.4 | ath-miR773a | 416 | 5′UTR | −87 | 80 | ||
| RD22 | NM_122472.4 | ath-miR398a-5p | 109 | 5′UTR | −89 | 81 | ABA-inducible protein—involved in drought and dehydration response |
| TIR1 | NM_116163.4 | ath-miR393a,b-5p | 1965 | CDS | −106 | 91 | Auxin receptor—mediates degradation of Aux/IAA repressors |
| NM_116163.4 | ath-miR854a-d | 218 | 5′UTR | −93 | 81 | ||
| NM_116163.4 | ath-miR854a-d | 224 | 5′UTR | −100 | 87 |
| Transcription Factors | Gene | NCBI Reference Sequence | miRNA | Start of Site, nt | Region | ΔG, kJ/ Mole | ΔG/ΔGm, % |
|---|---|---|---|---|---|---|---|
| AP2/ERF Family | DREB1A | NM_118680.2 | ath-miR414 | 1009 | CDS | −89 | 84 |
| DREB2A | NM_001036760.1 | ath-miR838 | 62 | 5′UTR | −87 | 84 | |
| ERF7 | NM_112922.3 | ath-miR418 | 154 | 5′UTR | −87 | 82 | |
| NAC Family | ANAC055 | NM_112418.4 | ath-miR393b-3p | 7 | 5′UTR | −89 | 84 |
| NAC083 | NM_121321.4 | ath-miR1886.3 | 1137 | CDS | −83 | 85 | |
| NM_121321.4 | ath-miR414 | 888 | CDS | −89 | 84 | ||
| NAC096 | NM_124029.3 | ath-miR863-3p | 874 | CDS | −85 | 82 | |
| NAC100 | NM_001345474.1 | ath-miR164a,b | 970 | CDS | −106 | 91 | |
| NAC102 | NM_001345612.1 | ath-miR158a-5p | 389 | CDS | −87 | 85 | |
| bZIP Family | ABF1 | NM_001198254.2 | ath-miR835-3p | 109 | 5′UTR | −87 | 80 |
| ABF3 | NM_001036708.3 | ath-miR169f-3p | 450 | CDS | −96 | 83 | |
| NM_001036708.3 | ath-miR866-3p | 677 | CDS | −83 | 83 | ||
| bZIP60 | NM_103458.3 | ath-miR414 | 388 | CDS | −93 | 88 | |
| NM_103458.3 | ath-miR414 | 391 | CDS | −89 | 84 | ||
| NM_103458.3 | ath-miR414 | 385 | CDS | −87 | 82 | ||
| NM_103458.3 | ath-miR414 | 394 | CDS | −87 | 82 | ||
| NM_103458.3 | ath-miR414 | 397 | CDS | −87 | 82 | ||
| bZIP68 | NM_102948.4 | ath-miR854a-d | 7 | 5′UTR | −96 | 83 | |
| MYB Family | MYB15 | NM_001035670.1 | ath-miR828 | 683 | CDS | −87 | 80 |
| NM_001035670.1 | ath-miR828 | 605 | CDS | −87 | 80 | ||
| MYB60 | NM_001331790.1 | ath-miR828 | 582 | CDS | −91 | 84 | |
| NM_001331790.1 | ath-miR414 | 965 | CDS | −87 | 82 | ||
| NM_001331790.1 | ath-miR414 | 959 | CDS | −87 | 82 | ||
| NM_001331790.1 | ath-miR414 | 971 | CDS | −85 | 80 | ||
| MYB96 | NM_125641.4 | ath-miR828 | 605 | CDS | −91 | 84 | |
| NM_125641.4 | ath-miR828 | 319 | CDS | −91 | 84 | ||
| MYB102 | NM_118264.3 | ath-miR828 | 439 | CDS | −96 | 88 | |
| NM_118264.3 | ath-miR829-5p | 978 | CDS | −85 | 83 | ||
| MYB108 | NM_111525.4 | ath-miR172b,e-5p | 1382 | CDS | −87 | 84 | |
| NM_111525.4 | ath-miR834 | 1011 | CDS | −91 | 81 | ||
| NM_111525.4 | ath-miR858a | 837 | CDS | −87 | 80 | ||
| MYB116 | NM_001036014.2 | ath-miR858a | 293 | CDS | −89 | 82 | |
| NM_001036014.2 | ath-miR858a | 498 | CDS | −89 | 82 | ||
| WRKY Family | WRKY18 | NM_001342115.1 | ath-miR781a | 1105 | 3′UTR | −85 | 83 |
| WRKY25 | NM_128578.4 | ath-miR403-5p | 658 | CDS | −87 | 82 | |
| WRKY33 | NM_129404.4 | ath-miR845a | 1167 | CDS | −89 | 81 | |
| WRKY40 | NM_106732.4 | ath-miR838 | 617 | CDS | −87 | 84 | |
| NM_106732.4 | ath-miR156g | 282 | CDS | −89 | 82 | ||
| NM_106732.4 | ath-miR868-5p | 437 | CDS | −87 | 82 | ||
| WRKY57 | NM_001334406.1 | ath-miR472-5p | 328 | CDS | −89 | 82 | |
| WRKY63 | NM_105331.4 | ath-miR855 | 224 | CDS | −89 | 81 | |
| WRKY75 | NM_121311.5 | ath-miR847 | 255 | CDS | −89 | 82 | |
| NM_121311.5 | ath-miR407 | 337 | CDS | −79 | 80 | ||
| HD-ZIP Family | ATHB7 | NM_001036473.1 | ath-miR865-3p | 786 | CDS | −83 | 83 |
| ATHB54 | NM_001198174.2 | ath-miR447a.2-3p | 224 | 5′UTR | −83 | 83 | |
| C2H2 Family | ZAT10 | NM_102538.3 | ath-miR778 | 623 | CDS | −91 | 83 |
| CAMTA Family | CAMTA3 | NM_001124889.2 | ath-miR844-3p | 2623 | CDS | −83 | 81 |
| NM_001124889.2 | ath-miR2112-3p | 1938 | CDS | −87 | 80 | ||
| CAMTA6 | NM_001338268.1 | ath-miR857 | 2161 | CDS | −83 | 81 | |
| NF-YA Family | NF-YA1 | NM_001343248.1 | miR169 family | 1147/1148 | 3′UTR | −98/−102 | 85/87 |
| NF-YA2 | NM_111443.4 | miR169 family | 1312/1313 | 3′UTR | −96/−100 | 82–89 | |
| NF-YA3 | NM_001036195.5 | miR169 family | 1413 | 3′UTR | −93/−98 | 80–87 | |
| NF-YA5 | NM_104294.3 | miR169 family | 1284 | 3′UTR | −96/−100 | 85/87 | |
| NF-YA6 | NM_001338098.1 | miR169 family | 1271 | 3′UTR | −93/−100 | 80–89 | |
| NF-YA8 | NM_001198095.1 | miR169 family | 1505 | 3′UTR | −98 | 87 | |
| NF-YA9 | NM_112983.5 | miR169 family | 1062/1063 | 3′UTR | −93/−96 | 80/83 | |
| NF-YA10 | NM_001342883.1 | miR169 family | 735/736 | 3′UTR | −96/−100 | 82/89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhakypbek, Y.; Rakhmetullina, A.; Kamarkhan, Z.; Tursbekov, S.; Shi, Q.; Xing, F.; Pyrkova, A.; Ivashchenko, A.; Kossalbayev, B.D.; Belkozhayev, A.M. In Silico Analysis of miRNA-mRNA Binding Sites in Arabidopsis thaliana as a Model for Drought-Tolerant Plants. Plants 2025, 14, 1800. https://doi.org/10.3390/plants14121800
Zhakypbek Y, Rakhmetullina A, Kamarkhan Z, Tursbekov S, Shi Q, Xing F, Pyrkova A, Ivashchenko A, Kossalbayev BD, Belkozhayev AM. In Silico Analysis of miRNA-mRNA Binding Sites in Arabidopsis thaliana as a Model for Drought-Tolerant Plants. Plants. 2025; 14(12):1800. https://doi.org/10.3390/plants14121800
Chicago/Turabian StyleZhakypbek, Yryszhan, Aizhan Rakhmetullina, Zhigerbek Kamarkhan, Serik Tursbekov, Qingdong Shi, Fei Xing, Anna Pyrkova, Anatoliy Ivashchenko, Bekzhan D. Kossalbayev, and Ayaz M. Belkozhayev. 2025. "In Silico Analysis of miRNA-mRNA Binding Sites in Arabidopsis thaliana as a Model for Drought-Tolerant Plants" Plants 14, no. 12: 1800. https://doi.org/10.3390/plants14121800
APA StyleZhakypbek, Y., Rakhmetullina, A., Kamarkhan, Z., Tursbekov, S., Shi, Q., Xing, F., Pyrkova, A., Ivashchenko, A., Kossalbayev, B. D., & Belkozhayev, A. M. (2025). In Silico Analysis of miRNA-mRNA Binding Sites in Arabidopsis thaliana as a Model for Drought-Tolerant Plants. Plants, 14(12), 1800. https://doi.org/10.3390/plants14121800






