Biosynthesis of Camphane Volatile Terpenes in Amomum villosum Lour: Involved Genes and Enzymes
Abstract
1. Introduction
2. Results
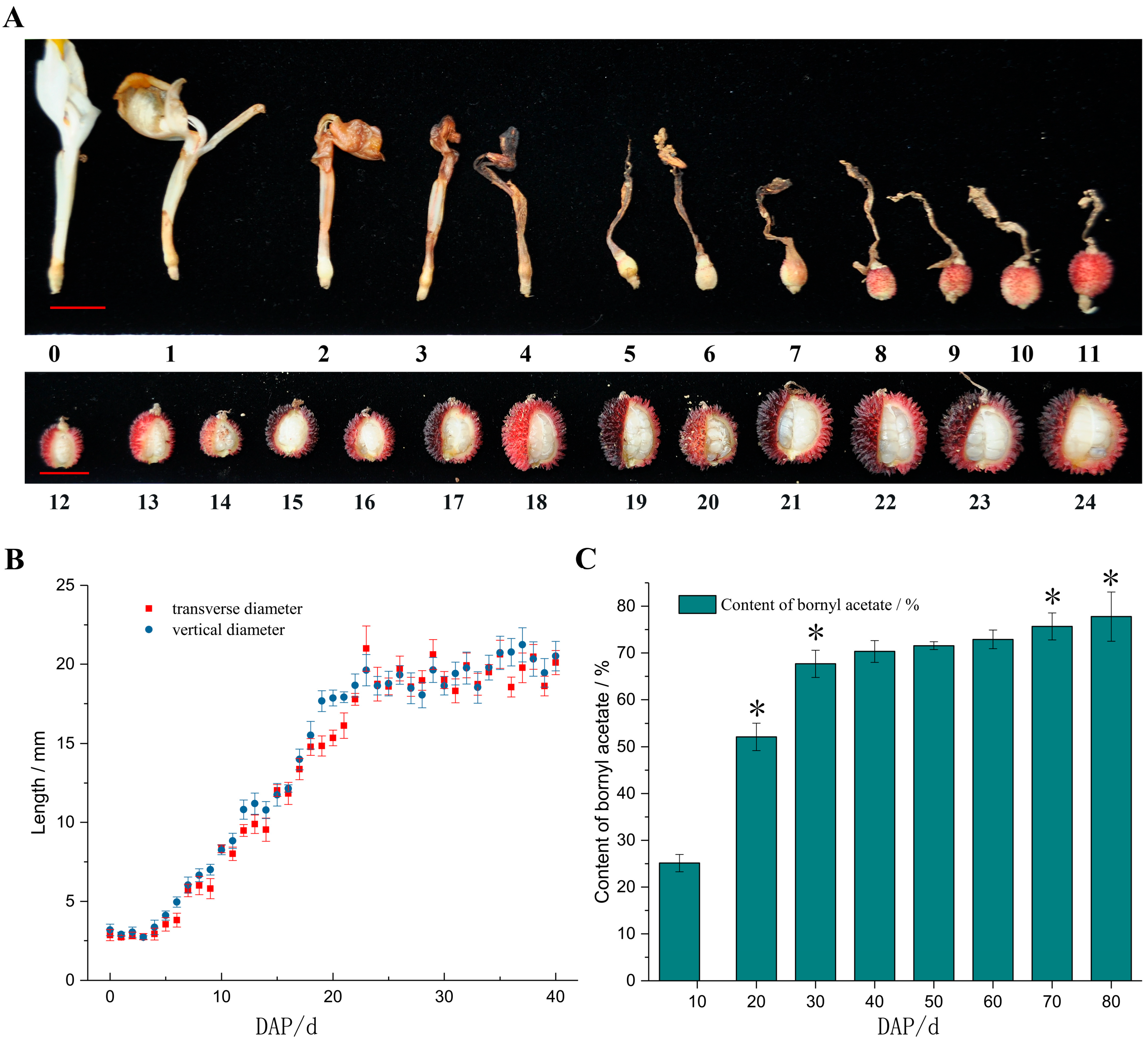
2.1. Morphological Characteristics and Quantitative Analysis of Bornyl Acetate of A. villosum Fruits
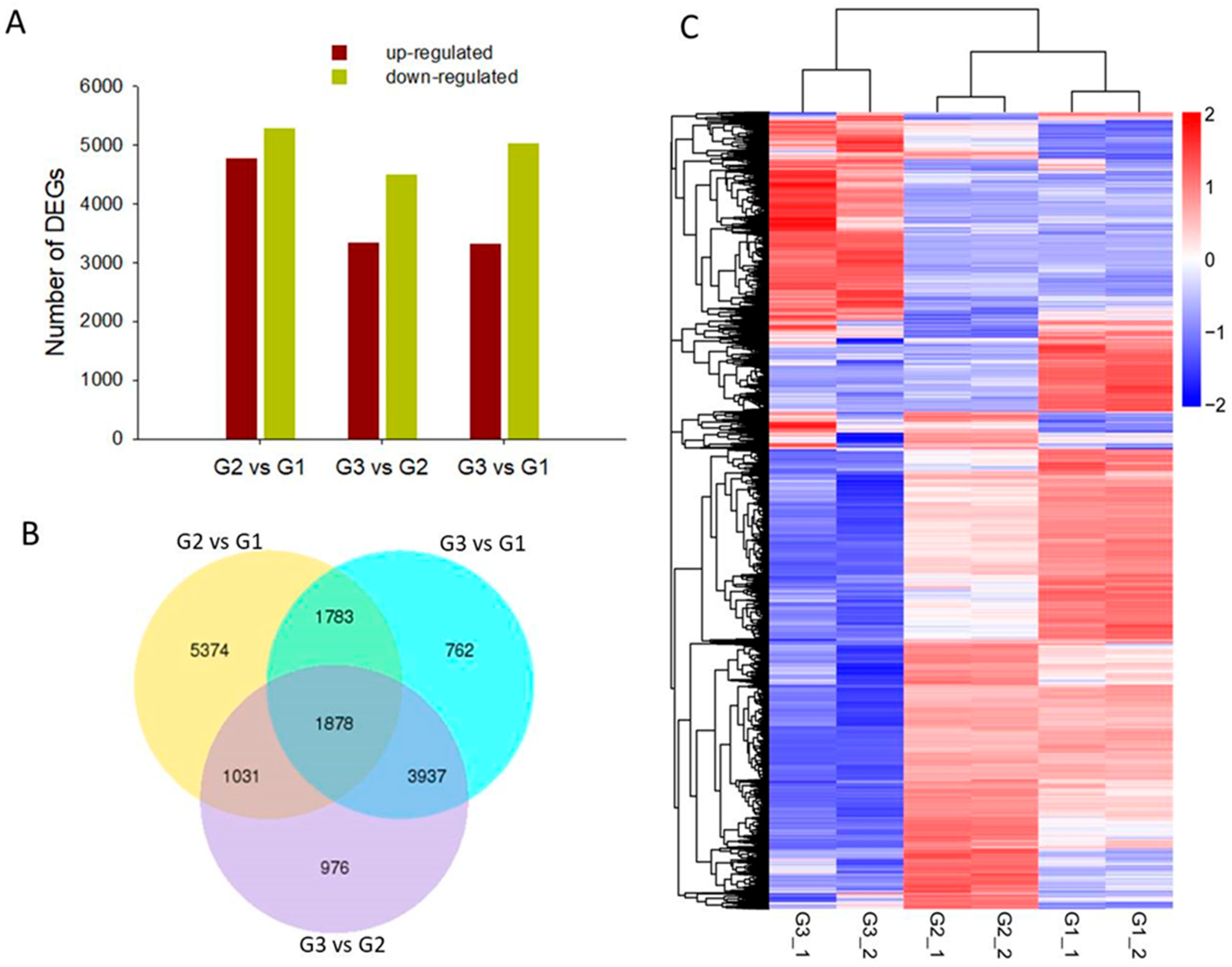
2.2. Differentially Expressed Gene (DEG) Identification
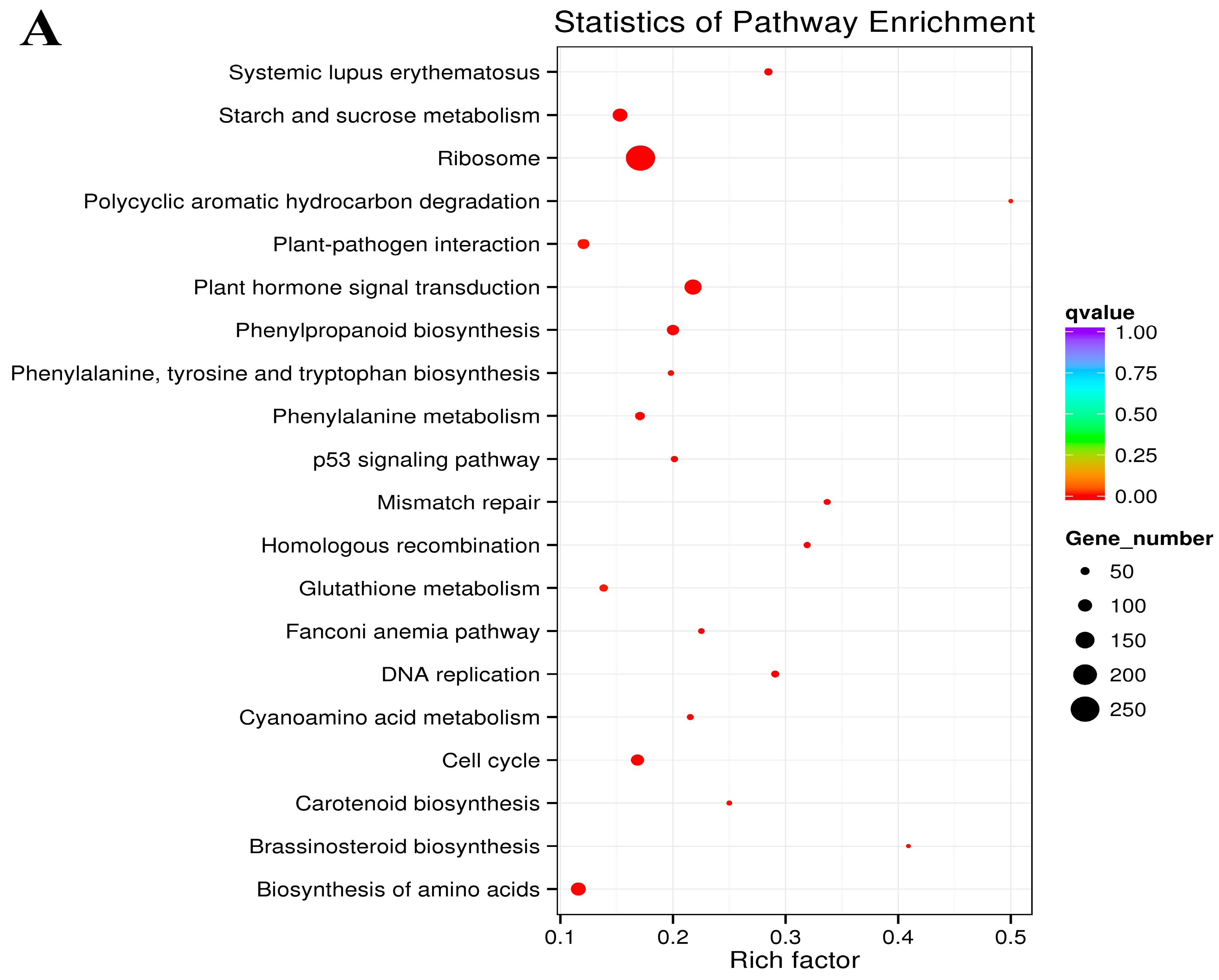
2.3. Functional Classification of the DEGs
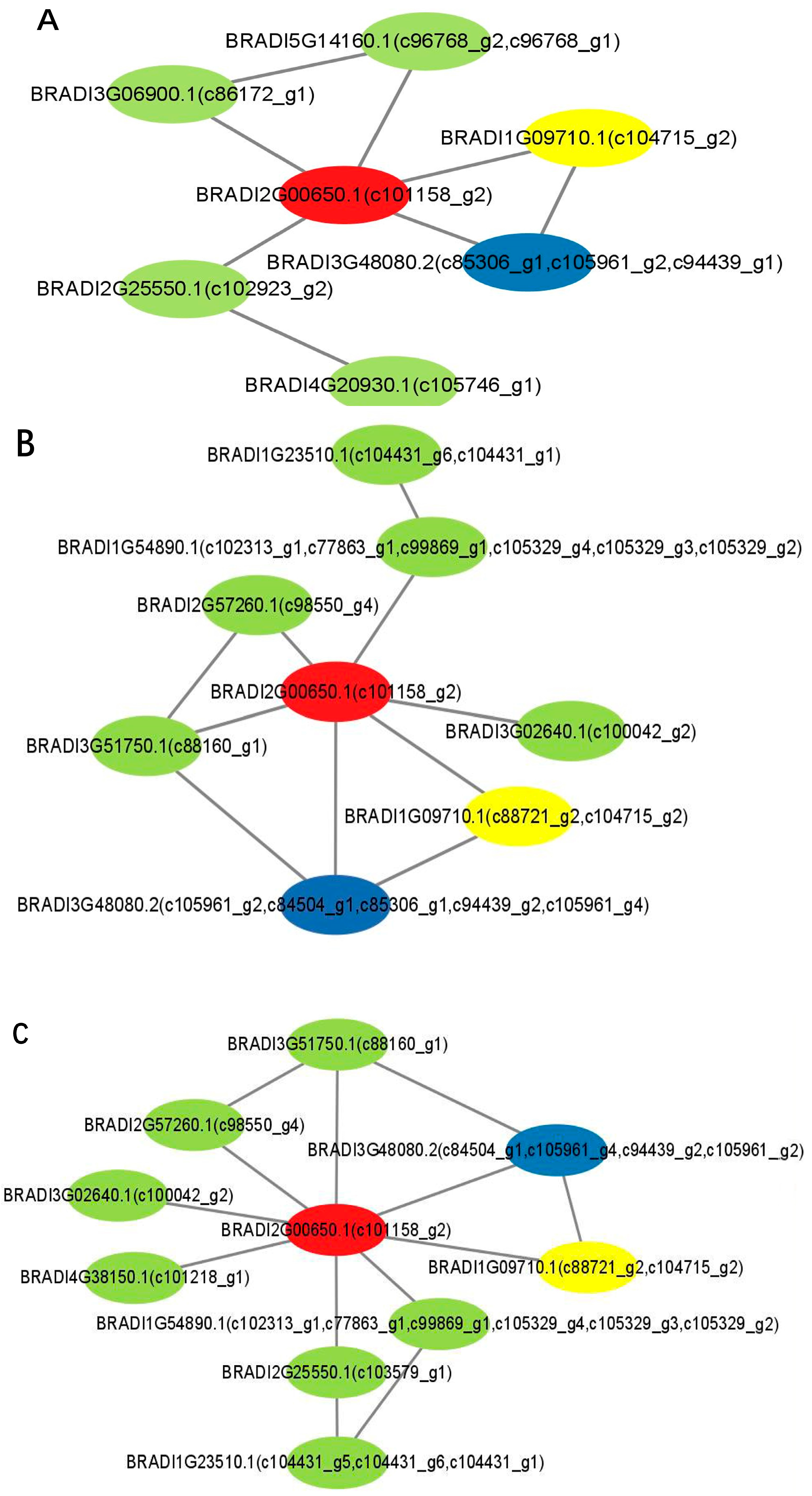
2.4. Network Analysis of Protein Interaction Based on the DEGs
2.5. Unigenes Involved in Backbone Biosynthesis of Camphane Volatile Terpenes
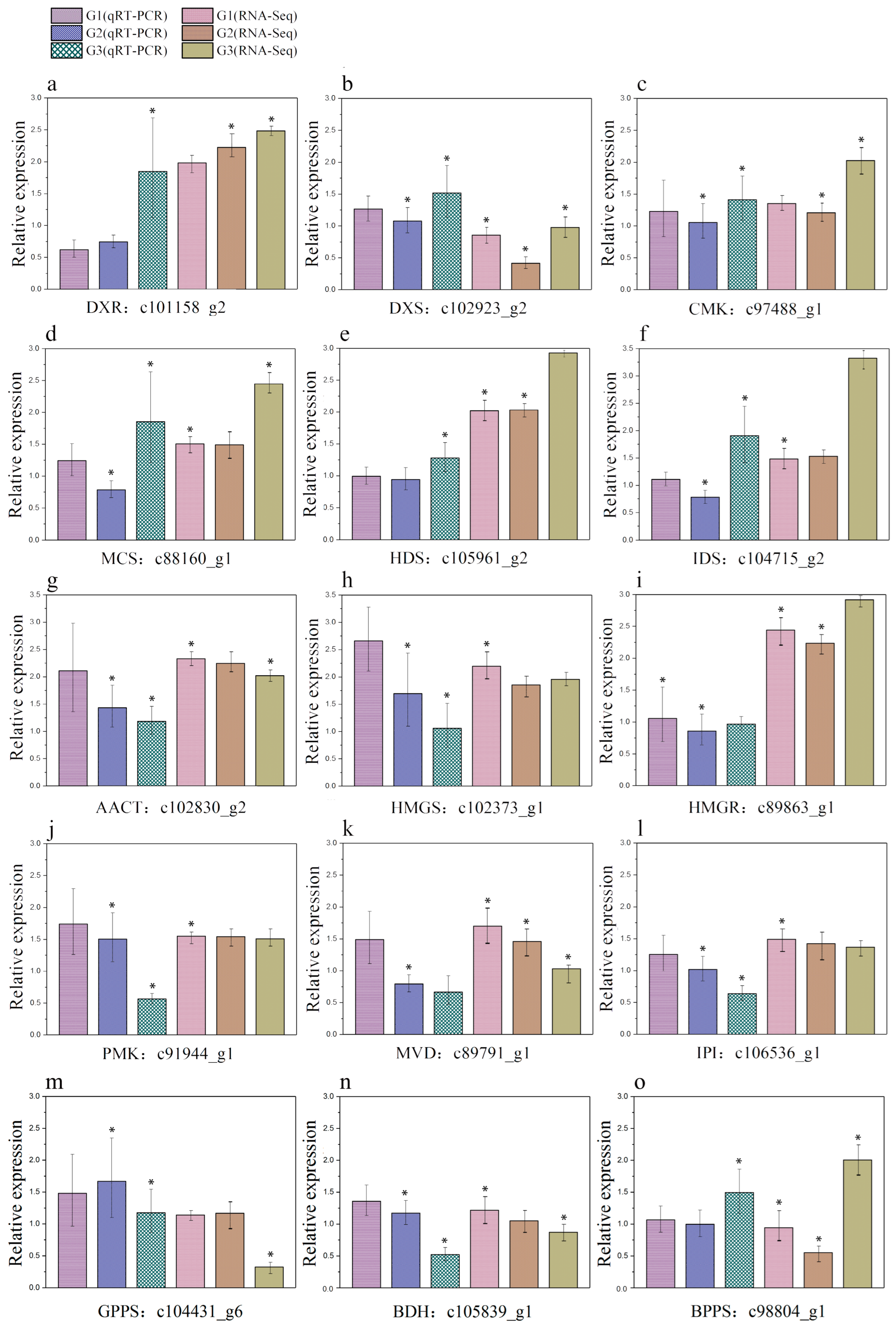
2.6. Verification of RNA-Seq Data by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Collection and Preparation of Samples
4.2. Growth Responses and Content Determination of Bornyl Acetate in A. villosum
4.3. Construction of Libraries, Sequence Reads Mapping, Assembly and Annotation
4.4. Gene Expression
4.5. RT-qPCR Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| DEGs | different expression genes |
| BAHD | family of enzymes found primarily in plants |
| DXR | 1-deoxy-D-xylulose-5-phosphate reductoisomerase |
| HDS | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| IDS | diphosphate reductase |
| PPI | protein–protein interaction |
References
- Zuo, X.; Wang, Y.; Zhao, H.; Li, G.; Wang, Y.; Li, G.; Zhang, L.; Gao, W. Allelopathic Effects of Amomum villosum Lour. Volatiles from Different Organs on Selected Plant Species and Soil Microbiota. Plants 2022, 11, 3550. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wu, H.; Qu, M.; Liu, Y.; Wang, D.; Yang, H.; Wang, Y.; Wang, X.; Blasi, F. Enhancement of Oxidative Stability of Deep-Fried Sunflower Oil by Addition of Essential Oil of Amomum villosum Lour. Antioxidants 2023, 12, 1429. [Google Scholar] [CrossRef]
- Zhao, H.; Li, M.; Zhao, Y.; Lin, X.; Liang, H.; Wei, J.; Wei, W.; Ma, D.; Zhou, Z.; Yang, J. A Comparison of Two Monoterpenoid Synthases Reveals Molecular Mechanisms Associated With the Difference of Bioactive Monoterpenoids Between Amomum villosum and Amomum longiligulare. Front. Plant Sci. 2021, 12, 695551. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Li, X.; Lan, L.; Wang, J.; Guo, P.; Sun, G. Simultaneous determination of eight components in Amomum villosum and its overall qualityconsistency evaluation by four-dimensional fingerprints assisted with antioxidant activity. J. Chromatogr. A 2022, 1674, 463135. [Google Scholar] [CrossRef]
- Tu, X.; Liu, Y.; Yao, Y.; Li, W.; Lou, P.; Du, L.; He, J.; Jian-Neng, L. Effects of four drying methods on Amomum villosum Lour. “Guiyan1” volatile organic compounds analyzed via headspace solid phase microextraction and gas chromatography-mass spectrometry coupled with OPLS-DA. RSC Adv. 2022, 12, 26485–26496. [Google Scholar] [CrossRef]
- Lai, Y.-F.; Chen, L.-X.; Chen, Y.-N.; Zhao, J.; Leong, F.; Li, X.-W.; Yang, Q.; Li, P.; Hu, H. Sustainable Development of Amomum villosum: A Systematic Investigation on Three Different Production Modes. Afr. J. Tradit. Complement. Altern. Med. 2016, 13, 97–104. [Google Scholar] [CrossRef]
- Gong, L.; Zhang, D.; Ding, X.; Huang, J.; Guan, W.; Qiu, X.; Huang, Z. DNA barcode reference library construction and genetic diversity and structure analysis of Amomum villosum Lour. (Zingiberaceae) populations in Guangdong Province. PeerJ 2021, 9, e12325. [Google Scholar] [CrossRef]
- Feng, L.; Wang, Z.; Lei, Z.; Zhang, X.; Zhai, B.; Sun, J.; Guo, D.; Wang, D.; Luan, F.; Zou, J.; et al. Amomum villosum Lour.: An insight into ethnopharmacological, phytochemical, and pharmacological overview. J. Ethnopharmacol. 2024, 33, 118615. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-R.; Paulrayer, A.; Kwon, Y.-G.; Ryu, D.-G.; Baek, D.-G.; Geum, J.-H.; Lee, J.-H.; Lee, G.-S.; Kwon, K.-B. Acute effects of Amomum villosum Lour. fruit extract on postprandial glycemia and insulin secretion: A single-blind, placebo-controlled, crossover study in healthy subjects. Saudi J. Biol. Sci. 2020, 27, 2968–2971. [Google Scholar] [CrossRef]
- Lee, Y.-G.; Park, J.-H.; Jeon, E.-S.; Kim, J.-H.; Lim, B.-K. Fructus Amomi Cardamomi Extract Inhibits Coxsackievirus-B3 Induced Myocarditis in a Murine Myocarditis Model. J. Microbiol. Biotechnol. 2016, 26, 2012–2018. [Google Scholar] [CrossRef]
- Yan, Y.; Li, X.; Wan, M.; Chen, J.; Li, S.; Cao, M.; Zhang, D. Effect of extraction methods on property and bioactivity of water-soluble polysaccharides from Amomum villosum. Carbohydr. Polym. 2015, 117, 632–635. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Qian, C.; Yang, D.; Tang, C.; Xu, X.; Liu, E.H.; Zhong, J.; Zhu, L.; Zhao, Z. Purification, structural characterization and immunomodulatory effects of polysaccharides from Amomumvillosum Lour. on RAW 264.7 macrophages. Molecules 2021, 26, 2672. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Li, J.; Li, Y.; Su, J.; Ding, X.; Yuan, Y.; Liu, S.; Mou, Y.; Li, G.; Zhang, L. Unveiling the potential mechanisms of Amomi fructus against gastric ulcers via integrating network pharmacology and in vivo experiments. J. Ethnopharmacol. 2024, 319, 117179. [Google Scholar] [CrossRef]
- Shengtan, Z.; Zhaoyu, W.; Tiesheng, W.; Miaoxia, L.; Jingming, L. Composition and Antimicrobial Activities of Essential Oil of Fructus Amomi. Nat. Prod. Res. Dev. 2011, 23, 464–472. [Google Scholar]
- National Pharmacopoeia Committee. Pharmacopoeia of the People’s Republic of China; China Medical Science Press: Beijing, China, 2015. [Google Scholar]
- Ao, H.; Wang, J.; Chen, L.; Li, S.; Dai, C. Comparison of Volatile Oil between the Fruits of Amomum villosum Lour. and Amomum villosum Lour. var. xanthioides T. L. Wu et Senjen Based on GC-MS and Chemometric Techniques. Molecules 2019, 24, 1663. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Luo, X.; Yao, Y.; Wang, Y.; Dai, Z.; Cheng, T.; Huang, X.; Bai, M.; He, J.; Wu, H. Integrated Analysis of Metabolomics, Flavoromics, and Transcriptomics for Evaluating New Varieties of Amomum villosum Lour. Plants 2024, 13, 2382. [Google Scholar] [CrossRef]
- Yang, Z.; Zhan, T.; Xie, C.; Huang, S.; Zheng, X. Genome-wide analyzation and functional characterization on the TPS family provide insight into the biosynthesis of mono-terpenes in the camphor tree. Plant Physiol. Biochem. 2023, 196, 55–64. [Google Scholar] [CrossRef]
- Barros, F.M.C.d.; Zambarda, E.d.O.; Heinzmann, B.M.; Mallmann, C.A. Variabilidade sazonal e biossíntese de terpenóides presentes no óleo essencial de Lippia alba (Mill.) N. E. Brown (Verbenaceae). Química Nova 2009, 32, 861–867. [Google Scholar] [CrossRef]
- Jiang, H.; Wang, X. Biosynthesis of monoterpenoid and sesquiterpenoid as natural flavors and fragrances. Biotechnol. Adv. 2023, 65, 108151. [Google Scholar] [CrossRef]
- Singh, S.; Apoorva; Saha, P.; Rai, N.; Kumari, S.; Pandey-Rai, S. Unravelling triterpenoid biosynthesis in plants for applications in bioengineering and large-scale sustainable production. Ind. Crop. Prod. 2023, 199, 116789. [Google Scholar] [CrossRef]
- Despinasse, Y.; Fiorucci, S.; Antonczak, S.; Moja, S.; Bony, A.; Nicolè, F.; Baudino, S.; Magnard, J.-L.; Jullien, F. Bornyl-diphosphate synthase from Lavandula angustifolia: A major monoterpene synthase involved in essential oil quality. Phytochemistry 2017, 137, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Adhikari, M.N.; Liu, H.; Xu, H.; He, G.; Zhan, R.; Wei, J.; Chen, W. Characterization and functional analysis of the genes encoding 1-deoxy-D-xylulose-5-phosphate reductoisomerase and 1-deoxy-D-xylulose-5-phosphate synthase, the two enzymes in the MEP pathway, from Amomum villosum Lour. Mol. Biol. Rep. 2012, 39, 8287–8296. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, J.; Köllner, T.G.; Gershenzon, J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 2009, 70, 15–16. [Google Scholar] [CrossRef]
- Yang, X.; Nambou, K.; Wei, L.; Cai, J. Heterologous production of α-pinene in E. coli via synthetic biology approaches. Metab. Eng. 2016, 38, 472–478. [Google Scholar]
- Kumar, K.R.; Cowley, M.J.; Davis, R.L. Next-Generation Sequencing and Emerging Technologies. Semin. Thromb. Hemost. 2024, 50, 1026–1038. [Google Scholar] [CrossRef]
- Pan, X.; Durrett, R.E.; Zhu, H.; Tanaka, Y.; Li, Y.; Zi, X.; Marjani, S.L.; Euskirchen, G.; Ma, C.; Lamotte, R.H.; et al. Two methods for full-length RNA sequencing for low quantities of cells and single cells. Proc. Natl. Acad. Sci. USA 2013, 110, 594–599. [Google Scholar] [CrossRef]
- Cheng, J.; Chen, J.; Liao, J.; Wang, T.; Shao, X.; Long, J.; Yang, P.; Li, A.; Wang, Z.; Lu, X.; et al. High-throughput transcriptional profiling of perturbations by Panax ginseng saponins and Panax notoginseng saponins using TCM-seq. J. Pharm. Anal. 2023, 13, 376–387. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Yan, Y.; Zhou, B.; Li, J.; Cui, L.; Guo, L.; Liu, W. Metabolomic and transcriptomic analyses highlight metabolic regulatory networks of Salvia miltiorrhiza in response to replant disease. BMC Plant Biol. 2024, 24, 575. [Google Scholar] [CrossRef]
- Yu, B.; Pan, Y.; Liu, Y.; Chen, Q.; Guo, X.; Tang, Z. A comprehensive analysis of transcriptome and phenolic compound profiles suggests the role of flavonoids in cotyledon greening in Catharanthus roseus seedling. Plant Physiol. Biochem. 2021, 167, 185–197. [Google Scholar] [CrossRef]
- Jiao, C.; Wei, M.; Fan, H.; Song, C.; Wang, Z.; Cai, Y.; Jin, Q. Transcriptomic analysis of genes related to alkaloid biosynthesis and the regulation mechanism under precursor and methyl jasmonate treatment in Dendrobium officinale. Front. Plant Sci. 2022, 13, 941231. [Google Scholar] [CrossRef]
- Adam, K.P.; Croteau, R. Monoterpene Biosynthesis in the Liverwort Conocephalum Conicum: Demonstration of Sabinene Synthase and Bornyl Diphosphate Synthase. Phytochemistry 1998, 49, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Laule, O.; Fürholz, A.; Chang, H.S.; Zhu, T.; Wang, X.; Heifetz, P.; Gruissem, W.; Lange, B.M. Crosstalk between Cytosolic and Plastidial Pathways of Isoprenoid Biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 6866–6871. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.K.; Sangwan, N.S.; Bose, S.K.; Sangwan, R.S. Biochemical characteristics of a novel vegetative tissue geraniol acetyltransferase from a monoterpene oil grass (Palmarosa, Cymbopogon martinii var. Motia) leaf. Plant Sci. 2013, 203, 63–73. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, B.; Luca, V.D. Chapter Nine Evolution of acyltransferase genes: Origin and diversification fo the BAHD superfamily of acyltransferases involved in secondary metabolism. Recent Adv. Phytochem. 2000, 34, 285–315. [Google Scholar]
- Tuominen, L.K.; Johnson, V.E.; Tsai, C.J. Differential phylogenetic expansions in BAHD acyltransferases across five angiosperm taxa and evidence of divergent expression among Populus paralogues. BMC Genom. 2011, 12, 236. [Google Scholar] [CrossRef]
- Sarker, L.S.; Galata, M.; Demissie, Z.A.; Mahmoud, S.S. Molecular cloning and functional characterization of borneol dehydrogenase from the glandular trichomes of Lavandula x intermedia. Arch. Biochem. Biophys. 2012, 528, 163–170. [Google Scholar] [CrossRef]
- Okamoto, S.; Yu, F.; Harada, H.; Okajima, T.; Hattan, J.; Misawa, N.; Utsumi, R. A short-chain dehydrogenase involved in terpene metabolism from Zingiber zerumbet. FEBS J. 2011, 278, 2892–2900. [Google Scholar] [CrossRef] [PubMed]
- Thumma, B.R.; Sharma, N.; Southerton, S.G. Transcriptome sequencing of Eucalyptus camaldulensis seedlings subjected to water stress reveals functional single nucleotide polymorphisms and genes under selection. BMC Genom. 2012, 13, 364. [Google Scholar] [CrossRef]
- Tang, Q.; Ma, X.; Mo, C.; Wilson, I.W.; Song, C.; Zhao, H.; Yang, Y.; Fu, W.; Qiu, D. An efficient approach to finding Siraitia grosvenorii triterpene biosynthetic genes by RNA-seq and digital gene expression analysis. BMC Genom. 2011, 12, 343. [Google Scholar] [CrossRef]
- Tyagi, P.; Singh, D.; Mathur, S.; Singh, A.; Ranjan, R. Upcoming progress of transcriptomics studies on plants: An overview. Front. Plant Sci. 2022, 13, 1030890. [Google Scholar] [CrossRef]
- Wei, S.; Ma, X.; Pan, L.; Miao, J.; Fu, J.; Bai, L.; Zhang, Z.; Guan, Y.; Mo, C.; Huang, H.; et al. Transcriptome Analysis of Taxillusi chinensis (DC.) Danser Seeds in Response to Water Loss. PLoS ONE 2017, 12, e0169177. [Google Scholar] [CrossRef] [PubMed]
- YU, A.; Wang, H.; HE, X.; Deng, K.; Zhan, R.; Yang, J. Screening of Reference Genes for Real-time Fluorescence Quantitative PCR in Amomum villosum Lour. J. Guangzhou Univ. Tradit. Chin. Med. 2014, 31, 814–820. [Google Scholar] [CrossRef]
- He, X.; Wang, H.; Yang, J.; Deng, K.; Wang, T. RNA sequencing on Amomum villosum Lour. induced by MeJA identifies the genes of WRKY and terpene synthases involved in terpene biosynthesis. Genome 2018, 61, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ding, G.; Li, B.; Guo, S.X. Transcriptome Analysis of Genes Involved in Dendrobine Biosynthesis in Dendrobium nobile Lindl. Infected with Mycorrhizal Fungus MF23 (Mycena sp.). Sci. Rep. 2017, 7, 316. [Google Scholar] [CrossRef]
- Gao, Y.; He, X.; Wu, B.; Long, Q.; Shao, T.; Wang, Z.; Wei, J.; Li, Y.; Ding, W. Time-Course Transcriptome Analysis Reveals Resistance Genes of Panax ginseng Induced by Cylindrocarpon destructans Infection Using RNA-Seq. PLoS ONE 2016, 11, e0149408. [Google Scholar] [CrossRef]
- Wang, R.; Xu, S.; Wang, N.; Xia, B.; Jiang, Y.; Wang, R. Transcriptome Analysis of Secondary Metabolism Pathway, Transcription Factors, and Transporters in Response to Methyl Jasmonate in Lycoris aurea. Front. Plant Sci. 2017, 7, 1971. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Luo, H.M.; Sun, C.; Song, J.Y.; Sun, Y.Z.; Wu, Q.; Wang, N.; Yao, H.; Steinmetz, A.; Chen, S.L. EST analysis reveals putative genes involved in glycyrrhizin biosynthesis. BMC Genom. 2010, 11, 268. [Google Scholar] [CrossRef]
- Yang, P.; Chen, X.M.; Liu, W.W.; Feng, Y.; Sun, T. Transcriptome analysis of sexually dimorphic Chinese white wax scale insects reveals key differences in developmental programs and transcription factor expression. Sci. Rep. 2015, 5, 8141. [Google Scholar] [CrossRef]
- Huang, Z.-Y.; Ye, R.-Y.; Yu, H.-L.; Li, A.-T.; Xu, J.-H. Mining methods and typical structural mechanisms of terpene cyclases. Bioresour. Bioprocess. 2021, 8, 1–27. [Google Scholar] [CrossRef]
- Wang, H.; Ma, D.; Yang, J.; Deng, K.; Li, M.; Ji, X.; Zhong, L.; Zhao, H. An Integrative Volatile Terpenoid Profiling and Transcriptomics Analysis for Gene Mining and Functional Characterization of AvBPPS and AvPS Involved in the Monoterpenoid Biosynthesis in Amomum villosum. Front. Plant Sci. 2018, 9, 846. [Google Scholar] [CrossRef]
- Fuchs, L.K.; Holland, A.H.; Ludlow, R.A.; Coates, R.J.; Armstrong, H.; Pickett, J.A.; Harwood, J.L.; Scofield, S. Genetic Manipulation of Biosynthetic Pathways in Mint. Front. Plant Sci. 2022, 13, 928178. [Google Scholar] [CrossRef] [PubMed]
- Major, D.T.; Weitman, M. Electrostatically guided dynamics—The root of fidelity in a promiscuous terpene synthase? J. Am. Chem. Soc. 2012, 134, 19454–19462. [Google Scholar] [CrossRef]
- Yang, C.Q.; Fang, X.; Wu, X.M.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Transcriptional regulation of plant secondary metabolism. J. Integr. Plant Biol. 2012, 54, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Spyropoulou, E.A.; Haring, M.A.; Schuurink, R.C. RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genom. 2014, 15, 402. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Patra, B.; Li, R.; Pattanaik, S.; Yuan, L. Promoter analysis reveals cis-regulatory motifs associated with the expression of the WRKY transcription factor CrWRKY1 in Catharanthus roseus. Planta 2013, 238, 1039–1049. [Google Scholar] [CrossRef]
- Shoji, T.; Kajikawa, M.; Hashimoto, T. Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 2010, 22, 3390–3409. [Google Scholar] [CrossRef]
- Lenka, S.K.; Nims, N.E.; Vongpaseuth, K.; Boshar, R.A.; Roberts, S.C.; Walker, E.L. Jasmonate-responsive expression of paclitaxel biosynthesis genes in Taxus cuspidata cultured cells is negatively regulated by the bHLH transcription factors TcJAMYC1, TcJAMYC2, and TcJAMYC4. Front. Plant Sci. 2015, 6, 115. [Google Scholar] [CrossRef]
- Yamada, Y.; Sato, F. Transcription Factors in Alkaloid Engineering. Biomolecules 2021, 11, 1719. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, G.; Liu, M.; Wang, L.; Lou, Y.; Baldwin, I.; Li, R. Multiomic analyses reveal key sectors of jasmonate-mediated defense responses in rice. Plant Cell 2024, 36, 3362–3377. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, J.; Xue, L.; Zhao, T.; Ding, W.; Han, Y.; Ye, H. A comparison of transcriptome analysis methods with reference genome. BMC Genom. 2022, 23, 232. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, T.; Zhang, C.; Ma, Z. RNA-seq based transcriptome analysis of ethanol extract of saffron protective effect against corticosterone-induced PC12 cell injury. BMC Complement. Med. Ther. 2022, 22, 29. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, 1–12. [Google Scholar] [CrossRef]
- Shen, X.N.; Wang, X.D.; Wan, F.H.; Lü, Z.C.; Liu, W.X. Gene Expression Analysis Reveals Potential Regulatory Factors Response to Temperature Stress in Bemisia tabaci Mediterranean. Genes 2023, 14, 1013. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Tang, D.; Huang, Q.; Wei, K.; Yang, X.; Wei, F.; Miao, J. Identification of Differentially Expressed Genes and Pathways Involved in Growth and Development of Mesona chinensis Benth Under Red- and Blue-Light Conditions. Front. Plant Sci. 2021, 12, 761068. [Google Scholar] [CrossRef]
- Chen, J.C.; Xie, T.A.; Lin, Z.Z.; Li, Y.Q.; Xie, Y.F.; Li, Z.W.; Guo, X.G. Identification of Key Pathways and Genes in SARS-CoV-2 Infecting Human Intestines by Bioinformatics Analysis. Biochem. Genet. 2022, 60, 1076–1094. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Zhao, Y.; Li, H.; Hou, W.; Tang, X.; Zhou, R. A simple and rapid method for isolating high-quality RNA from kenaf with high polysaccharide and polyphenol contents. Biotechniques 2023, 75, 218–226. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, L.; Meng, F.; Zhang, T.; Luo, J.; Chen, S.; Shi, H.; Liu, B.; Lv, Z. The Effect of Temperature on the Embryo Development of Cephalopod Sepiella japonica Suggests Crosstalk between Autophagy and Apoptosis. Int. J. Mol. Sci. 2023, 24, 15365. [Google Scholar] [CrossRef]
- Li, G.; Zhou, X.; Kong, L.; Wang, Y.; Zhang, H.; Zhu, H.; Mitchell, T.K.; Dean, R.A.; Xu, J.R. MoSfl1 is important for virulence and heat tolerance in Magnaporthe oryzae. PLoS ONE 2011, 6, e19951. [Google Scholar] [CrossRef]
- Zhao, J.; Sun, P.; Sun, Q.; Li, R.; Qin, Z.; Sha, G.; Zhou, Y.; Bi, R.; Zhang, H.; Zheng, L.; et al. The MoPah1 phosphatidate phosphatase is involved in lipid metabolism, development, and pathogenesis in Magnaporthe oryzae. Mol. Plant Pathol. 2022, 23, 720–732. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Li, Y.; Zhang, P.; Luo, Z.; Yin, J.; Ma, X.; Yuan, C. Biosynthesis of Camphane Volatile Terpenes in Amomum villosum Lour: Involved Genes and Enzymes. Plants 2025, 14, 1767. https://doi.org/10.3390/plants14121767
Guo Y, Li Y, Zhang P, Luo Z, Yin J, Ma X, Yuan C. Biosynthesis of Camphane Volatile Terpenes in Amomum villosum Lour: Involved Genes and Enzymes. Plants. 2025; 14(12):1767. https://doi.org/10.3390/plants14121767
Chicago/Turabian StyleGuo, Yuhua, Yamei Li, Pengfei Zhang, Zuliang Luo, Junmei Yin, Xiaojun Ma, and Chao Yuan. 2025. "Biosynthesis of Camphane Volatile Terpenes in Amomum villosum Lour: Involved Genes and Enzymes" Plants 14, no. 12: 1767. https://doi.org/10.3390/plants14121767
APA StyleGuo, Y., Li, Y., Zhang, P., Luo, Z., Yin, J., Ma, X., & Yuan, C. (2025). Biosynthesis of Camphane Volatile Terpenes in Amomum villosum Lour: Involved Genes and Enzymes. Plants, 14(12), 1767. https://doi.org/10.3390/plants14121767




