Phylogeography and Population Demography of Parrotia subaequalis, a Hamamelidaceous Tertiary Relict ‘Living Fossil’ Tree Endemic to East Asia Refugia: Implications from Molecular Data and Ecological Niche Modeling
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, DNA Extraction, cpDNA Sequencing, and EST-SSR Genotyping
2.2. Population Genetic, Phylogeographic, and Demographic Analyses of Chloroplast DNA Markers
2.3. Population Genetic Diversity, Structure, and Demographic Analyses of EST-SSRs
2.4. Ecological Niche Modeling
3. Results
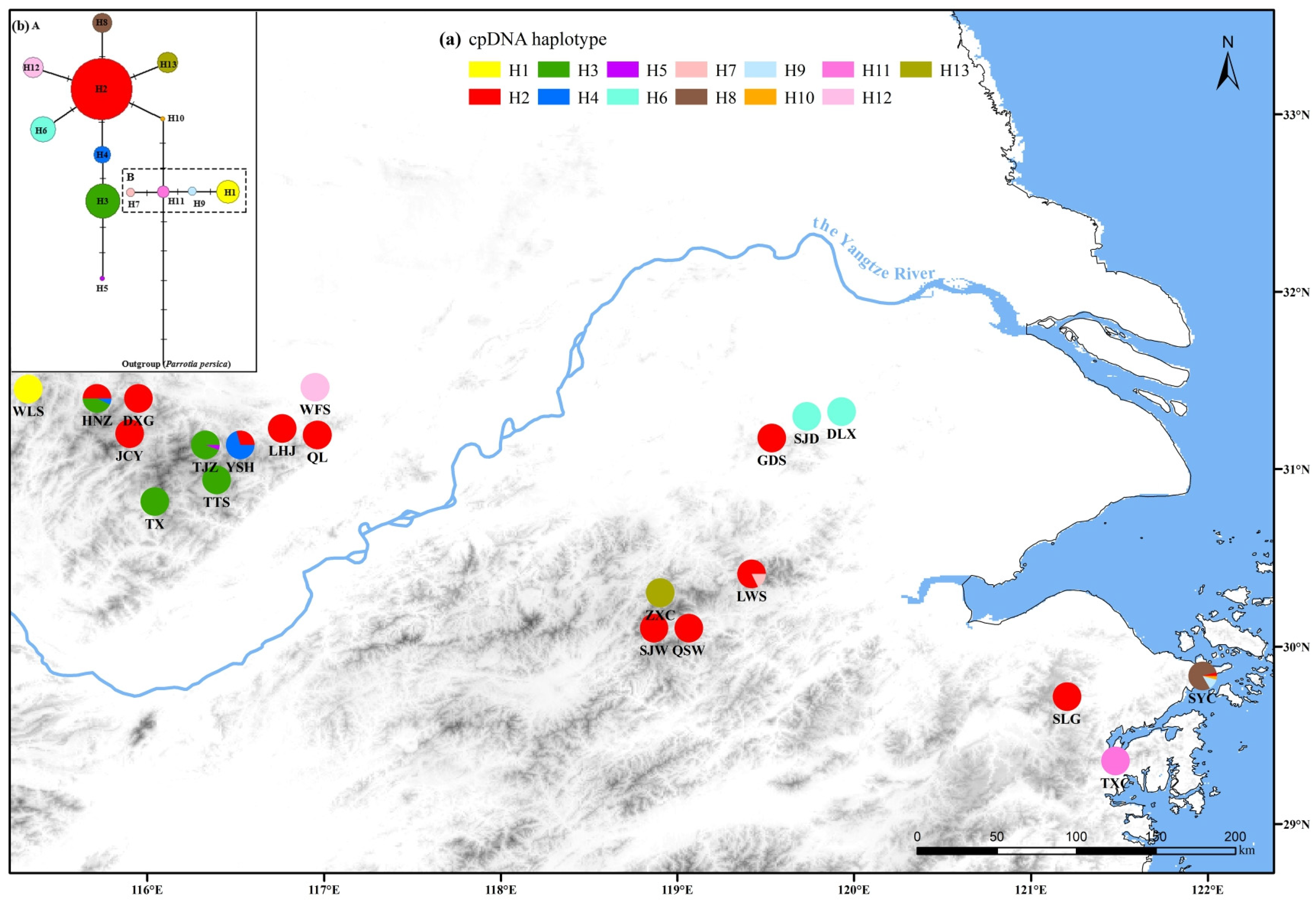
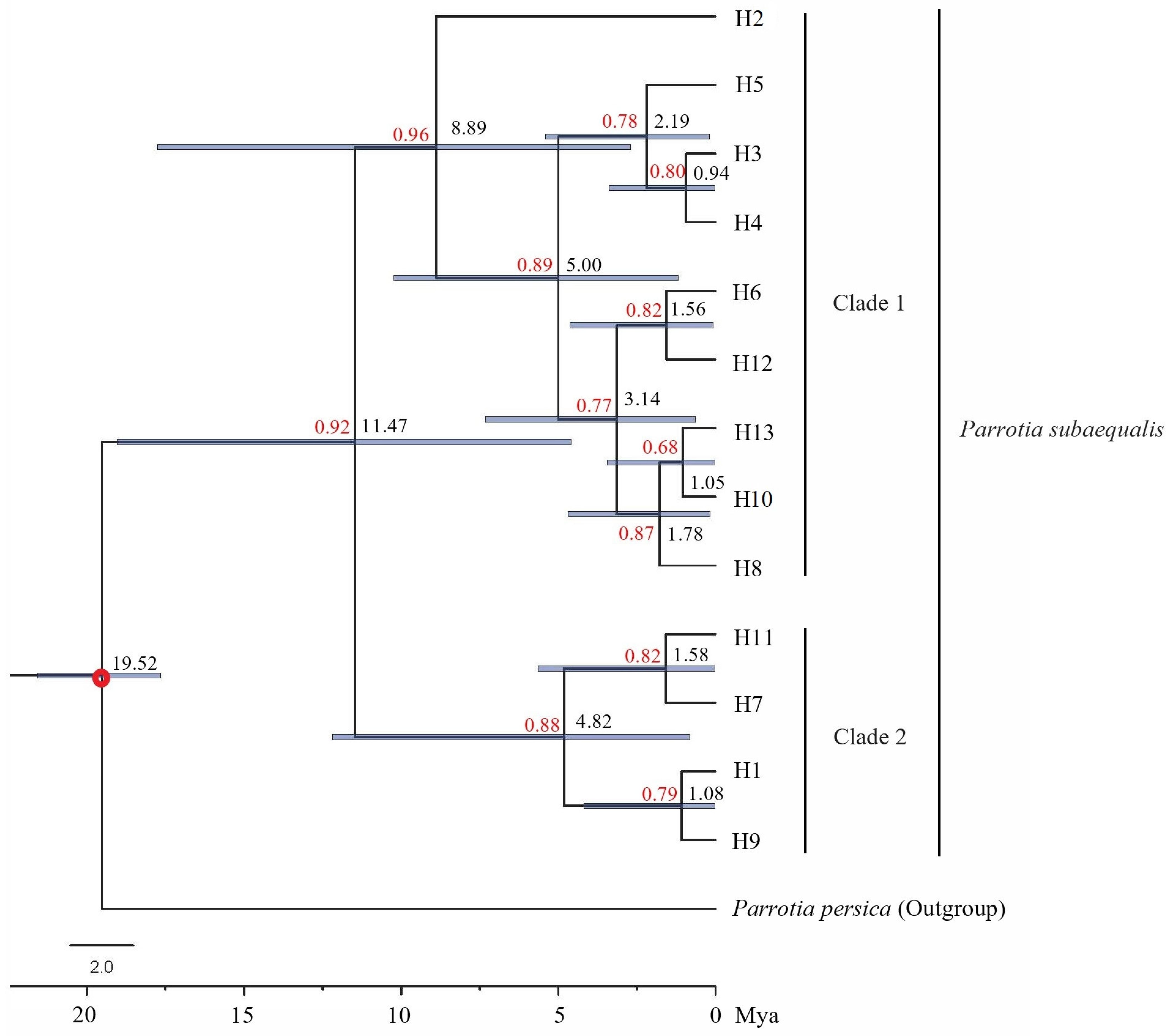
3.1. Chloroplast DNA Diversity and Population Structure of P. subaequalis
3.2. EST-SSR Diversity and Population Structure of P. subaequalis
3.3. Demographic History of P. subaequalis
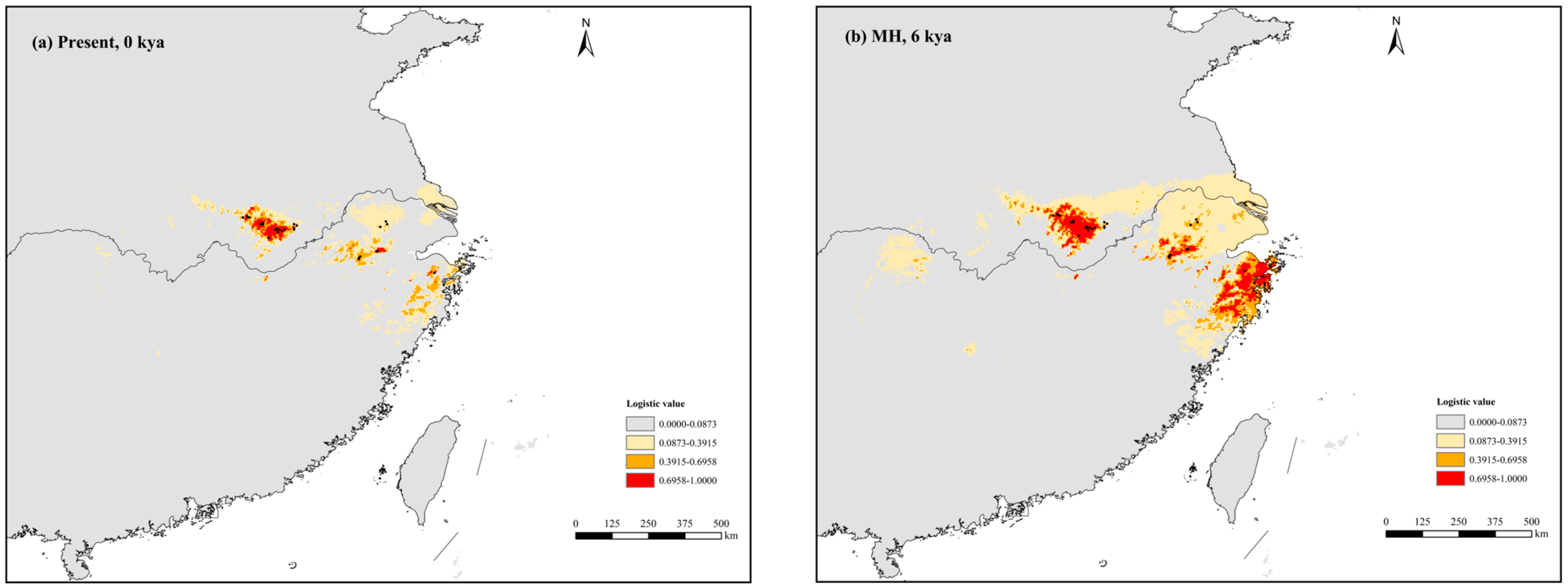
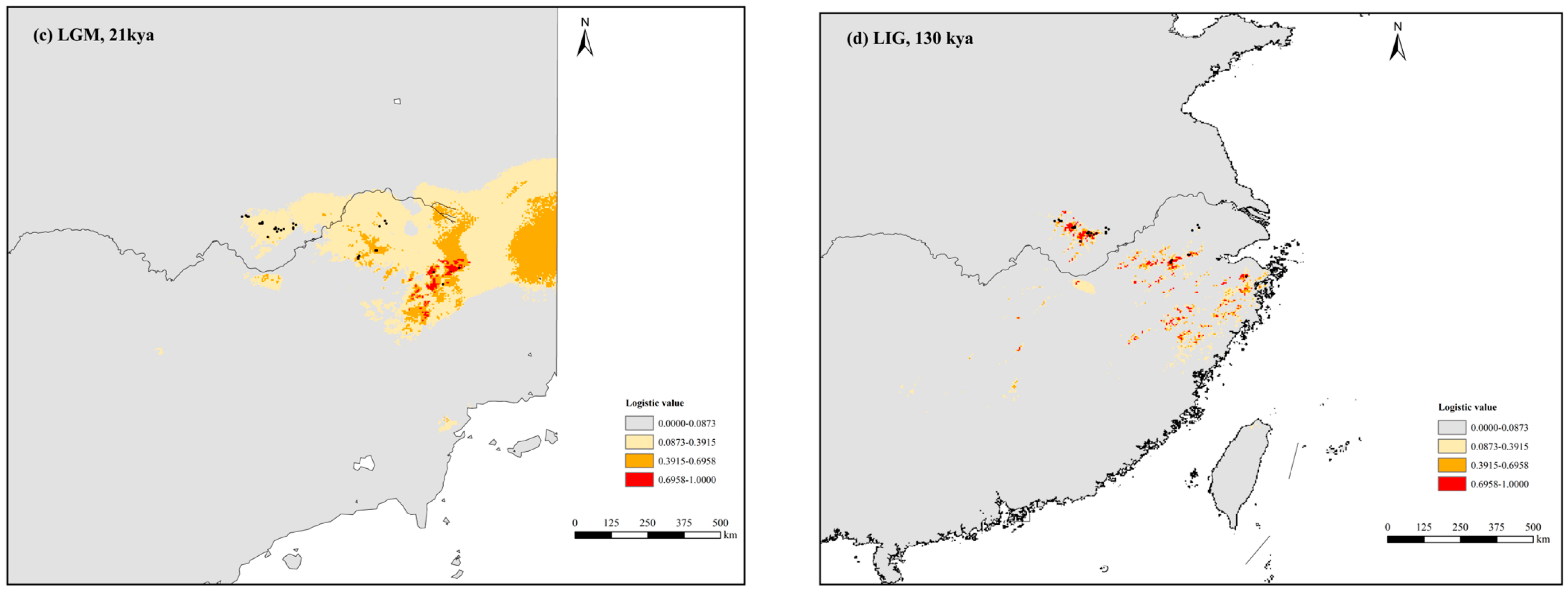
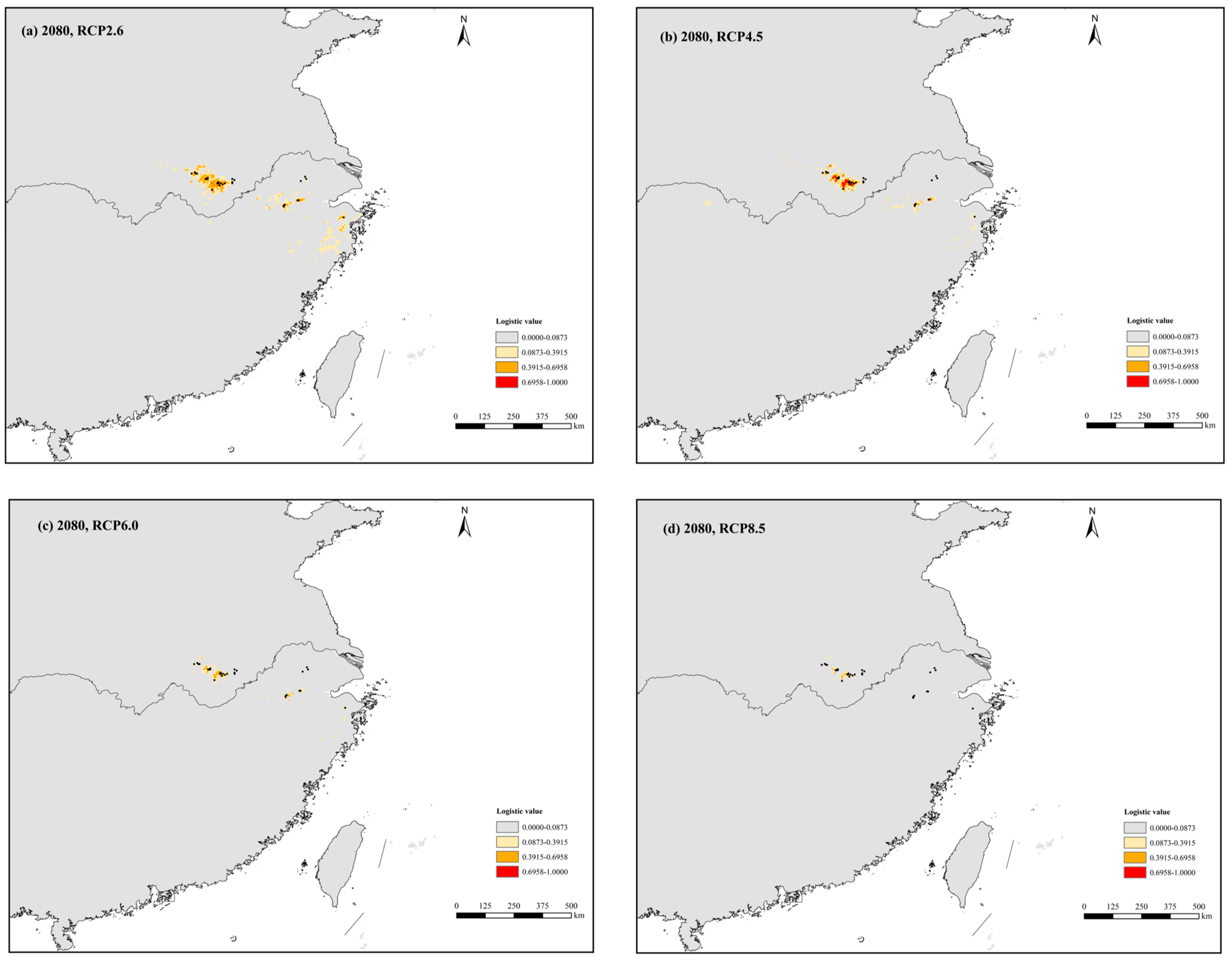
3.4. Present, Past, and Future Ecological Niches of P. subaequalis
4. Discussion
4.1. Genetic Diversity and Differentiation of P. subaequalis Populations
4.2. Population Demography and Multiple Glacial Refugia of P. subaequalis
4.3. Response to Climatic Oscillations and Conservation Implications for P. subaequalis
4.4. Conclusion and Prospect
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Milne, R.I.; Abbott, R.J. The origin and evolution of Tertiary relict floras. Adv. Bot. Res. 2002, 38, 281–314. [Google Scholar] [CrossRef]
- Grandcolas, P.; Nattier, R.; Trewick, S. Relict species: A relict concept? Trends Ecol. Evol. 2014, 29, 655–663. [Google Scholar] [CrossRef]
- Song, M.; Dodson, J.; Lu, F.; Yan, H. Central China as LGM plant refugia: Insights from biome reconstruction for palaeoclimate information. Sci. Total Environ. 2024, 942, 173783. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Deng, T.; Zhou, Z.; Sun, H. Is the East Asian flora ancient or not? Natl. Sci. Rev. 2018, 5, 920–932. [Google Scholar] [CrossRef]
- Guo, Z.T.; Ruddiman, W.F.; Hao, Q.Z.; Wu, H.B.; Qiao, Y.S.; Zhu, R.X.; Peng, S.Z.; Wei, J.J.; Yuan, B.Y.; Liu, T.S. Onset of Asian desertification by 22 Myr ago inferred from loess deposits in China. Nature 2002, 416, 159–163. [Google Scholar] [CrossRef]
- Xu, J.; Deng, M.; Jiang, X.L.; Westwood, M.; Song, Y.G.; Turkington, R. Phylogeography of Quercus glauca (Fagaceae), a dominant tree of East Asian subtropical evergreen forests, based on three chloroplast DNA interspace sequences. Tree Genet. Genomes 2015, 11, 805. [Google Scholar] [CrossRef]
- Tang, C.Q.; Matsui, T.; Ohashi, H.; Dong, Y.F.; Momohara, A.; Herrando-Moraira, S.; Qian, S.; Yang, Y.; Ohsawa, M.; Luu, H.T.; et al. Identifying long-term stable refugia for relict plant species in East Asia. Nat. Commun. 2018, 9, 4488. [Google Scholar] [CrossRef]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; Fonseca, G.A.B.D.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Ricklefs, R.E. Large-scale processes and the Asian bias in species diversity of temperate plants. Nature 2000, 407, 180–182. [Google Scholar] [CrossRef]
- Chen, K.; Abbott, R.J.; Milne, R.I.; Tian, X.M.; Liu, J. Phylogeography of Pinus tabulaeformis Carr. (Pinaceae), a dominant species of coniferous forest in northern China. Mol. Ecol. 2008, 17, 4276–4288. [Google Scholar] [CrossRef]
- Tian, B.; Liu, R.; Wang, L.; Qiu, Q.; Chen, K.; Liu, J. Phylogeographic analyses suggest that a deciduous species (Ostryopsis davidiana Decne., Betulaceae) survived in northern China during the Last Glacial Maximum. J. Biogeogr. 2009, 36, 2148–2155. [Google Scholar] [CrossRef]
- Stigall, A.L. Using ecological niche modelling to evaluate niche stability in deep time. J. Biogeogr. 2012, 39, 772–781. [Google Scholar] [CrossRef]
- Sork, V.L.; Aitken, S.N.; Dyer, R.J.; Eckert, A.J.; Legendre, P.; Neale, D.B. Putting the landscape into the genomics of trees: Approaches for understanding local adaptation and population responses to changing climate. Tree Genet. Genomes 2013, 9, 901–911. [Google Scholar] [CrossRef]
- Exposito-Alonso, M.; Burbano, H.A.; Bossdorf, O.; Nielsen, R.; Weigel, D. Natural selection on the Arabidopsis thaliana genome in present and future climates. Nature 2019, 573, 126. [Google Scholar] [CrossRef]
- Song, Y.; Li, Y.; Meng, H.H.; Fragnière, Y.; Kozlowski, G. Phylogeny, taxonomy, and biogeography of Pterocarya (Juglandaceae). Plants 2020, 9, 1524. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Del Tredici, P. The Chinese Parrotia: A sibling species of the Persian Parrotia. Arnoldia 2008, 66, 2–9. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Shi, E.; Yang, Z.P.; Geng, Q.F.; Qiu, Y.X.; Wang, Z.S. Development and application of genomic resources in an endangered palaeoendemic tree, Parrotia subaequalis (Hamamelidaceae) from eastern China. Front. Plant Sci. 2018, 9, 246. [Google Scholar] [CrossRef]
- Wu, X.T.; Shu, J.W.; Yin, S.X.; Sadowski, E.M.; Shi, G.L. Parrotia flower blooming in Miocene rainforest. J. Syst. Evol. 2024, 62, 449–456. [Google Scholar] [CrossRef]
- Mao, L.M.; Chen, X.J.; Wang, Y.H.; Liang, Y.S.; Zhou, Y.F. Parrotia (Hamamelidaceae) pollen morphology and a glimpse into the fossil record and historical biogeography. Rev. Palaeobot. Palyno. 2024, 324, 105038. [Google Scholar] [CrossRef]
- Werth, A.J.; Shear, W.A. The evolutionary truth about living fossils. Am. Sci. 2014, 102, 434–443. [Google Scholar]
- Wang, S.; Xie, Y. China Species Red List; Higher Education Press: Beijing, China, 2004; pp. 256–258. [Google Scholar]
- Yan, G.; Zhang, G. Predicting the potential distribution of endangered Parrotia subaequalis in China. Forests 2022, 13, 1595. [Google Scholar] [CrossRef]
- Zhong, Y.Q. Illustrations of Rare Plants in Jiangsu Province; Nanjing Normal University Press: Nanjing, China, 2006; pp. 162–164. [Google Scholar]
- Geng, Q.F.; Yao, Z.; Yang, J.; He, J.; Liu, H. Effect of Yangtze River on population genetic structure of the relict plant Parrotia subaequalis in eastern China. Ecol. Evol. 2015, 5, 4617–4627. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Zhang, M.Y.; Hu, Y.M.; Zhuang, X.; Xu, W.Q.; Li, P.F.; Wang, Z.S. Mining and characterization of novel EST-SSR markers of Parrotia subaequalis (Hamamelidaceae) from the first Illumina-based transcriptome datasets. PLoS ONE 2019, 14, e0215874. [Google Scholar] [CrossRef] [PubMed]
- Petit, R.J.; Aguinagalde, I.; de Beaulieu, J.L.; Bittkau, C.; Brewer, S.; Cheddadi, R.; Ennos, R.; Fineschi, S.; Grivet, D.; Lascoux, M.; et al. Glacial refugia: Hotspots but not melting pots of genetic diversity. Science 2003, 300, 1563–1565. [Google Scholar] [CrossRef]
- Schaal, B.A.; Hayworth, D.A.; Olsen, K.M.; Rauscher, J.T.; Smith, W.A. Phylogeographic studies in plants: Problems and prospects. Mol. Ecol. 1998, 7, 465–474. [Google Scholar] [CrossRef]
- Petit, R.J.; Duminil, J.; Fineschi, S.; Hampe, A.; Salvini, D.; Vendramin, G.G. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 2005, 14, 689–701. [Google Scholar] [CrossRef]
- Araújo, M.B.; Peterson, A.T. Uses and misuses of bioclimatic envelope modelling. Ecology 2012, 93, 1527–1539. [Google Scholar] [CrossRef]
- Li, J.H. Molecular Phylogenetics of Hamamelidaceae: Evidence from DNA Sequences of Nuclear and Chloroplast Genomes; Science Publisher: Washington, DC, USA, 2008. [Google Scholar]
- Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Caicedo, A.L.; Schaal, B.A. Population structure and phylogeography of Solanum pimpinellifolium inferred from a nuclear gene. Mol. Ecol. 2004, 13, 1871–1882. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. Dnasp 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Pons, O.; Petit, R.J. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 1996, 144, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, D.W.; Templeton, A.R. Correlation of pairwise genetic and geographic distance measures: Inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 1999, 53, 1898–1914. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.O.D.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Posada, D. Jmodeltest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Bobrov, A.V.; Roslov, M.S.; Romanov, M.S. Phylogenetic biogeography of Hamamelidaceae s. 1. based on molecular data. Earth Sci. 2020, 65, 224–244. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Harpending, H.C. Signature of ancient population-growth in a low-resolution mitochondrial-DNA mismatch distribution. Human Biol. 1994, 66, 591–600. [Google Scholar] [PubMed]
- Ray, N.; Currat, M.; Excoffier, L. Intra-deme molecular diversity in spatially expanding populations. Mol. Biol. Evol. 2003, 20, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT (Version 1.2): A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Kalinowski, S.T. Hp-rare 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- Rousset, F. Genepop′007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Res. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Miller, R.G. Simultaneous Statistical Inference; Springer: New York, NY, USA, 1981. [Google Scholar]
- Chapuis, M.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Res. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, M.; Rosenberg, N.A. Clumpp: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Wright, S. Isolation by distance. Genetics 1943, 28, 114–138. [Google Scholar] [CrossRef] [PubMed]
- Piry, S.; Luikart, G.; Cornuet, J.M. Bottleneck: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudik, M. Modeling of species distributions with maxent: New extensions and a comprehensive evaluation. Ecography 2008, 31, 161–175. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Fawcett, T. An introduction to ROC analysis. Pattern Recogn. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Swets, J.A. Measuring the accuracy of diagnostic systems. Science 1988, 240, 1285–1293. [Google Scholar] [CrossRef]
- Qi, X.S.; Chen, C.; Comes, H.P.; Sakaguchi, S.; Liu, Y.H.; Tanaka, N.; Sakio, H.; Qiu, Y.X. Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the East Asian Tertiary relict Cercidiphyllum (Cercidiphyllaceae). New Phytol. 2012, 196, 617–630. [Google Scholar] [CrossRef]
- Cao, Y.; Comes, H.P.; Sakaguchi, S.; Chen, L.Y.; Qiu, Y.X. Evolution of East Asia’s Arcto-Tertiary relict Euptelea (Eupteleaceae) shaped by Late Neogene vicariance and Quaternary climate change. BMC Evol. Biol. 2016, 16, 66. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Lin, F.R.; Huang, P.; Ye, X.M.; Lai, J.X.; Zheng, Y.Q. Phylogeographical structure of Liquidambar formosana Hance revealed by chloroplast phylogeography and species distribution models. Forests 2019, 10, 858. [Google Scholar] [CrossRef]
- Xiang, X.G.; Xiang, K.L.; Ortiz, R.D.C.; Jabbour, F.; Wang, W. Integrating palaeontological and molecular data uncovers multiple ancient and recent dispersals in the pantropical Hamamelidaceae. J. Biogeogr. 2019, 46, 2622–2631. [Google Scholar] [CrossRef]
- Chen, J.M.; Zhao, S.Y.; Liao, Y.Y.; Gichira, A.W.; Gituru, R.W.; Wang, Q.F. Chloroplast DNA phylogeographic analysis reveals significant spatial genetic structure of the relictual tree Davidia involucrata (Davidiaceae). Conserv. Genet. 2015, 16, 583–593. [Google Scholar] [CrossRef]
- Chen, D.M.; Zhang, X.X.; Kang, H.Z.; Sun, X.; Yin, S.; Du, H.M.; Yamanaka, N.; Gapare, W.; Wu, H.X.; Liu, C.J. Phylogeography of Quercus variabilis based on chloroplast DNA sequence in East Asia: Multiple glacial refugia and mainland-migrated island populations. PLoS ONE 2012, 7, e47268. [Google Scholar] [CrossRef]
- Ellis, J.R.; Burke, J.M. EST-SSRs as a resource for population genetic analyses. Heredity 2007, 99, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef]
- Meng, Y.H.; Xu, G.B.; Lu, M.Z.; Jiang, X.L.; Guo, F.L. Population genetic structure and demographic history of Disanthus cercidifolius var. longipes. Sci. Silvae Sin. 2020, 56, 55–62. [Google Scholar] [CrossRef]
- Chen, S.Y.; Dong, M.L.; Zhang, Y.; Qi, S.Z.; Liu, X.Z.; Zhang, J.F.; Zhao, J. Development and characterization of simple sequence repeat markers for, and genetic diversity analysis of Liquidambar formosana. Forests 2020, 11, 203. [Google Scholar] [CrossRef]
- Long, X.F.; Weng, Y.H.; Liu, S.Q.; Hao, Z.D.; Sheng, Y.; Guan, L.H.; Shi, J.S.; Chen, J.H. Genetic diversity and differentiation of relict plant Liriodendron populations based on 29 novel EST-SSR markers. Forests 2019, 10, 334. [Google Scholar] [CrossRef]
- Xu, W.Q.; Comes, H.P.; Feng, Y.; Zhang, Y.H.; Qiu, Y.X. A test of the centre-periphery hypothesis using population genetics in an East Asian Tertiary relict tree. J. Biogeogr. 2021, 48, 2853–2864. [Google Scholar] [CrossRef]
- Chen, S.F.; Li, M.W.; Hou, R.F.; Liao, W.B.; Zhou, R.C.; Fan, Q. Low genetic diversity and weak population differentiation in Firmiana danxiaensis, a tree species endemic to Danxia landform in northern Guangdong, China. Biochem. Syst. Ecol. 2014, 55, 66–72. [Google Scholar] [CrossRef]
- Yang, J.; Di, X.Y.; Meng, X.; Li, F.; Liu, Z.; Zhao, G.F. Phylogeography and evolution of two closely related oak species (Quercus) from north and northeast China. Tree Genet. Genomes 2016, 12, 89. [Google Scholar] [CrossRef]
- Royden, L.H.; Burchfiel, B.C.; van der Hilst, R.D. The geological evolution of the Tibetan plateau. Science 2008, 321, 1054–1058. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.F.; Bruch, A.A.; Mosbrugger, V.; Li, C.-S. Quantitative reconstruction of Miocene climate patterns and evolution in Southern China based on plant fossils. Palaeogeogr. Palaeoecol. 2011, 304, 291–307. [Google Scholar] [CrossRef]
- Sun, Y.X.; Moore, M.J.; Yue, L.L.; Feng, T.; Chu, H.J.; Chen, S.T.; Ji, Y.H.; Wang, H.C.; Li, J.Q. Chloroplast phylogeography of the East Asian Arcto-Tertiary relict Tetracentron sinense (Trochodendraceae). J. Biogeogr. 2014, 41, 1721–1732. [Google Scholar] [CrossRef]
- Stewart, J.R.; Lister, A.M.; Barnes, I.; Dalén, L. Refugia revisited: Individualistic responses of species in space and time. Proc. R. Soc. B Biol. Sci. 2010, 277, 661–671. [Google Scholar] [CrossRef]
- Keppel, G.; Vanniel, K.P.; Wardell-Johnson, G.W.; Yates, C.J.; Byrne, M.; Mucina, L.; Schut, A.G.T.; Hopper, S.D.; Franklin, S.E. Refugia: Identifying and understanding safe havens for biodiversity under climate change. Global Ecol. Biogeogr. 2012, 21, 393–404. [Google Scholar] [CrossRef]
- Gavin, D.G.; Fitzpatrick, M.C.; Gugger, P.F.; Heath, K.D.; Rodríguez-Sánchez, F.; Dobrowski, S.Z.; Hampe, A.; Hu, F.S.; Ashcroft, M.B.; Bartlein, P.J.; et al. Climate refugia: Joint inference from fossil records, species distribution models and phylogeography. New Phytol. 2014, 204, 37–54. [Google Scholar] [CrossRef]
- Da Rocha, D.G.; Kaefer, I.L. What has become of the refugia hypothesis to explain biological diversity in Amazonia? Ecol. Evol. 2019, 9, 4302–4309. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Chen, C.; Dobeš, C.; Fu, C.X.; Koch, M.A. Phylogeography of a living fossil: Pleistocene glaciations forced Ginkgo biloba L. (Ginkgoaceae) into two refuge areas in China with limited subsequent postglacial expansion. Mol. Phylogenet. Evol. 2008, 48, 1094–1105. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.K.; Huang, J.; Ding, W.N. The impact of major geological events on Chinese flora. Biodivers. Sci. 2017, 25, 125–132. [Google Scholar] [CrossRef]
- He, Y.; Wang, H.; Zhao, Y.; Cui, Y. Predicting the biodiversity hotspots of macrozoobenthos in the Yangtze River Basin. Ecol. Indic. 2021, 133, 108428. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Wang, I.J.; Comes, H.P.; Peng, H.; Qiu, Y.X. Contributions of historical and contemporary geographic and environmental factors to phylogeographic structure in a Tertiary relict species, Emmenopterys henryi (Rubiaceae). Sci. Rep. 2016, 6, 24041. [Google Scholar] [CrossRef]
- Moritz, C. Defining evolutionarily-significant-units for conservation. Trends Ecol. Evol. 1994, 9, 373–375. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Li, Z.; Chen, Q.; Wang, Y.; Wang, S.; Wang, G.; Li, P.; Liu, H.; Li, P.; Xu, C.; et al. Phylogeography and Population Demography of Parrotia subaequalis, a Hamamelidaceous Tertiary Relict ‘Living Fossil’ Tree Endemic to East Asia Refugia: Implications from Molecular Data and Ecological Niche Modeling. Plants 2025, 14, 1754. https://doi.org/10.3390/plants14121754
Zhang Y, Li Z, Chen Q, Wang Y, Wang S, Wang G, Li P, Liu H, Li P, Xu C, et al. Phylogeography and Population Demography of Parrotia subaequalis, a Hamamelidaceous Tertiary Relict ‘Living Fossil’ Tree Endemic to East Asia Refugia: Implications from Molecular Data and Ecological Niche Modeling. Plants. 2025; 14(12):1754. https://doi.org/10.3390/plants14121754
Chicago/Turabian StyleZhang, Yunyan, Zhiyuan Li, Qixun Chen, Yahong Wang, Shuang Wang, Guozheng Wang, Pan Li, Hong Liu, Pengfu Li, Chi Xu, and et al. 2025. "Phylogeography and Population Demography of Parrotia subaequalis, a Hamamelidaceous Tertiary Relict ‘Living Fossil’ Tree Endemic to East Asia Refugia: Implications from Molecular Data and Ecological Niche Modeling" Plants 14, no. 12: 1754. https://doi.org/10.3390/plants14121754
APA StyleZhang, Y., Li, Z., Chen, Q., Wang, Y., Wang, S., Wang, G., Li, P., Liu, H., Li, P., Xu, C., & Wang, Z. (2025). Phylogeography and Population Demography of Parrotia subaequalis, a Hamamelidaceous Tertiary Relict ‘Living Fossil’ Tree Endemic to East Asia Refugia: Implications from Molecular Data and Ecological Niche Modeling. Plants, 14(12), 1754. https://doi.org/10.3390/plants14121754






