The Influence of Bacterial Inoculants and a Biofertilizer on Maize Cultivation and the Associated Shift in Bacteriobiota During the Growing Season †
Abstract
1. Introduction
2. Results
2.1. Raw Data Analysis
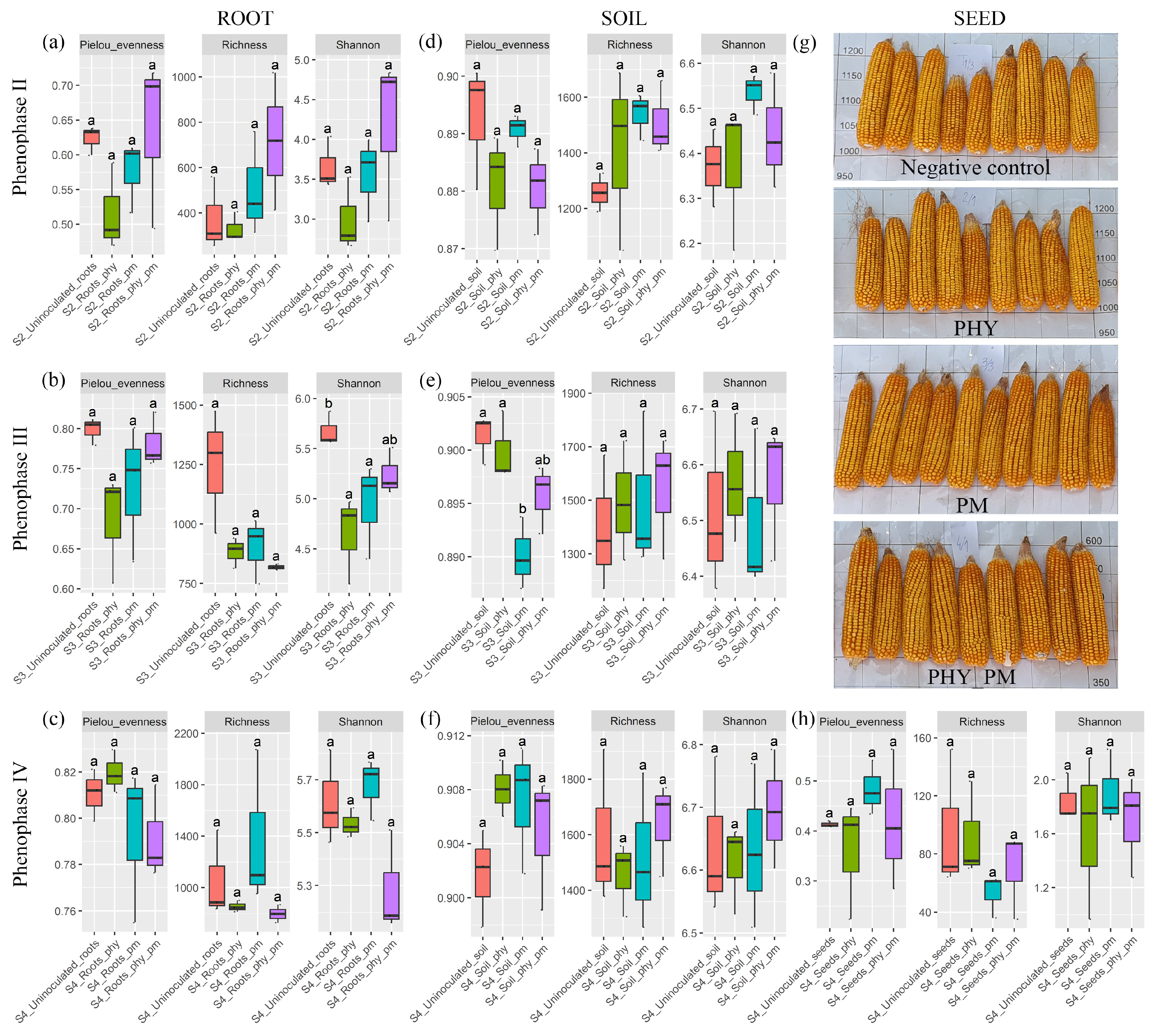
2.2. Alpha Diversity
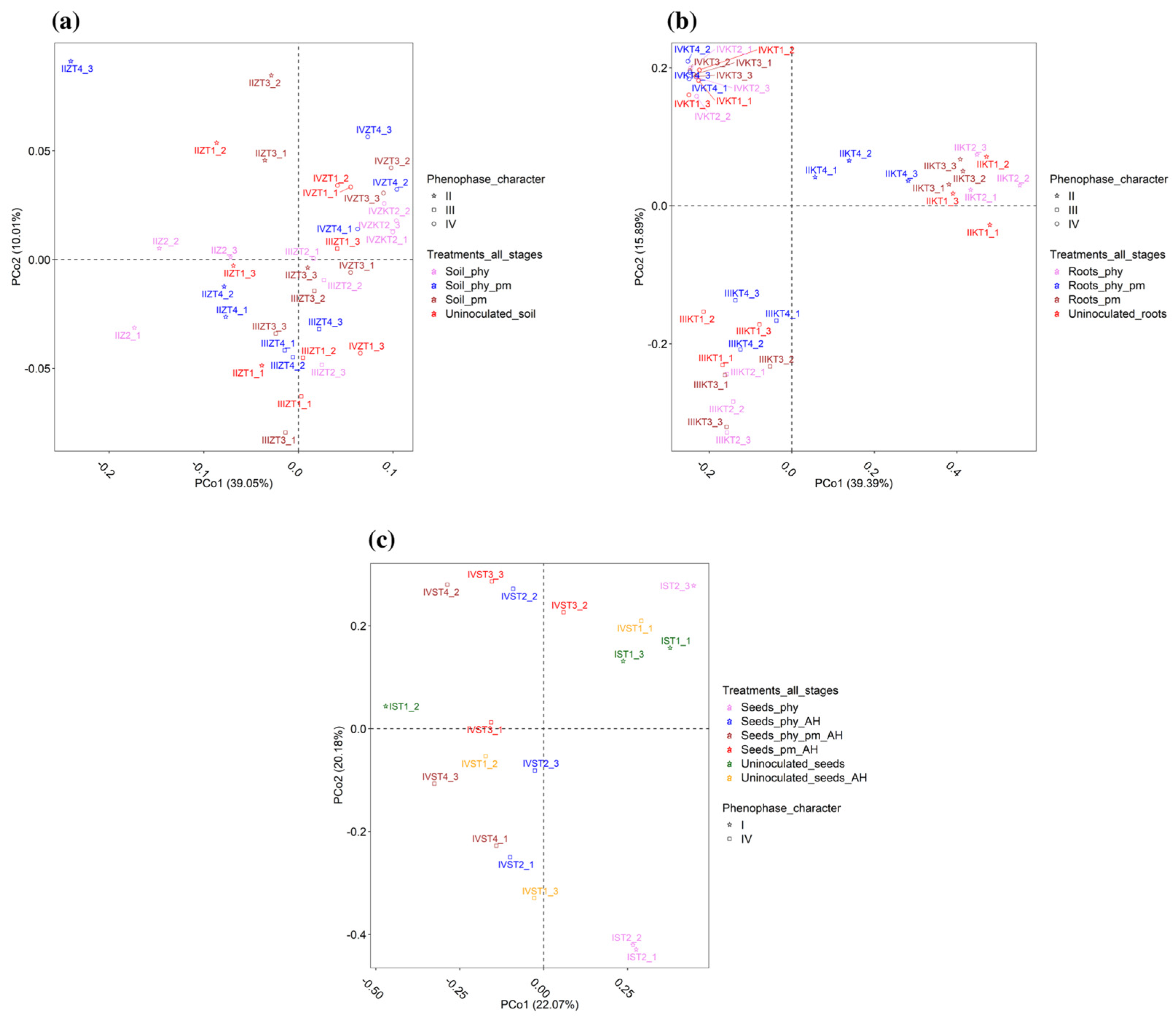
2.3. Beta Diversity
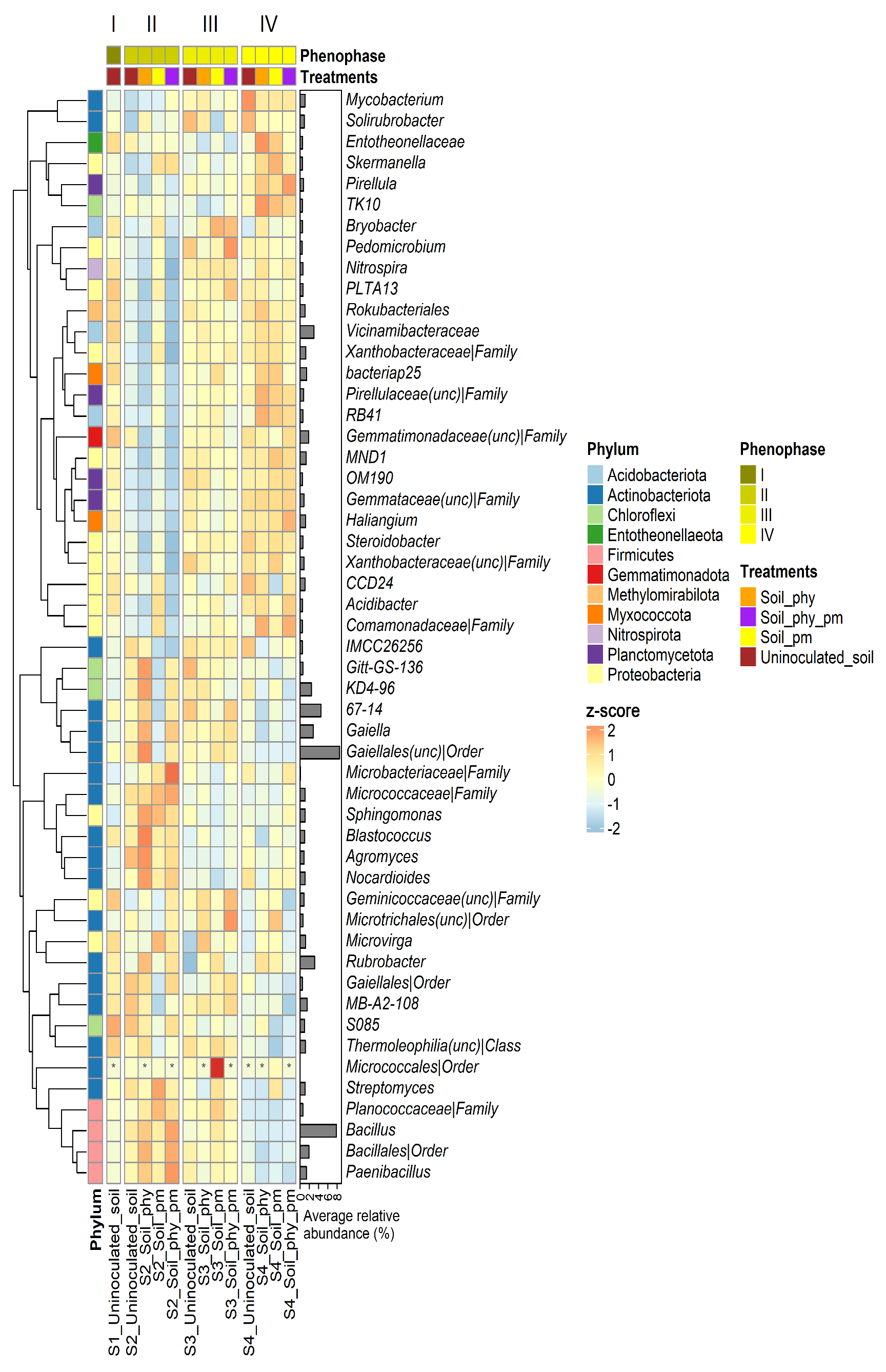
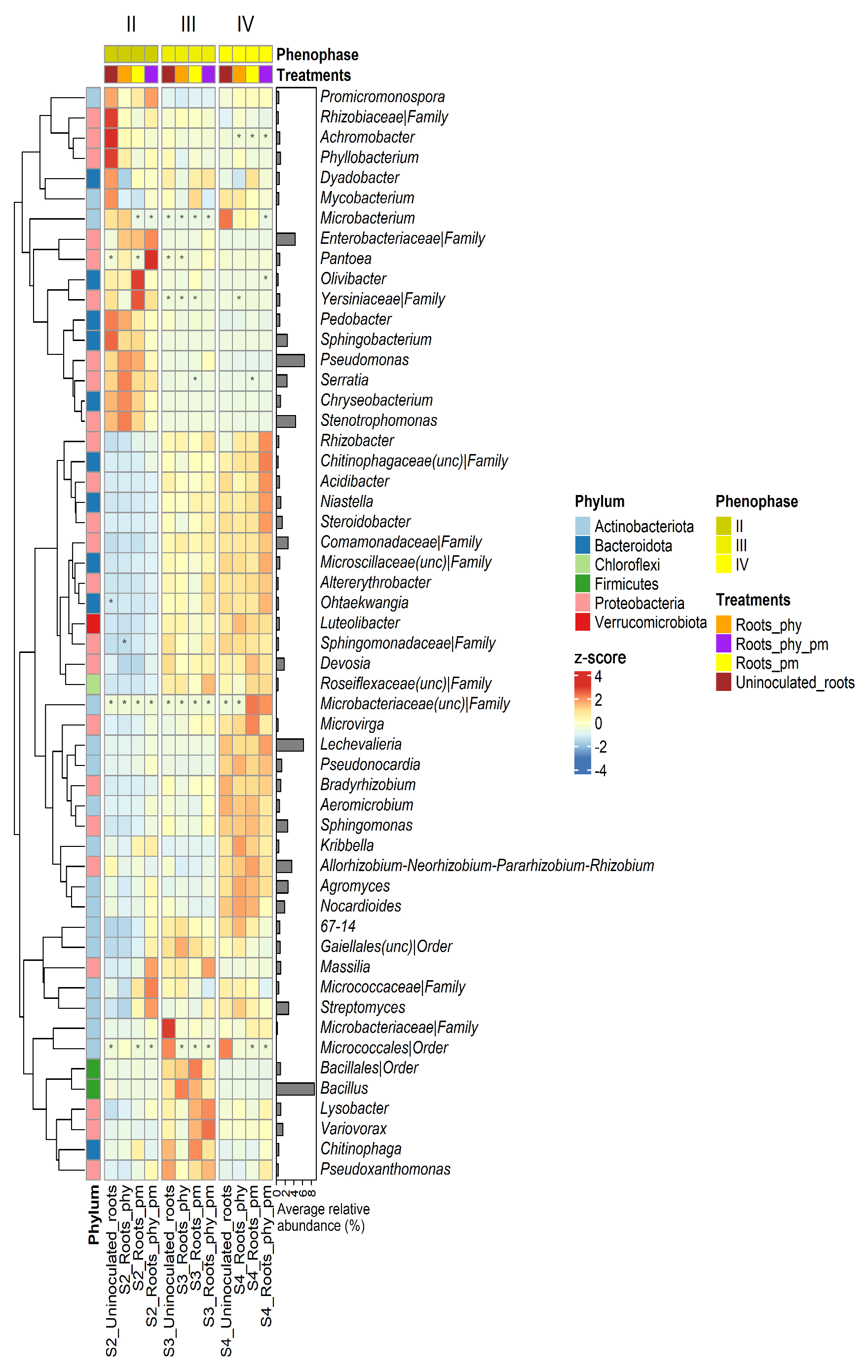
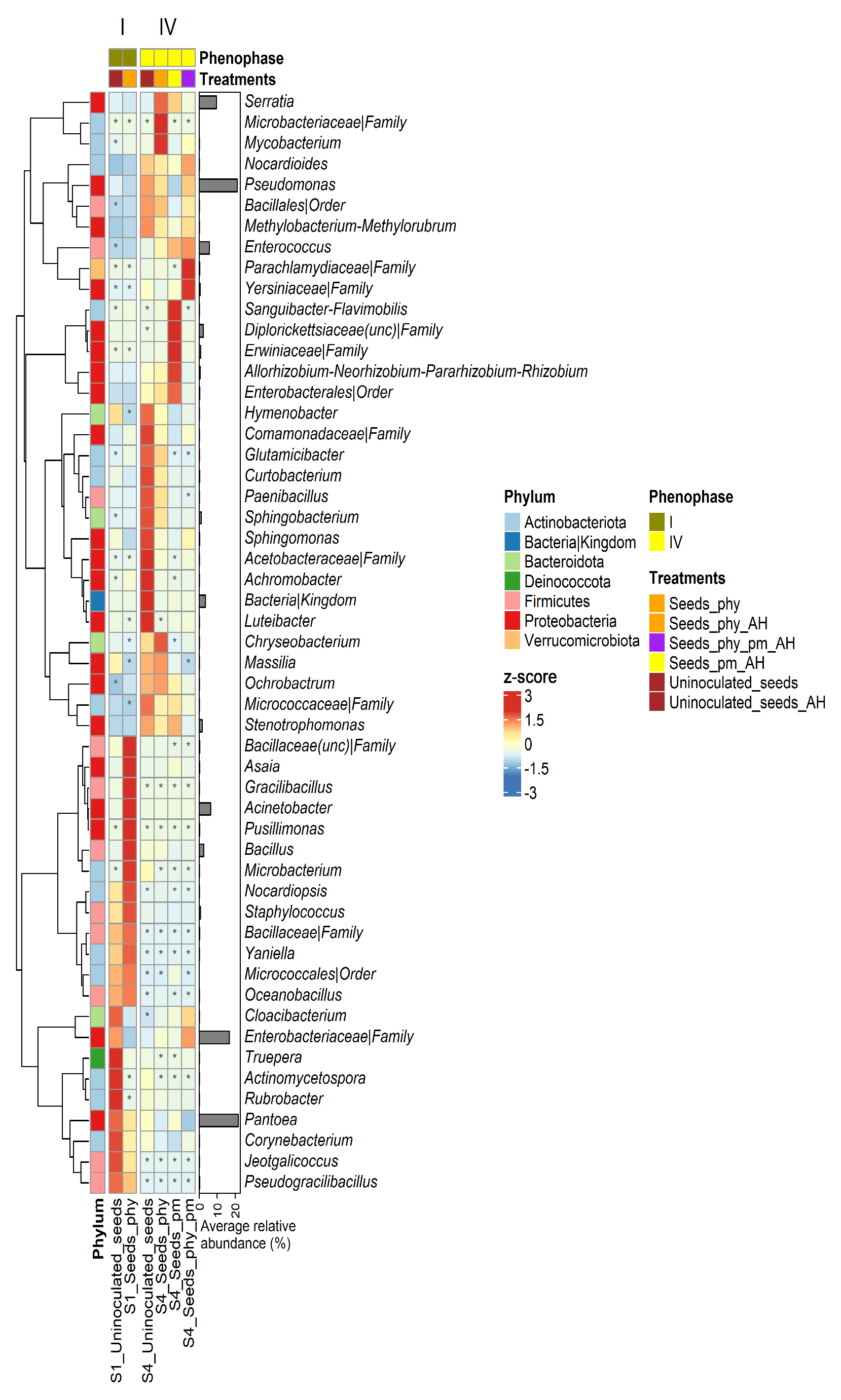
2.4. Taxonomy
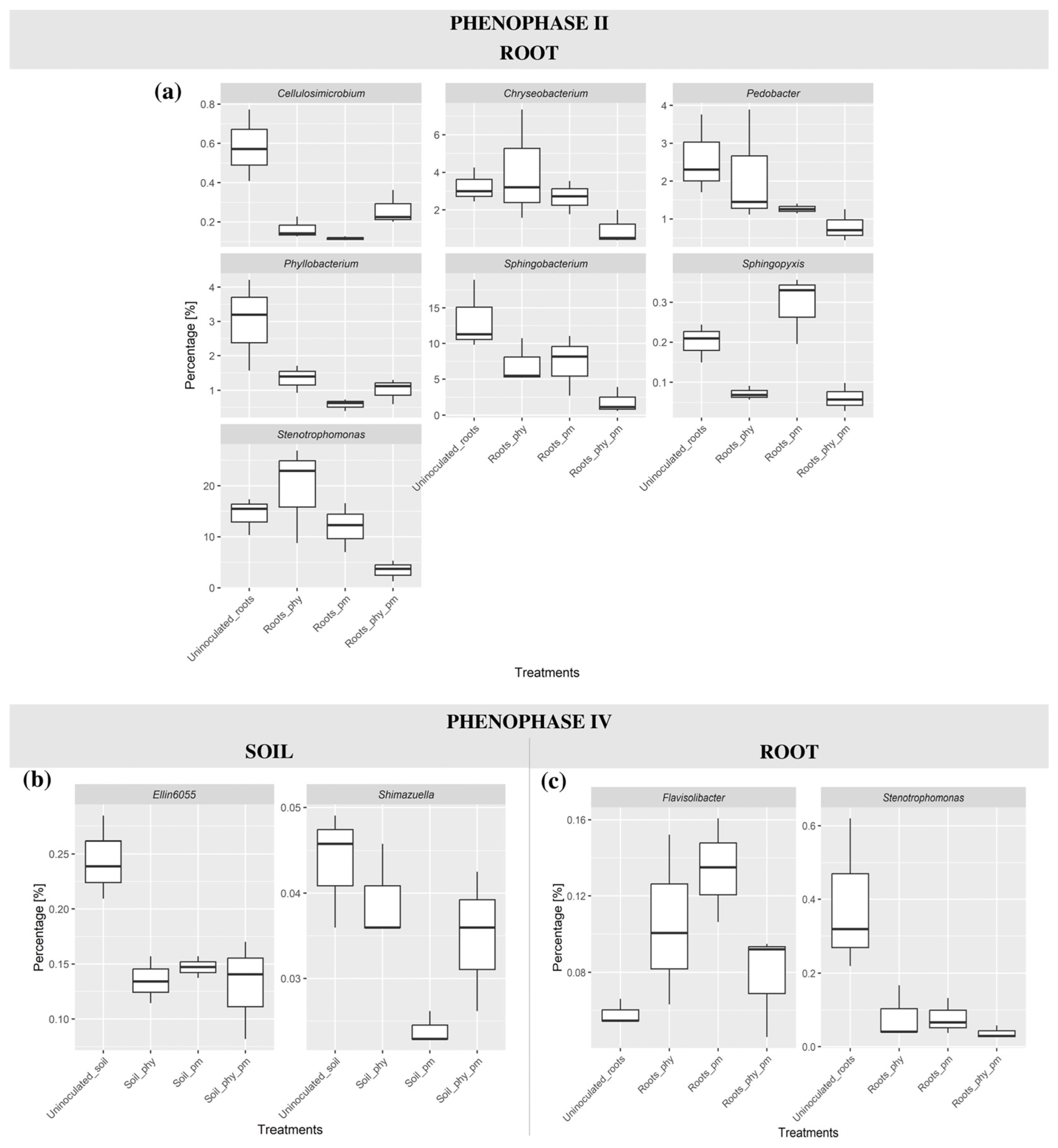
2.5. Differential Abundance Analysis
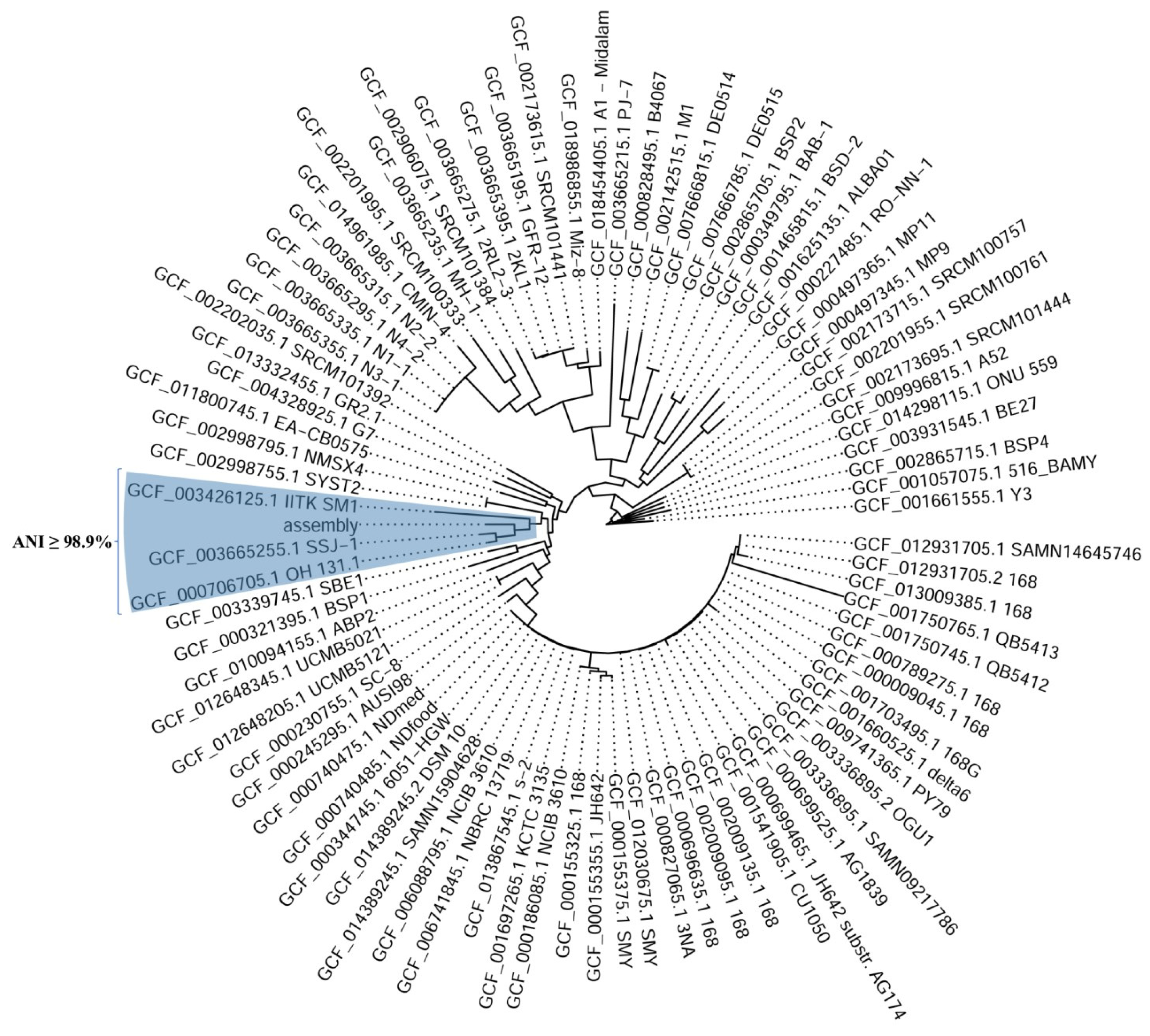
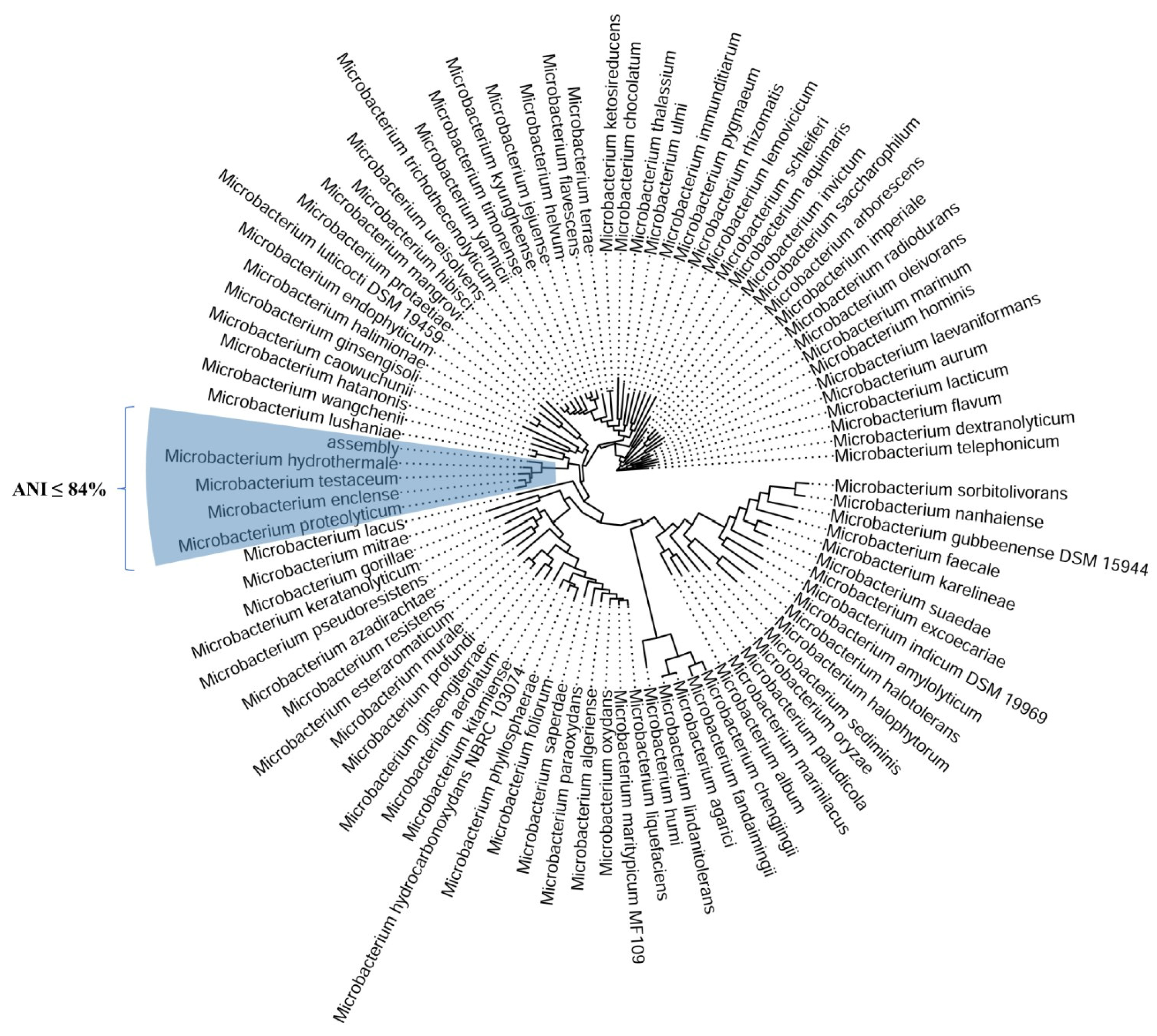
2.6. Whole-Genome Sequencing of Bacillus subtilis sp. subtilis and Microbacterium sp.
2.6.1. Bacillus subtilis sp. subtilis
2.6.2. Microbacterium sp.
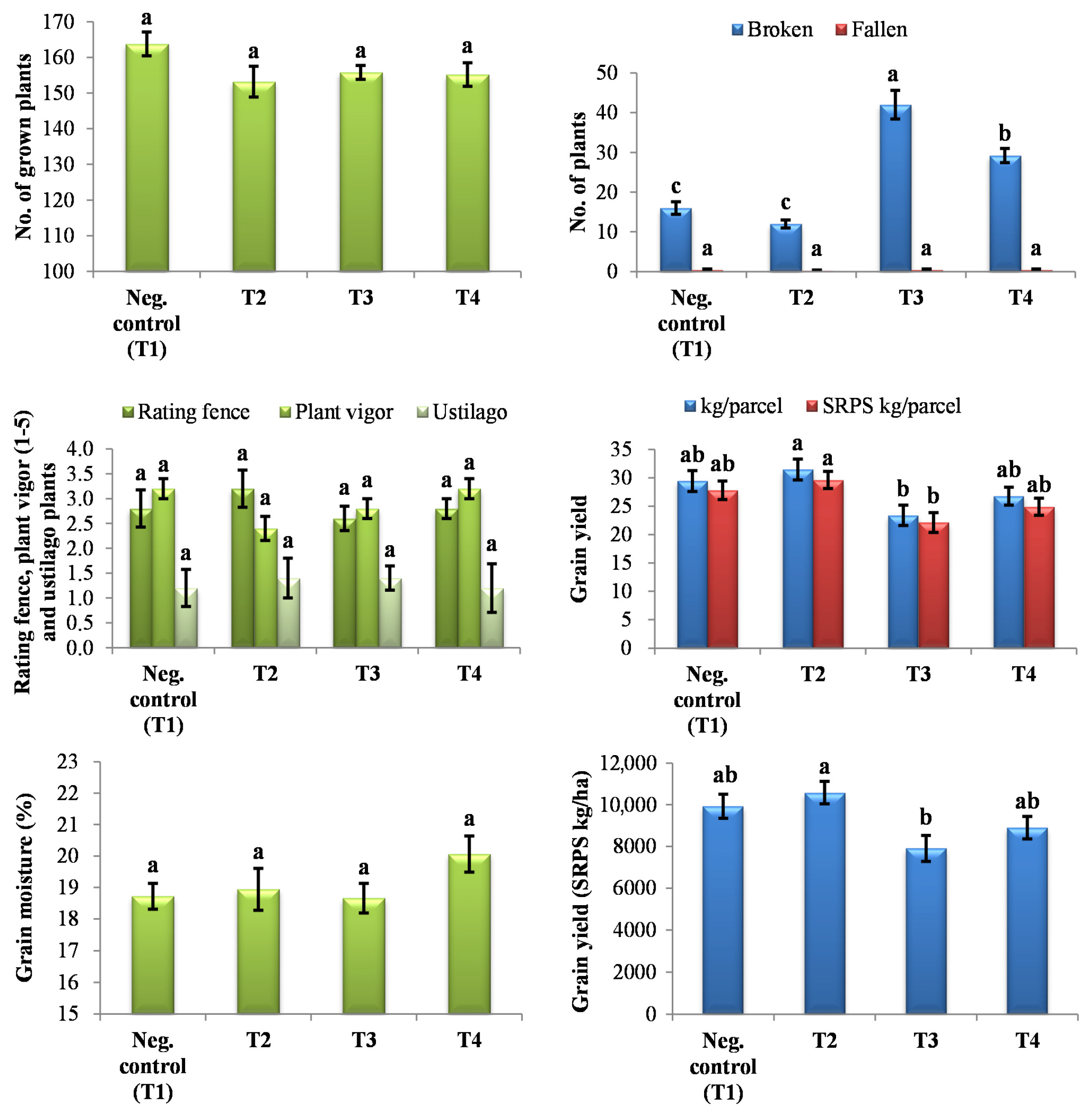
2.7. Evaluation of Yield Parameters
3. Discussion
4. Materials and Methods
4.1. Agricultural Field Conditions and Treatments
4.2. Sample Collection and Measurements
4.3. DNA Extraction and Amplicon Sequencing Preparation
4.4. Raw Data and Statistical Analyses
4.5. Bacterial Candidates’ Genome Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seyi-Amole, D.O.; Onilude, A.A. Microbiological control: A new age of maize production. In Cereal Grains—Volume 2; IntechOpen: London, UK, 2021. [Google Scholar]
- Gezahegn, A.M. Role of integrated nutrient management for sustainable maize production. Int. J. Agron. 2021, 2021, 9982884. [Google Scholar] [CrossRef]
- Khaliq, T.; Mahmood, T.; Kamal, J.; Masood, A. Effectiveness of farmyard manure, poultry manure and nitrogen for corn (Zea mays L.) productivity. Int. J. Agric. Biol. 2004, 2, 260–263. [Google Scholar]
- Avery, H. The role of organic fertilizers in transition to sustainable agriculture in the MENA Region. In New Generation of Organic Fertilizers; IntechOpen: London, UK, 2021. [Google Scholar]
- Lawal, T.E.; Babalola, O.O. Relevance of biofertilizers to agriculture. J. Hum. Ecol. 2014, 47, 35–43. [Google Scholar] [CrossRef]
- Hui, L.I.; Feng, W.T.; He, X.H.; Ping, Z.H.U.; Gao, H.J.; Nan, S.U.N.; XU, M.G. Chemical fertilizers could be completely replaced by manure to maintain high maize yield and soil organic carbon (SOC) when SOC reaches a threshold in the Northeast China Plain. J. Integr. Agric. 2017, 16, 937–946. [Google Scholar]
- Mahanty, T.; Bhattacharjee, S.; Goswami, M.; Bhattacharyya, P.; Das, B.; Ghosh, A.; Tribedi, P. Biofertilizers: A potential approach for sustainable agriculture development. Environ. Sci. Pollut. Res. 2017, 24, 3315–3335. [Google Scholar] [CrossRef]
- Sankar Ganesh, K.; Sundaramoorthy, P.; Nagarajan, M.; Lawrence Xavier, R. Role of organic amendments in sustainable agriculture. In Sustainable Agriculture Towards Food Security; Springer: Singapore, 2017; pp. 111–124. [Google Scholar]
- Arif, I.; Batool, M.; Schenk, P.M. Plant microbiome engineering: Expected benefits for improved crop growth and resilience. Trends Biotechnol. 2020, 38, 1385–1396. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, Y.; Zhang, S.; Wang, Y. What could promote farmers to replace chemical fertilizers with organic fertilizers? J. Clean. Prod. 2018, 199, 882–890. [Google Scholar] [CrossRef]
- Kumar, R.R.; Park, B.J.; Cho, J.Y. Application and environmental risks of livestock manure. J. Korean Soc. Appl. Biol. Chem. 2013, 56, 497–503. [Google Scholar] [CrossRef]
- Rayne, N.; Aula, L. Livestock manure and the impacts on soil health: A review. Soil. Syst. 2020, 4, 64. [Google Scholar] [CrossRef]
- Kolawole, G. Effect of time of poultry manure application on the performance of maize in Ogbomoso, Oyo State, Nigeria. J. Appl. Agric. Res. 2014, 6, 253–258. [Google Scholar]
- Adeyemo, A.J.; Akingbola, O.O.; Ojeniyi, S.O. Effects of poultry manure on soil infiltration, organic matter contents and maize performance on two contrasting degraded alfisols in southwestern Nigeria. Int. J. Recycl. Org. Waste Agric. 2019, 8, 73–80. [Google Scholar] [CrossRef]
- Riaz, U.; Mehdi, S.M.; Iqbal, S.; Khalid, H.I.; Qadir, A.A.; Anum, W.; Ahmad, M.; Murtaza, G. Bio-fertilizers: Eco-friendly approach for plant and soil environment. In Bioremediation and Biotechnology: Sustainable Approaches to Pollution Degradation; Springer: Cham, Switzerland, 2020; pp. 189–213. [Google Scholar]
- Mitter, E.K.; Tosi, M.; Obregón, D.; Dunfield, K.E.; Germida, J.J. Rethinking crop nutrition in times of modern microbiology: Innovative biofertilizer technologies. Front. Sustain. Food Syst. 2021, 5, 606815. [Google Scholar] [CrossRef]
- Seenivasagan, R.; Babalola, O.O. Utilization of microbial consortia as biofertilizers and biopesticides for the production of feasible agricultural product. Biology 2021, 10, 1111. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-H. The combined use of chemical and organic fertilizers and/or biofertilizer for crop growth and soil fertility. In International Workshop on Sustained Management of the Soil-Rhizosphere System for Efficient Crop Production and Fertilizer Use; Land Development Department: Bangkok, Thailand, 2006; pp. 1–11. [Google Scholar]
- Basu, A.; Prasad, P.; Das, S.N.; Kalam, S.; Sayyed, R.; Reddy, M.; El Enshasy, H. Plant growth promoting rhizobacteria (PGPR) as green bioinoculants: Recent developments, constraints, and prospects. Sustainability 2021, 13, 1140. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Adesemoye, A.O. Improvement of crop protection and yield in hostile agroecological conditions with PGPR-based biofertilizer formulations. In Bioformulations: For Sustainable Agriculture; Springer: New Delhi, India, 2016; pp. 199–211. [Google Scholar]
- Vessey, J.K. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil. 2003, 255, 571–586. [Google Scholar] [CrossRef]
- Riaz, U.; Murtaza, G.; Anum, W.; Samreen, T.; Sarfraz, M.; Nazir, M.Z. Plant growth-promoting rhizobacteria (PGPR) as biofertilizers and biopesticides. In Microbiota and Biofertilizers: A Sustainable Continuum for Plant and Soil Health; Springer: Cham, Switzerland, 2021; pp. 181–196. [Google Scholar]
- Meena, M.; Swapnil, P.; Divyanshu, K.; Kumar, S.; Harish; Tripathi, Y.N.; Zehra, A.; Marwal, A.; Upadhyay, R.S. PGPR-mediated induction of systemic resistance and physiochemical alterations in plants against the pathogens: Current perspectives. J. Basic. Microbiol. 2020, 60, 828–861. [Google Scholar] [CrossRef]
- Hassen, A.I.; Bopape, F.; Sanger, L. Microbial inoculants as agents of growth promotion and abiotic stress tolerance in plants. In Microbial Inoculants in Sustainable Agricultural Productivity: Vol. 1: Research Perspectives; Springer: New Delhi, India, 2016; pp. 23–36. [Google Scholar]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Poveda, J.; González-Andrés, F. Bacillus as a source of phytohormones for use in agriculture. Appl. Microbiol. Biotechnol. 2021, 105, 8629–8645. [Google Scholar] [CrossRef]
- Ghazy, N.; El-Nahrawy, S. Siderophore production by Bacillus subtilis MF497446 and Pseudomonas koreensis MG209738 and their efficacy in controlling Cephalosporium maydis in maize plant. Arch. Microbiol. 2021, 203, 1195–1209. [Google Scholar] [CrossRef]
- Rivas, R.; Trujillo, M.E.; Sanchez, M.; Mateos, P.F.; Martínez-Molina, E.; Velazquez, E. Microbacterium ulmi sp. nov., a xylanolytic, phosphate-solubilizing bacterium isolated from sawdust of Ulmus nigra. Int. J. Syst. Evol. Microbiol. 2004, 54, 513–517. [Google Scholar] [CrossRef]
- Zheng, J.; Liao, Y.; Li, Y.; Li, D.; Sun, Y.; Xiao, Z. Description of Microbacterium dauci sp. nov., a plant growth hormone indoleacetic acid-producing and nitrogen-fixing bacterium isolated from carrot rhizosphere soil. Arch. Microbiol. 2024, 206, 102. [Google Scholar] [CrossRef] [PubMed]
- National Library of Medicine—National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/bioproject/495416 (accessed on 5 March 2025).
- National Library of Medicine—National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA221154 (accessed on 5 March 2025).
- National Library of Medicine—National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA485230 (accessed on 5 March 2025).
- National Library of Medicine—National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA527766 (accessed on 5 March 2025).
- National Library of Medicine—National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJDB3283 (accessed on 5 March 2025).
- National Library of Medicine—National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA323170 (accessed on 5 March 2025).
- National Library of Medicine—National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA546708 (accessed on 5 March 2025).
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef] [PubMed]
- Gouda, S.; Kerry, R.G.; Das, G.; Paramithiotis, S.; Shin, H.-S.; Patra, J.K. Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol. Res. 2018, 206, 131–140. [Google Scholar] [CrossRef]
- Junges, E.; Toebe, M.; Santos, R.F.d.; Finger, G.; Muniz, M.F.B. Effect of priming and seed-coating when associated with Bacillus subtilis in maize seeds. Rev. Ciência Agronômica 2013, 44, 520–526. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Garcia Arredondo, M.; Kew, W.; Chu, R.; Jones, M.E.; Boiteau, R.M.; Cardon, Z.G.; Keiluweit, M. Differential Exudation Creates Biogeochemically Distinct Microenvironments during Rhizosphere Evolution. Environ. Sci. Technol. 2024, 58, 18713–18722. [Google Scholar] [CrossRef]
- Zhu, F.; Kamiya, T.; Fujiwara, T.; Hashimoto, M.; Gong, S.; Wu, J.; Nakanishi, H.; Fujimoto, M. A Comparison of Rice Root Microbial Dynamics in Organic and Conventional Paddy Fields. Microorganisms 2024, 13, 41. [Google Scholar] [CrossRef]
- Bourceret, A.; Guan, R.; Dorau, K.; Mansfeldt, T.; Omidbakhshfard, A.; Medeiros, D.B.; Fernie, A.R.; Hofmann, J.; Sonnewald, U.; Mayer, J. Maize field study reveals covaried microbiota and metabolic changes in roots over plant growth. MBio 2022, 13, e0258421. [Google Scholar] [CrossRef]
- Sant’Anna, G.S.L.; de Carvalho, L.A.L.; da Silva, M.S.R.d.A.; Gonçalves, J.V.d.S.; Pinheiro, D.G.; Zonta, E.; Coelho, I.d.S. Short-Term Effects of Poultry Litter and Cattle Manure on Soil’s Chemical Properties and Bacterial Community. Agronomy 2024, 14, 1382. [Google Scholar] [CrossRef]
- Ochai, M.I.; Aremu, J.K.; Agada, E.E. Effects of poultry litter on soil physico-chemical properties for crop production in Kauru local government area, Kaduna State, Nigeria. Fudma J. Sci. 2024, 8, 111–117. [Google Scholar] [CrossRef]
- Perruchon, C.; Tagkalidou, N.; Kalogiouri, N.; Katsivelou, E.; Karas, P.A.; Menkissoglu-Spiroudi, U.; Vasileiadis, S.; Karpouzas, D.G. Isolation of a novel Sphingomonas strain able to degrade the pleuromutilin tiamulin: Omic analysis reveals its transformation pathway. bioRxiv 2024. bioRxiv:2024.12.16.628731. [Google Scholar]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.-K. Production, characterization and antioxidant potential of protease from Streptomyces sp. MAB18 using poultry wastes. BioMed Res. Int. 2013, 2013, 496586. [Google Scholar] [CrossRef] [PubMed]
- Alamoudi, S.A.; Khalel, A.F.; Alghamdi, M.A.; Alshehri, W.A.; Alsubeihi, G.K.; Alsolmy, S.A.; Hakeem, M.A. Isolation, identification and characterization of keratin-degrading Streptomyces rochei AM8. J. Pure Appl. Microbiol. 2022, 16, 2045–2054. [Google Scholar] [CrossRef]
- Saha, S.; Dhanasekaran, D.; Shanmugapriya, S.; Latha, S. Nocardiopsis sp. SD 5: A potent feather degrading rare actinobacterium isolated from feather waste in Tamil Nadu, India. J. Basic. Microbiol. 2013, 53, 608–616. [Google Scholar] [CrossRef]
- Yang, Q.; Yang, X.; Huang, X.; Ye, W.; Wang, T.; Cheng, Z.; Shi, J.; Li, Y.; Xu, J.; He, Y. Seed coating with fungicide causes a beneficial shift in root-associated microbiomes of mature soybean. Soil. Sci. Soc. Am. J. 2023, 87, 43–62. [Google Scholar] [CrossRef]
- Al Methyeb, M.; Ruppel, S.; Eichler-Löbermann, B.; Vassilev, N. The Combined Applications of Microbial Inoculants and Organic Fertilizer Improve Plant Growth under Unfavorable Soil Conditions. Microorganisms 2023, 11, 1721. [Google Scholar] [CrossRef]
- Bang, D.; Chung, W.; Chang, S. Influence of Effective Microbial Additives Inoculation on Indigenous Bacterial Community Dynamics and Co-Occurrence Patterns During the Composting of Mixed Food Waste and Livestock Manure. Agronomy 2024, 14, 2973. [Google Scholar] [CrossRef]
- Cao, R.; Huang, Y.; Li, R.; Li, K.; Ren, Z.; Wu, J. Regulation of nitrogen transformation and microbial community by inoculation during livestock manure composting. Environ. Microbiol. Rep. 2024, 16, e13256. [Google Scholar] [CrossRef]
- Jin, H.; Zhang, D.; Yan, Y.; Yang, C.; Fang, B.; Li, X.; Shao, Y.; Wang, H.; Yue, J.; Wang, Y. Short-term application of chicken manure under different nitrogen rates alters structure and co-occurrence pattern but not diversity of soil microbial community in wheat field. Front. Microbiol. 2022, 13, 975571. [Google Scholar] [CrossRef]
- Cai, F.; Pang, G.; Li, R.-X.; Li, R.; Gu, X.-L.; Shen, Q.-R.; Chen, W. Bioorganic fertilizer maintains a more stable soil microbiome than chemical fertilizer for monocropping. Biol. Fertil. Soils 2017, 53, 861–872. [Google Scholar] [CrossRef]
- Mehta, S.; Singh, B.; Patra, A.; Tripathi, A.; Easwaran, M.; Choudhary, J.R.; Choudhary, M.; Aggarwal, S. Maize microbiome: Current insights for the sustainable agriculture. In Microbiomes and Plant Health; Elsevier: Amsterdam, The Netherlands, 2021; pp. 267–297. [Google Scholar]
- Macedo-Raygoza, G.M.; Valdez-Salas, B.; Prado, F.M.; Prieto, K.R.; Yamaguchi, L.F.; Kato, M.J.; Canto-Canché, B.B.; Carrillo-Beltrán, M.; Di Mascio, P.; White, J.F. Enterobacter cloacae, an endophyte that establishes a nutrient-transfer symbiosis with banana plants and protects against the black Sigatoka pathogen. Front. Microbiol. 2019, 10, 804. [Google Scholar] [CrossRef] [PubMed]
- Dimkić, I.; Janakiev, T.; Petrović, M.; Degrassi, G.; Fira, D. Plant-associated Bacillus and Pseudomonas antimicrobial activities in plant disease suppression via biological control mechanisms—A review. Physiol. Mol. Plant Pathol. 2022, 117, 101754. [Google Scholar] [CrossRef]
- Ali, S.; Saldias, S.; Weerasuriya, N.; Delaney, K.; Kandasamy, S.; Lazarovits, G. Corn microbial diversity and its relationship to yield. Can. J. Microbiol. 2020, 66, 457–473. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-C.; Zhang, Y.-L.; Guo, X.-J.; Luo, M.; Li, S.-D.; Guo, R.-J. Combination of Bacillus and low fertigation input promoted the growth and productivity of Chinese cabbage and enriched beneficial rhizosphere bacteria Lechevalieria. Biology 2023, 12, 1130. [Google Scholar] [CrossRef]
- Glaeser, S.P.; McInroy, J.A.; Busse, H.-J.; Kämpfer, P. Pseudogracilibacillus auburnensis gen. nov., sp. nov., isolated from the rhizosphere of Zea mays. Int. J. Syst. Evol. Microbiol. 2014, 64, 2442–2448. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, M.-K.; Yun, B.-R.; Han, J.-H.; Kim, S.B. Pseudogracilibacillus endophyticus sp. nov., a moderately thermophilic and halophilic species isolated from plant root. Int. J. Syst. Evol. Microbiol. 2018, 68, 165–169. [Google Scholar] [CrossRef]
- Jin, C.-Z.; Lee, J.M.; Kim, C.-J.; Lee, H.-G.; Shin, K.-S. Genomic Insight into Shimazuella soli sp. Nov. isolated from soil and its putative novel class II lasso peptide. Bioengineering 2022, 9, 812. [Google Scholar] [CrossRef]
- Hou, F.; Du, J.; Yuan, Y.; Wu, X.; Zhao, S. Analysis of microbial communities in aged refuse based on 16S sequencing. Sustainability 2021, 13, 4111. [Google Scholar] [CrossRef]
- Peng, T.; Meng, L.; Wang, Y.; Jin, L.; Jin, H.; Yang, T.; Yao, Y. Alterations of the rhizosphere soil microbial community composition and metabolite profiles of Angelica sinensis seedlings by co-application of Nitrogen fixing bacteria and amino acids. Plant Soil 2023, 493, 535–554. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Eida, A.A.; Bougouffa, S.; Alam, I.; Saad, M.M.; Hirt, H. Complete genome sequence of the endophytic bacterium Cellulosimicrobium sp. JZ28 isolated from the root endosphere of the perennial desert tussock grass Panicum turgidum. Arch. Microbiol. 2020, 202, 1563–1569. [Google Scholar] [CrossRef] [PubMed]
- Flores-Félix, J.D.; Velázquez, E.; Martínez-Molina, E.; González-Andrés, F.; Squartini, A.; Rivas, R. Connecting the lab and the field: Genome analysis of Phyllobacterium and Rhizobium strains and field performance on two vegetable crops. Agronomy 2021, 11, 1124. [Google Scholar] [CrossRef]
- Viso, N.P.; Rizzo, P.; Young, B.; Gabioud, E.; Bres, P.; Riera, N.; Merino, L.; Farber, M.; Crespo, D. Loss of Bacterial Genetic Diversity Together with the Increment of Eutrophic and Phytotoxic Risk, the Side Effect of Using Raw Poultry Waste as Soil Amendment. 2022. Available online: https://www.researchsquare.com/article/rs-1507219/v1 (accessed on 1 May 2025).
- Xie, P.; Yang, S.; Liu, X.; Zhang, T.; Zhao, X.; Wen, T.; Zhang, J.; Xue, C.; Shen, Q.; Yuan, J. Learning from seed microbes: Trichoderma coating intervenes in rhizosphere microbiome assembly. Microbiol. Spectr. 2023, 11, e0309722. [Google Scholar] [CrossRef]
- Deng, Z.; Wang, J.; He, Y.; Tu, Z.; Tian, F.; Li, H.; Wu, Z.; An, X. Biochar-based Bacillus subtilis inoculant for enhancing plants disease prevention: Microbiota response and root exudates regulation. Biochar 2023, 5, 81. [Google Scholar] [CrossRef]
- Brooke, J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012, 25, 2–41. [Google Scholar] [CrossRef] [PubMed]
- Bjerketorp, J.; Levenfors, J.J.; Nord, C.; Guss, B.; Öberg, B.; Broberg, A. Selective isolation of multidrug-resistant Pedobacter spp., producers of novel antibacterial peptides. Front. Microbiol. 2021, 12, 642829. [Google Scholar] [CrossRef]
- Mwanza, E.P.; Hugo, A.; Charimba, G.; Hugo, C.J. Pathogenic potential and control of Chryseobacterium species from clinical, fish, food and environmental sources. Microorganisms 2022, 10, 895. [Google Scholar] [CrossRef]
- Rojas, A.; Holguin, G.; Glick, B.R.; Bashan, Y. Synergism between Phyllobacterium sp.(N2-fixer) and Bacillus licheniformis (P-solubilizer), both from a semiarid mangrove rhizosphere. FEMS Microbiol. Ecol. 2001, 35, 181–187. [Google Scholar] [CrossRef]
- Chen, P.; Zhang, F.; Zhang, L.; Liu, H.; Zhang, Q.; Xing, Z.; Zhao, T. Characterization of a novel salt-tolerant strain Sphingopyxis sp. CY-10 capable of heterotrophic nitrification and aerobic denitrification. Bioresour. Technol. 2022, 358, 127353. [Google Scholar] [CrossRef]
- Ye, Y.-f.; Min, H.; Du, Y.-f. Characterization of a strain of Sphingobacterium sp. and its degradation to herbicide mefenacet. J. Environ. Sci. 2004, 16, 343–347. [Google Scholar]
- Tebele, S.M.; Marks, R.A.; Farrant, J.M. Exploring the root-associated microbiome of the resurrection plant Myrothamnus flabellifolia. Plant Soil 2024, 500, 53–68. [Google Scholar] [CrossRef]
- Kumar, A.; Soni, R.; Kanwar, S.S.; Pabbi, S. Stenotrophomonas: A versatile diazotrophic bacteria from the rhizospheric soils of Western Himalayas and development of its liquid biofertilizer formulation. Vegetos 2019, 32, 103–109. [Google Scholar] [CrossRef]
- Kusstatscher, P.; Wicaksono, W.A.; Thenappan, D.P.; Adam, E.; Müller, H.; Berg, G. Microbiome management by biological and chemical treatments in maize is linked to plant health. Microorganisms 2020, 8, 1506. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Hashem, A.; Abd_Allah, E.F. Bacillus: A biological tool for crop improvement through bio-molecular changes in adverse environments. Front. Physiol. 2017, 8, 667. [Google Scholar] [CrossRef]
- Ouertani, R.; Ouertani, A.; Mahjoubi, M.; Bousselmi, Y.; Najjari, A.; Cherif, H.; Chamkhi, A.; Mosbah, A.; Khdhira, H.; Sghaier, H. New plant growth-promoting, chromium-detoxifying microbacterium species isolated from a tannery wastewater: Performance and genomic insights. Front. Bioeng. Biotechnol. 2020, 8, 521. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Q.; Sun, W.; Guo, J.; Liu, W.; Zhao, M. Microbial communities in the rhizosphere soil of Ambrosia artemisiifolia facilitate its growth. Plant Soil 2023, 492, 353–365. [Google Scholar] [CrossRef]
- Dobrzyński, J.; Wierzchowski, P.S.; Stępień, W.; Górska, E.B. The Reaction of Cellulolytic and Potentially Cellulolytic Spore-Forming Bacteria to Various Types of Crop Management and Farmyard Manure Fertilization in Bulk Soil. Agronomy 2021, 11, 772. [Google Scholar] [CrossRef]
- Francis, A.J.; Dodge, C.J.; Meinken, G. Biotransformation of pertechnetate by Clostridia. Radiochim. Acta 2002, 90, 791–797. [Google Scholar] [CrossRef]
- Rabbi, S.M.; Warren, C.R.; Swarbrick, B.; Minasny, B.; McBratney, A.B.; Young, I.M. Microbial decomposition of organic matter and wetting–drying promotes aggregation in artificial soil but porosity increases only in wet-dry condition. Geoderma 2024, 447, 116924. [Google Scholar] [CrossRef]
- Kielak, A.M.; Cipriano, M.A.; Kuramae, E.E. Acidobacteria strains from subdivision 1 act as plant growth-promoting bacteria. Arch. Microbiol. 2016, 198, 987–993. [Google Scholar] [CrossRef]
- Dong, Q.; Zhao, X.; Zhou, D.; Liu, Z.; Shi, X.; Yuan, Y.; Jia, P.; Liu, Y.; Song, P.; Wang, X. Maize and peanut intercropping improves the nitrogen accumulation and yield per plant of maize by promoting the secretion of flavonoids and abundance of Bradyrhizobium in rhizosphere. Front. Plant Sci. 2022, 13, 957336. [Google Scholar] [CrossRef] [PubMed]
- Schmalenberger, A.; Hodge, S.; Bryant, A.; Hawkesford, M.J.; Singh, B.K.; Kertesz, M.A. The role of Variovorax and other Comamonadaceae in sulfur transformations by microbial wheat rhizosphere communities exposed to different sulfur fertilization regimes. Environ. Microbiol. 2008, 10, 1486–1500. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, C.; Xu, Q.; Ahmad, S.; Zhang, H.; Pang, Y.; Aikemu, A.; Liu, Y.; Yan, H. Characterization and genomic analysis of an efficient dibutyl phthalate degrading bacterium Microbacterium sp. USTB-Y. World J. Microbiol. Biotechnol. 2021, 37, 212. [Google Scholar] [CrossRef] [PubMed]
- Han, J.W.; Choi, G.J.; Kim, B.S. Antimicrobial aromatic polyketides: A review of their antimicrobial properties and potential use in plant disease control. World J. Microbiol. Biotechnol. 2018, 34, 163. [Google Scholar] [CrossRef]
- Kim, B.S.; Moon, S.S.; Hwang, B.K. Structure Elucidation and Antifungal Activity of an Anthracycline Antibiotic, Daunomycin, Isolated from Actinomadura r oseola. J. Agric. Food Chem. 2000, 48, 1875–1881. [Google Scholar] [CrossRef]
- Yan, H.; Li, Y.; Zhang, X.Y.; Zhou, W.Y.; Feng, T.J. A new cytotoxic and anti-fungal C-glycosylated benz [α] anthraquinone from the broth of endophytic Streptomyces blastomycetica strain F4-20. J. Antibiot. 2017, 70, 301–303. [Google Scholar] [CrossRef]
- Schönbichler, A.; Díaz-Moreno, S.M.; Srivastava, V.; McKee, L.S. Exploring the potential for fungal antagonism and cell wall attack by Bacillus subtilis natto. Front. Microbiol. 2020, 11, 521. [Google Scholar] [CrossRef]
- Wang, T.; Liu, X.-h.; Wu, M.-B.; Ge, S. Molecular insights into the antifungal mechanism of bacilysin. J. Mol. Model. 2018, 24, 118. [Google Scholar] [CrossRef]
- Dertz, E.A.; Xu, J.; Stintzi, A.; Raymond, K.N. Bacillibactin-mediated iron transport in bacillus s ubtilis. J. Am. Chem. Soc. 2006, 128, 22–23. [Google Scholar] [CrossRef]
- Singh, P.; Khan, A.; Kumar, R.; Ojha, K.K.; Singh, V.K.; Srivastava, A. In silico analysis of comparative affinity of phytosiderophore and bacillibactin for iron uptake by YSL15 and YSL18 receptors of Oryza sativa. J. Biomol. Struct. Dyn. 2023, 41, 2733–2746. [Google Scholar] [CrossRef]
- Zhu, C.; Zhang, G.; Shen, C.; Chen, S.; Tang, Y.; Mei, B.; Song, R. Expression of bacterial glutamine synthetase gene in Arabidopsis thaliana increases the plant biomass and level of nitrogen utilization. Biologia 2015, 70, 1586–1596. [Google Scholar] [CrossRef]
- Bittner, F.; Mendel, R.R. Molybdenum cofactor biosynthesis and cross talk with iron–sulfur. In Encyclopedia of Inorganic and Bioinorganic Chemistry; Wiley: Hoboken, NJ, USA, 2011; pp. 1–13. [Google Scholar]
- Adhikary, H.; Sanghavi, P.B.; Macwan, S.R.; Archana, G.; Naresh Kumar, G. Artificial citrate operon confers mineral phosphate solubilization ability to diverse fluorescent pseudomonads. PLoS ONE 2014, 9, e107554. [Google Scholar] [CrossRef]
- Nanda, A.K.; Andrio, E.; Marino, D.; Pauly, N.; Dunand, C. Reactive oxygen species during plant-microorganism early interactions. J. Integr. Plant Biol. 2010, 52, 195–204. [Google Scholar] [CrossRef]
- Dubbs, J.M.; Mongkolsuk, S. Peroxiredoxins in bacterial antioxidant defense. In Peroxiredoxin Systems: Structures and Functions; Springer: Dordrecht, The Netherlands, 2007; pp. 143–193. [Google Scholar]
- Saleem, H. Plant-bacteria partnership: Phytoremediation of hydrocarbons con-taminated soil and expression of catabolic genes. Bullet Environ. Stud. 2016, 1, 19. [Google Scholar]
- Hartmann, A.; Singh, M.; Klingmüller, W. Isolation and characterization of Azospirillum mutants excreting high amounts of indoleacetic acid. Can. J. Microbiol. 1983, 29, 916–923. [Google Scholar] [CrossRef]
- Jankiewicz, U.; Bielawski, W. The properties and functions of bacterial aminopeptidases. Acta Microbiol. Pol. 2003, 52, 217–231. [Google Scholar]
- Löwe, M.; Jürgens, K.; Zeier, T.; Hartmann, M.; Gruner, K.; Müller, S.; Yildiz, I.; Perrar, M.; Zeier, J. N-hydroxypipecolic acid primes plants for enhanced microbial pattern-induced responses. Front. Plant Sci. 2023, 14, 1217771. [Google Scholar] [CrossRef]
- Morcillo, R.J.; Manzanera, M. The effects of plant-associated bacterial exopolysaccharides on plant abiotic stress tolerance. Metabolites 2021, 11, 337. [Google Scholar] [CrossRef] [PubMed]
- Palmer, G.C.; Jorth, P.A.; Whiteley, M. The role of two Pseudomonas aeruginosa anthranilate synthases in tryptophan and quorum signal production. Microbiology 2013, 159, 959–969. [Google Scholar] [CrossRef]
- Singh, V.K.; Utaida, S.; Jackson, L.S.; Jayaswal, R.K.; Wilkinson, B.J.; Chamberlain, N.R. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology 2007, 153, 3162–3173. [Google Scholar] [CrossRef]
- Vanghele, L.M.; Ionescu, M.; Coste, H.; Ganea, E. Induction of DnaK and GroEL in Brucella ovis Under Various Stress Conditions. Int. J. Vet. Med. Res. Rep. 2013, 2013, 1–11. [Google Scholar] [CrossRef][Green Version]
- Sedláček, V.; Kučera, I. Functional and mechanistic characterization of an atypical flavin reductase encoded by the pden_5119 gene in Paracoccus denitrificans. Mol. Microbiol. 2019, 112, 166–183. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Shin, B.; Park, W. Protective role of bacterial alkanesulfonate monooxygenase under oxidative stress. Appl. Environ. Microbiol. 2020, 86, e00692-20. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, E.; van der Ploeg, J.R.; Leisinger, T. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J. Biol. Chem. 1999, 274, 26639–26646. [Google Scholar] [CrossRef]
- Speck, J.J.; James, E.K.; Sugawara, M.; Sadowsky, M.J.; Gyaneshwar, P. An alkane sulfonate monooxygenase is required for symbiotic nitrogen fixation by Bradyrhizobium diazoefficiens (syn. Bradyrhizobium japonicum) USDA110T. Appl. Environ. Microbiol. 2019, 85, e01552-19. [Google Scholar] [CrossRef]
- Avalos, J.; Carmen Limón, M. Biological roles of fungal carotenoids. Curr. Genet. 2015, 61, 309–324. [Google Scholar] [CrossRef]
- Jadhav, H.; Shaikh, S.; Sayyed, R. Role of hydrolytic enzymes of rhizoflora in biocontrol of fungal phytopathogens: An overview. In Rhizotrophs: Plant Growth Promot. Bioremediation; Springer: Singapore, 2017; pp. 183–203. [Google Scholar]
- Mitrofanova, O.; Mardanova, A.; Evtugyn, V.; Bogomolnaya, L.; Sharipova, M. Effects of Bacillus serine proteases on the bacterial biofilms. BioMed Res. Int. 2017, 2017, 8525912. [Google Scholar] [CrossRef]
- Peng, W.; Wang, Y.; Fu, Y.; Deng, Z.; Lin, S.; Liang, R. Characterization of the tellurite-resistance properties and identification of the core function genes for tellurite resistance in Pseudomonas citronellolis SJTE-3. Microorganisms 2022, 10, 95. [Google Scholar] [CrossRef]
- Johari, N.A.F.; Abidin, A.A.Z.; Ismail, N.F.N.; Yusof, Z.N.B. Endophytic Bacteria Induce Thiamine (Vitamin B1) Production in Oil Palm (Elaeis guineensis). Trop. Life Sci. Res. 2024, 35, 1. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Yourstone, S.; Mieczkowski, P.; Jones, C.D.; Dangl, J.L. Practical innovations for high-throughput amplicon sequencing. Nat. Methods 2013, 10, 999–1002. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Robeson, M.S.; O’Rourke, D.R.; Kaehler, B.D.; Ziemski, M.; Dillon, M.R.; Foster, J.T.; Bokulich, N.A. RESCRIPt: Reproducible sequence taxonomy reference database management. PLoS Comput. Biol. 2021, 17, e1009581. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Tanizawa, Y.; Fujisawa, T.; Kaminuma, E.; Nakamura, Y.; Arita, M. DFAST and DAGA: Web-based integrated genome annotation tools and resources. Biosci. Microbiota Food Health 2016, 35, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Sun, S.; Sun, N.; Lu, L.; Zhang, C.; Shi, W.; Zhao, Y.; Jia, S. HMMER-Extractor: An auxiliary toolkit for identifying genomic macromolecular metabolites based on Hidden Markov Models. Int. J. Biol. Macromol. 2024, 283, 137666. [Google Scholar] [CrossRef]
- Liang, Q.; Liu, C.; Xu, R.; Song, M.; Zhou, Z.; Li, H.; Dai, W.; Yang, M.; Yu, Y.; Chen, H. fIDBAC: A platform for fast bacterial genome identification and typing. Front. Microbiol. 2021, 12, 723577. [Google Scholar] [CrossRef]
- Asnicar, F.; Thomas, A.M.; Beghini, F.; Mengoni, C.; Manara, S.; Manghi, P.; Zhu, Q.; Bolzan, M.; Cumbo, F.; May, U. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 2020, 11, 2500. [Google Scholar] [CrossRef]


| Strain | Source and Year of Isolation | Contigs (N) | N50 (bp) | Genome Size (bp) | GC Content (%) | Genes | Protein Coding Sequences (CDSs) | Reference |
|---|---|---|---|---|---|---|---|---|
| Bacillus subtilis sp. subtilis BM-15a | Soil, Serbia, 2019 | 73 | 447,052 | 3,955,365 | 43.9 | 4024 | 3956 | This work |
| Bacillus subtilis sp. subtilis SSJ1 GCF_003665255.1 | Cheonggukjang, Republic of Korea, 2016 | 1 | 4,200,000 | 4,200,000 | 43.5 | 4418 | 4187 | [30] |
| Bacillus subtilis sp. subtilis OH 131.1 GCF_000706705.1 | Wheat anther, USA, 2014 | 1 | 4,000,000 | 4,000,000 | 44 | 4171 | 4061 | [31] |
| Bacillus subtilis sp. subtilis IITK SM1 GCF_003426125.1 | Food waste compost, India, 2018 | 43 | 238,200 | 4,100,000 | 43.5 | 4255 | 4005 | [32] |
| Related Genome | ANI | Matched | Fragments | Subspecies |
|---|---|---|---|---|
| BM-15a (our strain) | 100 | 1305 | 1305 | Bacillus subtilis sp. subtilis |
| GCF_000706705.1 | 99.4345 | 1276 | 1305 | OH 131.1 |
| GCF_003665255.1 | 99.3279 | 1281 | 1305 | SSJ-1 |
| GCF_003426125.1 | 98.9039 | 1282 | 1305 | IITK SM1 |
| GCF_003339745.1 | 98.8583 | 1245 | 1305 | SBE1 |
| GCF_000230755.1 | 98.8149 | 1272 | 1305 | SC-8 |
| GCF_001625135.1 | 98.1527 | 1252 | 1305 | ALBA01 |
| GCF_000349795.1 | 98.1265 | 1257 | 1305 | BAB-1 |
| GCF_000497365.1 | 98.0839 | 971 | 1305 | MP11 |
| GCF_003665215.1 | 98.0186 | 1160 | 1305 | PJ-7 |
| GCF_000227485.1 | 97.8605 | 1244 | 1305 | RO-NN-1 |
| Strain | Source and Year of Isolation | Contigs (N) | N50 (bp) | Genome Size (bp) | GC Content (%) | Genes | Protein Coding Sequences (CDSs) | Reference |
|---|---|---|---|---|---|---|---|---|
| Microbacterium sp. AL-11a | Soil, Serbia, 2019 | 104 | 44,752 | 2,953,076 | 70.08 | 2748 | 2698 | This work |
| Microbacterium hydrothermale (GCF_004854025.1) | Mirabilis jalapa, India, 2019 | 18 | 503,505 | 3,580,540 | 71 | 3379 | 3317 | [33] |
| Microbacterium testaceum (GCF_006539145.1) | Chinese paddy, Japan, 2019 | 28 | 249,634 | 3,590,404 | 69.5 | 3402 | 3339 | [34] |
| Microbacterium enclense (GCF_900096885.1) | N/A | 15 | 302,128 | 3,666,504 | 70.5 | 3403 | 3322 | [35] |
| Microbacterium proteolyticum (GCF_014192415.1) | N/A | 10 | 1,816,927 | 3,482,219 | 70 | 3261 | 3198 | [36] |
| Related Genome | ANI | Matched | Fragments | Species |
|---|---|---|---|---|
| AL-11a (our strain) | 100 | 930 | 935 | Microbacterium sp. |
| GCF_004854025.1 | 84.036 | 686 | 935 | Microbacterium hydrothermale |
| GCF_900096885.1 | 83.698 | 680 | 935 | Microbacterium enclense |
| GCF_006539145.1 | 83.276 | 649 | 935 | Microbacterium testaceum |
| GCF_014192415.1 | 83.055 | 675 | 935 | Microbacterium proteolyticum |
| GCF_014779795.1 | 80.743 | 482 | 935 | Microbacterium helvum |
| GCF_000956465.1 | 80.501 | 478 | 935 | Microbacterium trichothecenolyticum |
| GCF_008017415.1 | 80.496 | 524 | 935 | Microbacterium hatanonis |
| GCF_019511665.1 | 80.466 | 503 | 935 | Microbacterium jejuense |
| GCF_006783905.1 | 80.441 | 483 | 935 | Microbacterium kyungheense |
| GCF_015278255.1 | 80.439 | 482 | 935 | Microbacterium hibisci |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kruščić, K.; Jelušić, A.; Hladnik, M.; Janakiev, T.; Anđelković, J.; Bandelj, D.; Dimkić, I. The Influence of Bacterial Inoculants and a Biofertilizer on Maize Cultivation and the Associated Shift in Bacteriobiota During the Growing Season. Plants 2025, 14, 1753. https://doi.org/10.3390/plants14121753
Kruščić K, Jelušić A, Hladnik M, Janakiev T, Anđelković J, Bandelj D, Dimkić I. The Influence of Bacterial Inoculants and a Biofertilizer on Maize Cultivation and the Associated Shift in Bacteriobiota During the Growing Season. Plants. 2025; 14(12):1753. https://doi.org/10.3390/plants14121753
Chicago/Turabian StyleKruščić, Katarina, Aleksandra Jelušić, Matjaž Hladnik, Tamara Janakiev, Jovana Anđelković, Dunja Bandelj, and Ivica Dimkić. 2025. "The Influence of Bacterial Inoculants and a Biofertilizer on Maize Cultivation and the Associated Shift in Bacteriobiota During the Growing Season" Plants 14, no. 12: 1753. https://doi.org/10.3390/plants14121753
APA StyleKruščić, K., Jelušić, A., Hladnik, M., Janakiev, T., Anđelković, J., Bandelj, D., & Dimkić, I. (2025). The Influence of Bacterial Inoculants and a Biofertilizer on Maize Cultivation and the Associated Shift in Bacteriobiota During the Growing Season. Plants, 14(12), 1753. https://doi.org/10.3390/plants14121753








