Abstract
Peanut (Arachis hypogaea L.), a vital oilseed and cash crop, faces yield limitations due to abiotic stresses. The 9-cis-epoxycarotenoid dioxygenase (NCED) enzyme, a key enzyme in abscisic acid (ABA) biosynthesis regulating plant development and stress responses, remains mechanistically uncharacterized in peanut abiotic stress tolerance. In this study, we isolated a novel gene, AhNCED4, from the salt-tolerant mutant M24. The expression of AhNCED4 was strongly induced by NaCl, PEG6000, and ABA in peanut huayu20. Overexpression of AhNCED4 enhanced salt and drought tolerance in Arabidopsis. Transgenic overexpression of AhNCED4 improved salt and stress resistance through upregulated ROS-scavenging genes superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) with elevated enzymatic activities while reducing malondialdehyde (MDA), superoxide anion (O2−), and hydrogen peroxide (H2O2) accumulation compared to wild-type plants. Further research showed that the chlorophyll fluorescence parameters of transgenic lines were significantly increased, while light damage was significantly reduced. These findings establish AhNCED4 as a critical regulator of stress adaptation and an excellent candidate gene for resistance breeding in peanut.
1. Introduction
Peanut (Arachis hypogaea L.), an important oil and cash crop, is extensively cultivated in more than 100 countries and is highly valued by consumers globally [1,2,3]. As well as serving as an excellent source of edible oils, peanut also act as a high-quality ingredient in food industry applications. In 2023, global peanut cultivation spanned approximately 30.92 million hectares, producing around 54.27 million metric tons, with China alone contributing over 19.23 million metric tons (without being shelled) [4]. Drought and salinity represent critical abiotic stresses that induce osmotic/oxidative stress, disrupt cellular ion/redox homeostasis, and increasingly threaten global food sustainability [5,6,7,8,9,10,11]. To cope with these stresses, plants employ defense mechanisms involving gene expression regulation, with enzymes and proteins serving as key mediators of abiotic stress tolerance through reactive oxygen species (ROS) scavenging, ion balance maintenance, and cell membrane integrity preservation [12,13].
The plant enzyme 9-cis-epoxycarotenoid dioxygenase (NCED), a pivotal component in abscisic acid (ABA) biosynthesis, catalyzes the stereospecific cleavage of 9-cis-epoxycarotenoids to generate xanthoxin—the essential ABA precursor [14,15,16,17]. NCED mediates the upregulation of endogenous ABA biosynthesis to enhance plant stress resistance under adverse environmental conditions [18,19,20,21,22]. NCEDs are members of a multigene family detected in many plant species. NCED homologs have been identified and studied in many plant species [23,24,25,26]. The first NCED gene was identified through the characterization of specific oxidative cleavage in the maize vp14 mutant [27]. In Brassica napus, the expression of BnNCED3 enhances stress tolerance by enhancing endogenous ABA biosynthesis and the production of NO and ROS in transgenic Arabidopsis [28]. In cotton (Gossypium hirsutum L.), the expression of GhNCED5, GhNCED6, and GhNCED13 was similar to changes in ABA content, confirming their functional roles in ABA synthesis [25]. In rice, overexpression of OsNCED3 results in reduced relative water loss, delayed seed germination, and greater drought tolerance relative to that of wild-type plants [29]. In wheat, heterologous expression of TaNCED1 in tobacco significantly improved its drought tolerance, and it exhibited a higher germination rate and higher relative water content when compared with WT plants [30].
Research on the regulation of ABA biosynthesis has focused on stress responses in vegetative-stage plants across various species, as both environmental stimuli and developmental cues critically influence abscisic acid production in plants. The equilibrium between ABA biosynthesis and catabolism tightly regulates endogenous ABA concentrations. Notably, 9-cis-epoxycarotenoid dioxygenase (NCED) is recognized as a pivotal regulatory enzyme in ABA production, as its transcriptional activity strongly correlates with cellular ABA levels, and its overexpression directly induces ABA overaccumulation [31]. In Arabidopsis plants, overexpressing AtNCED3 displays enhanced water stress tolerance accompanied by increased ABA levels [32]. After drought treatment, overexpression of OsNCED3 results in increased accumulation of ABA [29]. Most importantly, the nced3 mutant shows defective ABA biosynthesis [15]. While the functions of ABA metabolic genes are well-characterized in model plant systems, their identification and functional validation in peanuts are still important.
Peanut, a globally crucial oilseed crop, displays a certain level of tolerance to abiotic stress, though productivity remains significantly constrained by drought and salinity. NCED genes are established regulators of plant stress responses; however, their functional roles in peanut remain uncharacterized. The NCED gene family is rate-limiting in abscisic acid (ABA) biosynthesis, a central regulator of plant stress responses. However, the evolutionary conservation and functional divergence of NCED homologs in peanut remain poorly characterized, particularly under abiotic stresses. Analysis of the peanut transcriptome dataset comparing ‘Huayu20’ and the salt-tolerant mutant M24 [33] revealed that NCED4 exhibited the most significant expression changes, suggesting its potential involvement in salt stress tolerance. This study aimed to identify the NCED gene associated with salt and drought tolerance in peanut and validate its role in ROS scavenging through heterologous expression. This study elucidates the molecular characteristic of AhNCED4 under salt and drought stress conditions in peanut.
2. Results
2.1. The Expression Analysis of the AhNCED4 Gene in Peanut
In peanut, a novel NCED gene was isolated from a salt-tolerant peanut mutant M24 through reanalysis of transcriptome data (9-cis-epoxycarotenoid dioxygenase gene 4). Subsequent expression analysis in ‘Huayu20’ under NaCl, 30% PEG6000, and ABA treatments showed distinct induction patterns. In roots, AhNCED4 expression peaked at 6 h, 1 h, and 24 h after NaCl, PEG, and ABA treatments, respectively (Figure 1A). Leaves displayed more pronounced responses, with expression levels reaching 67.28-fold, 47.37-fold, and 5.14-fold increases over controls following 6 h NaCl, 30% PEG6000, and 100 μM ABA treatments (Figure 1B). These results demonstrate that AhNCED4 might play an important role in the response to salt, drought, and ABA stress.
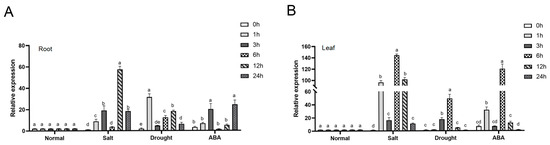
Figure 1.
Expression analysis of AhNCED4 gene in Huayu20. The expression levels of AhNCED4 were assessed in roots (A) and leaves (B) before and after treatment with H2O, 30% PEG6000, 300 mM NaCl, and 100 μM ABA. Data are presented as mean ± SE (n = 3). Different letters indicate significant differences between treatment groups at the same time point.
2.2. Cloning and Sequence Analysis of AhNCED4
AhNCED4, which showed strong induction under salt and drought stresses, was cloned from the salt-tolerant peanut mutant M24. The DNA sequence of this gene was 2970 bp, and it contained two exons and one intron. The full-length cDNA of AhNCED4 was 1779 bp, and it featured a complete open reading frame encoding a 593 amino acid polypeptide with a predicted molecular weight of 148.46 kDa and a pI of 4.88 (Table S1). The encoding protein of AhNCED4 contained a conserved EXT signature domain, as demonstrated through multiple sequence alignment with NCED4 homologs from other species (Figure 2A,B). In addition, based on phylogenetic analysis, this novel protein shared the highest identity with AtNCED4 and was named AhNCED4 (Figure 2C).
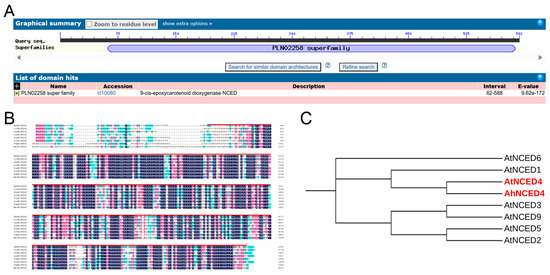
Figure 2.
Sequence alignment and phylogenetic analysis of AhNCED4 proteins. (A) In the NCED protein sequence 62–588aa, there was a conserved domain called the PLN02258 superfamily. (B) Sequence alignment: Homologous NCEDs from seven different plant species, including Cajanus cajan, Gastrolobium bilobum, Lupinus angustifolius, Populus alba, Stylosanthes scabra, Tripterygium wilfordii, and Morella rubra. The conserved domain is marked by red lines. (C) Alignment of the amino acid sequences of AhNCED4 and its 7 orthologs.
2.3. Overexpression of AhNCED4 Enhanced Salt and Drought Tolerance in Arabidopsis
To investigate the function of AhNCED4 in plants, the expressed vector of this gene was transformed into WT Arabidopsis. A total of twelve independent homozygous Arabidopsis lines of AhNCED4 overexpressing were obtained. T3 seeds from two confirmed transgenic lines (designated OE-1 and OE-2) were selected for subsequent analysis under salt and drought stress conditions. Under normal conditions, the expression level of AhNCED4 was significantly elevated in these two overexpressing lines. Moreover, under both salt and drought stress conditions, its expression was even more markedly higher than that in the wild-type plants (Figure 3).
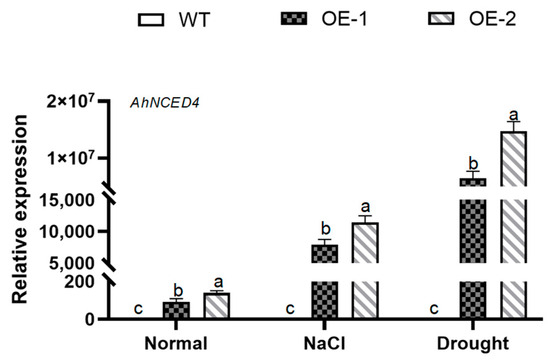
Figure 3.
Expression analysis of the AhNCED4 gene was performed in WT and overexpressing lines. The expression levels of AhNCED4 were assessed in leaves before and after treatment with H2O, 30% PEG6000, and 300 mM NaCl. Data are presented as mean ± SE (n = 3). Different letters indicate significant differences between the same treatment groups.
To determine the germination rate of AhNCED4 under salt and drought stresses, 1/2 MS medium containing 125 mM NaCl or 325 mM mannitol was employed for salt or drought stresses for seven days, respectively. Under control conditions, all the seeds from the WT and transgenic lines could be fully germinated after sowing, with no significant difference in germination rate (Figure 4A–C). However, the germination and cotyledon greening rates were significantly higher in the transgenic lines than in the WT plants under the stress treatments (Figure 4A–C). Based on the results of the root elongation experiment, no significant differences in the root length and fresh weight were observed between the WT plants and the two transgenic lines (Figure 4D–F). In contrast, the transgenic seedlings maintained significantly longer roots and greater biomass than the WT plants under salt and drought stresses (Figure 4D–F).
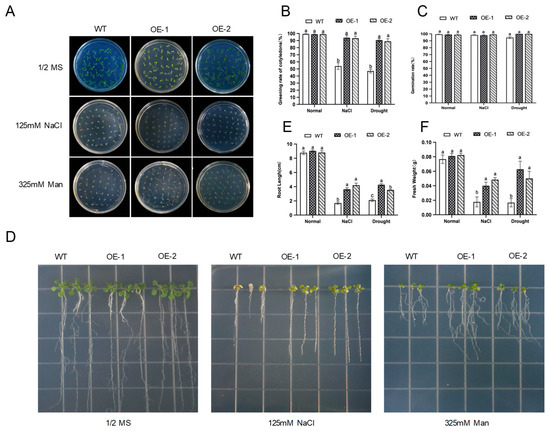
Figure 4.
The seed germination phenotype, seedling surface morphology, and physiological trait variations in WT and transgenic Arabidopsis under normal, salt stress, and drought stress conditions. (A) The seed germination phenotype of WT and transgenic Arabidopsis under 125 mM NaCl and 325 mM mannitol treatments. (B) The phenotypes of transgenic lines and WT seedling plants after one week of growth on 1/2 MS medium supplemented with 125 mM NaCl and 325 mM mannitol. (C,D) The cotyledon greening rate (C) and germination rates (D) were calculated between WT and transgenic Arabidopsis under salt and drought treatments, respectively. (E,F) The primary root length (E) and fresh weight (F) of transgenic lines and WT plants were measured, respectively. Different letters indicate significant differences between the same treatment groups.
2.4. Transgenic Arabidopsis Plants Display Better Chlorophyll Fluorescence Parameters Under Salt and Drought Stresses
To further investigate the function of AhNCED4 in mature soil growth plants, 4-week-old seedlings of the transgenic and WT lines were subjected to salt and drought treatments. The tested plants all showed similar growth trends under normal conditions (Figure 5A). Under salt stress conditions, the wild-type (WT) plants exhibited nearly complete leaf yellowing, while the transgenic lines maintained over 50% of green foliage (Figure 5A). When subjected to drought stress (without irrigation), the WT plants showed extensive wilting of rosette leaves, contrasting with the transgenic AhNCED4-overexpressing lines, where senescence was primarily limited to mature basal leaves (Figure 5A). Notably, the WT plants failed to survive after a 2-day re-watering period, while the transgenic plantlets successfully recovered (Figure 5A). These phenotypic data indicated that AhNCED4 overexpression enhances salt and drought tolerance in Arabidopsis.
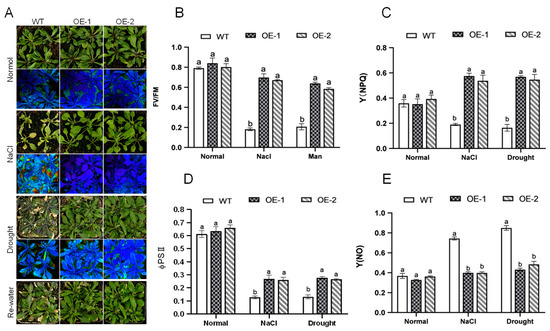
Figure 5.
AhNCED4 enhances tolerance to salt and drought stress in Arabidopsis. (A) The responses of AhNCED4 transgenic and WT Arabidopsis plants were identified after two weeks of cultivation in pots under normal conditions, 300 mM NaCl stress, or drought stress followed by re-watering for three days. The maximal photochemical efficiency of photosystem II (PSII) was measured as (Fv/Fm). The false color code depicted at the bottom of the image ranges from 0 (red) to 1 (purple). (B–E) The chlorophyll fluorescence parameters, including Fv/Fm (B), Y(NPQ) (C), φPII quantum efficiency II (PSII) index (D), and Y(NO) (E) for WT and overexpressing Arabidopsis plants under normal, salt, and drought conditions. Data are presented as mean ± SE (n = 3). Different letters indicate significant differences between the same treatment groups.
To estimate photosynthetic responses under salt and drought stress, several chlorophyll fluorescence parameters, including Fv/Fm, ΦPSII, NPQ, and NO were compared between the WT and AhNCED4 overexpressing Arabidopsis lines. We used the same transgenic lines grown in potting soil mixture to investigate abiotic stress tolerance. After treatment with normal conditions for two weeks, there were no significant differences in the chlorophyll fluorescence between the WT and transgenic lines. Following treatment with salt and drought stress, more leaves died in the WT plantlets than in the transgenic plantlets (Figure 5A). The survival rate, chlorophyll content, Fv/Fm, ΦPSII, and NPQ of the transgenic lines were significantly higher compared with the WT plants (Figure 5B–D). Conversely, the Y(NO) of overexpression lines was significantly lower than those of the WT plants (Figure 5E). These results demonstrate that AhNCED4 overexpression preserves photosynthetic efficiency during abiotic stress through enhanced photoprotective capacity.
2.5. Overexpression of AhNCED4 Activated the ROS Scavenging System Under Salt and Drought Stress
Salt and drought stresses induce oxidative damage in plants through ROS accumulation. The ABA content was markedly higher in the transgenic lines than in the WT plantlets after stress treatment, and the content of the phytohormone did not significantly differ among the different lines without stress treatment (Figure 6B). In addition, the accumulations of two representative reactive oxygen species (ROS), H2O2 and O2−, were detected by DAB staining and NBT staining. Under normal conditions, the leaves of both the WT and AhNCED4-overexpressing lines exhibited only slight and comparable staining (Figure 6A), indicating that H2O2 and O2− accumulated at low and similar levels in the leaves of both the WT and transgenic plants. However, after treatment with salt and drought stresses, significant staining differences became apparent between the WT and transgenic lines. The leaves of the WT plants developed more dense plaques compared to those of the overexpressing plants (Figure 6). These results indicate that the leaves of transgenic lines accumulated less H2O2 and O2− than those of the WT plants.
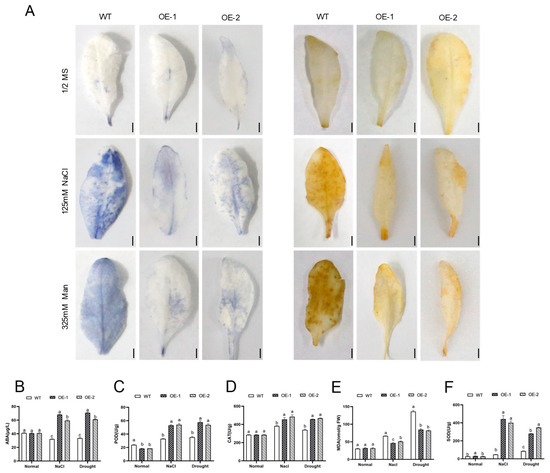
Figure 6.
Detection of H2O2 and O2− accumulation in transgenic and WT Arabidopsis plants under normal, salt, and drought stresses. (A) Leaves of AhNCED4 transgenic and WT Arabidopsis plants were stained with NBT (left) and DAB (right) after three weeks of cultivation under normal conditions and NaCl and drought treatments. (B) The ABA hormone content. (C–F) The activity of enzyme SOD activity (C), POD activity (D), CAT (E), and MDA content (F) in WT and overexpression lines under normal conditions and NaCl and drought treatments. Data are presented as mean ± SE (n = 3). Different letters indicate significant differences between the same treatment groups.
In addition, to evaluate the physiological changes in the plants under salt and drought conditions, four physiological parameters, superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and malondialdehyde (MDA) levels, were measured. Under normal conditions, the overexpressing lines showed a similar accumulation of ROS compared to the WT plants (Figure 6C–F). However, after stress treatment, the transgenic lines showed significantly higher activities of SOD, POD, and CAT, as well as a lower MDA content compared with the WT plants (Figure 6C–F). Salt and drought stresses increased the H2O2 content in all types of plants but to a lesser extent in the overexpression lines. These results indicate that AhNCED4-overexpressing plants have an enhanced ROS-scavenging capacity.
2.6. AhNCED4 Activated Stress-Response-Related Genes
To further investigate how AhNCED4 responds to drought and salt, the expression levels of certain ROS genes and stress-responsive genes were analyzed. Under normal conditions, no significant differences were observed in the expression levels of these genes between the transgenic and control lines (Figure 7A–D). Conversely, the expression of AhNCED4 increased significantly following salt and drought treatment. Moreover, the expression of ROS genes (AtSOD, AtPOD), ABA, or proline-biosynthesis-associated genes (AtABA1, AtP5CS) was notably higher in the transgenic lines compared to the control lines (Figure 7A–D). These findings demonstrate that overexpression of AhNCED4 enhanced the salt and drought stress tolerance of the plants by regulating the expression of stress-responsive genes related to ROS scavenging.
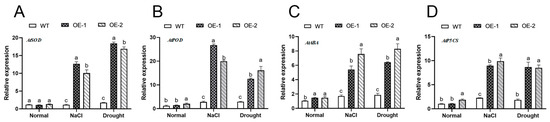
Figure 7.
The relative expression levels of AhNCED4 and abiotic-stress-responsive genes in the WT and transgenic Arabidopsis plants under normal salt and drought conditions. Expression levels of AtSOD (A), AtPOD (B), AtABA (C) and AtP5CS (D). Different letters indicate significant differences between the same treatment groups.
3. Discussion
The enzymatic activity of NCED family members mediates the conversion of cis-epoxycarotenoids to xanthoxin, a critical biochemical process that serves as a key regulatory node in the abscisic acid biosynthesis pathways of plants [31]. In the present study, a cDNA encoding AhNCED4 was isolated from the salt-tolerant mutant M24. The expression of AhNCED4 was strongly induced by NaCl, PEG6000, and ABA in peanut. AhNCED4-overexpressing transgenic plants demonstrated reduced malondialdehyde and peroxide levels, along with enhanced ROS scavenging capacity. Therefore, our results demonstrate that AhNCED4 is crucial for peanut tolerance to salt and drought stresses.
In recent years, salinity and drought have increasingly constrained agricultural productivity and regional economies, disrupting plant physiological processes and limiting crop yields. As a key rate-limiting enzyme in abscisic acid biosynthesis, NCED proteins have been extensively studied for their regulatory roles in plant development and stress adaptation [34,35]. Despite this importance, the NCED gene family remains uncharacterized in peanut. In this study, a novel AhNCED gene was successfully identified and functionally characterized, demonstrating its critical involvement in salt and drought stress responses.
NCED serves as a critical regulator of abscisic acid (ABA) biosynthesis, directly activating downstream ABA signaling components [32,34]. In maize, the reduced ABA content in all the mutants is likely a result of downregulated NCED4 expression, indicating a key role of NCED4 in ABA biosynthesis and seed dormancy maintenance [36]. Previous studies have shown that overexpression of OsNCED5 in Oryza sativa could increase ABA levels, enhanced tolerance to drought stress, and accelerated leaf senescence [37]. ABA-mediated stress responses involve synergistic interactions with jasmonic acid (JA) and salicylic acid (SA) pathways [38]. The overexpression of BnNCED3 facilitates ABA accumulation, as well as the production of NO and ROS in transgenic Arabidopsis plants, consequently enhancing the plants’ tolerance to abiotic stress [28]. Consistent with these conserved mechanisms, in this study, overexpressing AhNCED4 consistently increased tolerance to salt and drought stresses. The phenotypes observed in the transgenic lines were accompanied by significantly higher levels of ABA in AhNCED4-overexpressing transgenic plants. Therefore, the AhNCED4 gene plays an important role in stress response.
In plants, salinity and drought induce oxidative stress in plants through reactive oxygen species (ROS) overaccumulation [39]. The excessive accumulation of ROS disrupts cellular homeostasis by impairing ion balance, damaging membrane integrity, inhibiting enzymatic activity, and degrading photosynthetic efficiency [5,40,41,42]. ROS-scavenging enzymes are major contributors to alleviating ROS damage, especially under various stresses [43,44,45,46]. Maintaining ROS equilibrium is critical for stress adaptation [47,48,49,50]. In our study, higher enzyme activities of SOD, POD, and CAT were detected in Arabidopsis AhNCED4-overexpressing lines when subjected to salt and drought treatment (Figure 6B–D). The transgenic lines exhibited lower levels of O2− and MDA compared to the control plants, which indicates less membrane damage from excessive ROS (Figure 6E). ROS scavenging system-related enzymes were upregulated in overexpression lines under salt and drought treatment through qRT-PCR analysis. Furthermore, ABA modulates the activity of antioxidant enzymes (e.g., SOD, CAT) to regulate ROS levels. When ABA-induced ROS accumulation exceeds the scavenging capacity, it exacerbates thylakoid membrane damage, thereby further suppressing photosynthetic electron transport. Consequently, the enhanced capacity for ROS-scavenging likely contributes to the improved tolerance to salt and drought in the transgenic peanut and Arabidopsis lines.
Under stress treatment, ABA may influence chlorophyll metabolism through dual mechanisms: by promoting chlorophyllase activity while simultaneously suppressing the expression of chlorophyll-synthesis-related genes, ultimately leading to chlorophyll degradation. This process directly reduces the light-harvesting efficiency of photosystem II (PSII). Salt- and drought-stress-induced excessive reactive oxygen species (ROS) pose a threat to the photosynthetic apparatus, thereby impeding plant growth and development. Therefore, activation of ROS-scavenging mechanisms can effectively safeguard the photosynthetic machinery and the resistance of plants to salt and drought stress [44]. In our study, the chlorophyll content, Fv/Fm, ΦPSII, and NPQ of the transgenic lines were significantly higher compared with the WT plants under salt and drought stress. These findings demonstrate that AhNCED4 overexpression strengthened the antioxidant defense system, consequently preserving photosystem integrity under salt and drought conditions.
4. Materials and Methods
4.1. Plant Materials
The salt-tolerant mutant M24 was developed through the Pingyangmycin mutagenesis of the “Huayu23” population followed by NaCl-directed screening. The wild-type (WT) Arabidopsis thaliana used in our study was ecotype Columbia (Col). The WT plant was cultivated under a light incubator (22 °C, 16 h light/8 h dark cycle) or on a half-strength MS medium for AhNCED4 functional characterization through genetic transformation.
4.2. AhNCED4 Expression Analysis in Peanut
The total RNA was extracted from 30-day-old leaves and roots of huayu20 using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) followed by purification using a Qiagen RNeasy Kit (Qiagen, Hilden, Nordrhein-Westfalen, Germany). First-strand cDNA synthesis was conducted using a Superscript™ II 1st Strand cDNA Synthesis Kit (TaKaRa, Beijing, China). To analyze the expression of AhNCED4 under different stress conditions, the 3-week-old seedlings of huayu20 were treated with 300 mM NaCl, 30% PEG6000, and 100 μM ABA at different times (0, 1, 3, 6, 12, and 24 h). The expression levels were analyzed using a CFX96 Real-Time System (Bio-Rad, Hercules, CA, USA) with AhActin as the internal reference control. The variation in expression was estimated from three biological replicates, analyzing the relative gene expression data by the 2−ΔΔCt method.
4.3. Cloning and Sequences Analysis of AhNCED4
According to the sequence alignment, AhNCED4-specific primers AhNCED4-F/R were designed from the AhNCED4 coding sequence. Similarities in protein sequences were analyzed using the BLAST program on the NCBI website, according to the GenBank database (https://www.ncbi.nlm.nih.gov//Blast.cgi, accessed on 15 March 2023). Multiple sequence alignment of NCED4 from different plants was conducted with the DNAMAN 7 software (Table S2). For the phylogenetic tree construction, MEGA11 was used with the neighbor-joining method [51].
4.4. Vector Construction and Arabidopsis Transformation
The full-length coding sequence (CDS) of AhNCED4 was amplified from salt-tolerant mutant M24 and cloned into the SuperpCAMBIA1300 vector via the KpnⅠ/BamHⅠ restriction sites to generate the SuperpCAMBIA1300-AhNCED4 overexpression construct. This recombinant vector was introduced into wide-type Arabidopsis using the Agrobacterium-mediated floral dip [52]. Transgenic Arabidopsis seeds were planted on 1/2 Murashige and Skoog (MS) media containing 25 mg/L hygromycin, with two T3 homozygous lines (OE-1 and OE-2) selected for phenotypic analysis.
4.5. Drought and Salt Tolerance Analysis of Arabidopsis Transgenic Lines
To perform the germination assay, WT and transgenic Arabidopsis seeds (these seeds were cultivated under a light incubator 22 °C 16 h light/8 h dark cycle) and were planted on 1/2 MS medium containing either 125 mM NaCl or 325 mM mannitol, respectively. A seed was regarded as germinated when the radicle tip emerged from the seed coat, and each culture dish contained 50 seeds. The germination rate and green rate of cotyledons were counted within 7 days. Seven-day-old Arabidopsis seedlings of transgenic and WT lines were transferred to 1/2 MS medium supplemented with 100 mM NaCl or 350 mM mannitol, and the root length and fresh weight were measured after 10 days. Transgenic Arabidopsis and WT plantlets were grown in soil (a mixed substrate composed of substrate soil–vermiculite–perlite at a volume ratio of 2:2:1) for further growth for three weeks. The experiments were conducted in plastic pots with cubic dimensions of 7 cm (length) × 7 cm (width) × 7 cm (height). Uniform seedlings were then watered with 300 mM NaCl solution (200 mL of NaCl solution was applied for salinization) every two days for three weeks or were not watered for three weeks followed by re-watering for two days, respectively.
4.6. Histochemical Detection of ROS
ROS accumulation in Arabidopsis leaves under normal/stress conditions was visualized through 3,3′-diaminobenzidine (DAB) and nitroblue tetrazolium chloride (NBT) staining following established protocols [53]. Fresh leaves were collected from salt-/drought-treated WT and transgenic Arabidopsis plants. The activity of superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) was measured according to the manufacturer’s instruction (SOD: AKAO001M, boxbio, Beijing, China; POD: AKAO005M, boxbio, Beijing, China; CAT: AKAO003-2M; boxbio, Beijing, China), and the content of MDA was measured using the relevant chemical kits (Suzhou Grace Biotechnology Co, Suzhou, China). The detailed experimental procedures were conducted as follows: The nitrotetrazolium (NBT) photochemical reduction method: SOD inhibited the photochemical reduction reaction of NBT under light by catalyzing the disproportionation of superoxide anion radicals (O2−) into H2O2 and O2−. The NBT reduction product had a characteristic absorption peak at 560 nm, and the absorbance was negatively correlated with SOD activity. The guaiacylphenol method: In the presence of H2O2, POD catalyzed the oxidation of guaiacol to generate red–brown products. The absorbance change was measured at 470 nm, and the increase in absorbance was positively correlated with the enzyme activity. CAT decomposed H2O2 to generate H2O and O2. The enzyme activity was calculated by monitoring the absorbance attenuation rate (ΔA/min) of H2O2 at 240 nm. The thiobarbituric acid (TBA) method: MDA was heated with TBA under acidic conditions (95 °C, 30 min) to form a reddish-brown product (Mikagawa). The absorbance was measured at 532 nm, and the non-specific absorption was measured at 600 nm, and the background was subtracted simultaneously. The MDA content was calculated based on the molar extinction coefficient.
4.7. Determination of Chlorophyll Fluorescence in Arabidopsis
Chlorophyll fluorescence parameters were measured afterwards using the IMAG-K7 chlorophyll fluorescence imaging system (WALZ, Bavaria, Nuremberg, Germany). The maximal photochemical efficiency of photosystem II (PSII) (Fv/Fm), the quantum efficiency of PSII photochemistry (ΦPSII), the non-photochemical quenching (NPQ), and the photodamage index (NO) were calculated according to the formulas described by Kramer et al. (2004) [54]. The parameter calculation formula is as follows: Fv/FM = (FM − Fo)/FM5; ΦPSII = (FM’ − Fs)/FM’ = ΔF/FM’; NPQ = (FM − FM’)/FM’ = FM/FM’ − 1; qP = (FM − FS)/(FM’ − Fo’).
4.8. Expression Analysis of Stress-Related Genes
The leaves of AhNCED4 transgenic Arabidopsis plants were collected to detect gene expression under normal, salt, and drought conditions. qRT-PCR quantification assessed expression patterns of ROS-scavenging genes and stress-response genes. The AhActin gene was used as an internal control.
4.9. Primers
The primers used in this study are listed in Table S3.
4.10. Statistical Analysis
All experiments were repeated three times, and data are presented as means ± SE. Two-tailed Student’s t-tests were performed using SPSS version 17 (SPSS Inc., Chicago, IL, USA). Statistical significance was set at p < 0.05 and p < 0.01.
5. Conclusions
Peanuts are an important economic and oil crop, providing oil and protein for human nutrition. In this study, the AhNCED4 gene was shown to play an important role in salt and drought tolerance. Transgenic plants of AhNCED4 exhibited lower malondialdehyde and peroxide contents as well as a higher ability to remove ROS. These findings establish that AhNCED4 confers stress tolerance through coordinated upregulation of antioxidant enzymes and stress-responsive genes. This result may unveil that a novel gene, AhNCED4, has the potential to improve the abiotic stress tolerance of peanut, although the molecular mechanisms underlying salt and drought tolerance warrant further investigation. Therefore, our results demonstrate that AhNCED4 is crucial for peanut tolerance to these stresses. Moreover, the potential of AhNCED4 in genetically improving crops to better withstand the challenges posed by climate change cannot be overstated.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants14121741/s1, Table S1: The full-length AhNCED4 gene sequence with its coding and protein sequences in peanut; Table S2: Protein sequences of homologous NCED4 from eight different plant species; Table S3: Primes used in this study.
Author Contributions
Data curation, W.W., M.Z., S.X., Z.H., X.L., S.W. and K.Z.; funding acquisition, L.Q. and Y.T.; supervision, L.Q. and Y.T.; validation, C.Z.; writing—original draft, W.W.; writing—review and editing, W.W., M.Z., L.Q. and Y.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Key R&D Program of Shandong Province, China, grant number 2024LZGC031, Salt-Alkali Agriculture Industry System of Shandong Province, grant number SDAIT-29-03, and Shandong Province College Students’ innovation and entrepreneurship training program, grant number S202410435023.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Cuc, L.M.; Mace, E.S.; Crouch, J.H.; Quang, V.D.; Long, T.D.; Varshney, R.K. Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea). BMC Plant Biol. 2008, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Miao, P.; Meng, X.; Li, Z.; Sun, S.; Chen, C.Y.; Yang, X. Mapping Quantitative Trait Loci (QTLs) for Hundred-Pod and Hundred-Seed Weight under Seven Environments in a Recombinant Inbred Line Population of Cultivated Peanut (Arachis hypogaea L.). Genes 2023, 14, 1792. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Gao, G.; He, L.; Han, Z.; Shan, S.; Zhong, R.; Zhou, C.; Jiang, J.; Li, Y.; Zhuang, W. Genetic diversity in cultivated groundnut based on SSR markers. J. Genet. Genom. 2007, 34, 449–459. [Google Scholar] [CrossRef]
- FAOSTAT. FAOSTAT Statistical Database; FAO Food and Agriculture Organization of the United Nations: Rome, Italy, 2023. [Google Scholar]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.M.; Horie, T. Genomics, Physiology, and Molecular Breeding Approaches for Improving Salt Tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef]
- Surendran, U.; Kumar, V.; Ramasubramoniam, S.; Raja, P. Development of drought indices for semi-arid region using drought indices calculator (DrinC)—A case study from Madurai District, a semi-arid region in India. Water Resour. Manag. 2007, 31, 3593–3605. [Google Scholar] [CrossRef]
- Kader, M.A.; Lindberg, S. Cytosolic calcium and pH signaling in plants under salinity stress. Plant Signal. Behav. 2010, 5, 233–238. [Google Scholar] [CrossRef]
- Ramachandra Reddy, A.; Chaitanya, K.V.; Vivekanandan, M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J. Plant Physiol. 2004, 161, 1189–1202. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Z.H.; Feng, L.; Zheng, Y.; Li, D.D.; Li, X.B. Genome-wide functional analysis of cotton (Gossypium hirsutum) in response to drought. PLoS ONE 2013, 8, e80879. [Google Scholar] [CrossRef]
- Wang, T.; Li, X.K.; Liu, X.; Yang, X.Q.; Li, Y.J.; Hou, B.K. Rice glycosyltransferase gene UGT2 functions in salt stress tolerance under the regulation of bZIP23 transcription factor. Plant Cell Rep. 2023, 42, 17–28. [Google Scholar] [CrossRef]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Env. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, Q.; Yan, C.; Sun, Q.; Wang, J.; Li, C.; Yuan, C.; Mou, Y.; Shan, S. The bHLH transcription factor AhbHLH121 improves salt tolerance in peanut. Int. J. Biol. Macromol. 2024, 256, 128492. [Google Scholar] [CrossRef]
- García-Gómez, B.E.; Salazar, J.A.; Nicolás-Almansa, M.; Razi, M.; Rubio, M.; Ruiz, D.; Martínez-Gómez, P. Molecular Bases of Fruit Quality in Prunus Species: An Integrated Genomic, Transcriptomic, and Metabolic Review with a Breeding Perspective. Int. J. Mol. Sci. 2020, 22, 333. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef] [PubMed]
- García-Pastor, M.E.; Giménez, M.J.; Serna-Escolano, V.; Guillén, F.; Valero, D.; Serrano, M.; García-Martínez, S.; Terry, L.A.; Alamar, M.C.; Zapata, P.J. Oxalic Acid Preharvest Treatment Improves Colour and Quality of Seedless Table Grape ‘Magenta’ Upregulating on-Vine Abscisic Acid Metabolism, Relative VvNCED1 Gene Expression, and the Antioxidant System in Berries. Front. Plant Sci. 2021, 12, 740240. [Google Scholar] [CrossRef] [PubMed]
- Ke, D.; Guo, J.; Li, K.; Wang, Y.; Han, X.; Fu, W.; Miao, Y.; Jia, K.P. Carotenoid-derived bioactive metabolites shape plant root architecture to adapt to the rhizospheric environments. Front. Plant Sci. 2022, 13, 986414. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Qin, X.; Zeevaart, J.A. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 2003, 131, 1591–1601. [Google Scholar] [CrossRef]
- Voisin, A.S.; Reidy, B.; Parent, B.; Rolland, G.; Redondo, E.; Gerentes, D.; Tardieu, F.; Muller, B. Are ABA, ethylene or their interaction involved in the response of leaf growth to soil water deficit? An analysis using naturally occurring variation or genetic transformation of ABA production in maize. Plant Cell Env. 2006, 29, 1829–1840. [Google Scholar] [CrossRef]
- Ren, H.; Gao, Z.; Chen, L.; Wei, K.; Liu, J.; Fan, Y.; Davies, W.J.; Jia, W.; Zhang, J. Dynamic analysis of ABA accumulation in relation to the rate of ABA catabolism in maize tissues under water deficit. J. Exp. Bot. 2007, 58, 211–219. [Google Scholar] [CrossRef]
- Tan, B.C.; Schwartz, S.H.; Zeevaart, J.A.; McCarty, D.R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 1997, 94, 12235–12240. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Jackson, A.C.; Parker, R.A.; Morpeth, D.R.; Burbidge, A.; Taylor, I.B. Abscisic acid biosynthesis in tomato: Regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant. Mol. Biol. 2000, 42, 833–845. [Google Scholar] [CrossRef]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.; Li, Q.B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef]
- Li, Q.; Yu, X.; Chen, L.; Zhao, G.; Li, S.; Zhou, H.; Dai, Y.; Sun, N.; Xie, Y.; Gao, J.; et al. Genome-wide identification and expression analysis of the NCED family in cotton (Gossypium hirsutum L.). PLoS ONE 2021, 16, e0246021. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, F.; Shi, X.; Wang, X.; Ji, X.; Wang, Z.; Wang, B.; Zheng, X.; Wang, H. Evolution and expression of NCED family genes in Vitis vinifera. Chin. Bull. Bot. 2019, 54, 474–485. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.A.; McCarty, D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 1997, 276, 1872–1874. [Google Scholar] [CrossRef]
- Xu, P.; Cai, W. Functional characterization of the BnNCED3 gene in Brassica napus. Plant Sci. 2017, 256, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.G.; Chen, H.C.; Huang, W.Y.; Chu, Y.C.; Shii, C.T.; Cheng, W.H. Ectopic expression of rice OsNCED3 in Arabidopsis increases ABA level and alters leaf morphology. Plant Sci. 2010, 178, 12–22. [Google Scholar] [CrossRef]
- Son, S.; Chitnis, V.R.; Liu, A.; Gao, F.; Nguyen, T.N.; Ayele, B.T. Abscisic acid metabolic genes of wheat (Triticum aestivum L.): Identification and insights into their functionality in seed dormancy and dehydration tolerance. Planta 2016, 244, 429–447. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef]
- Chernys, J.T.; Zeevaart, J.A. Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol. 2000, 124, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Jiang, Y.; Guo, Y.; Huang, J.; Zhou, M.; Tang, Y.; Sui, J.; Wang, J.; Qiao, L. A novel salt inducible WRKY transcription factor gene, AhWRKY75, confers salt tolerance in transgenic peanut. Plant Physiol. Biochem. 2021, 160, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Frey, A.; Effroy, D.; Lefebvre, V.; Seo, M.; Perreau, F.; Berger, A.; Sechet, J.; To, A.; North, H.M.; Marion-Poll, A. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 2012, 70, 501–512. [Google Scholar] [CrossRef]
- Lang, J.; Fu, Y.; Zhou, Y.; Cheng, M.; Deng, M.; Li, M.; Zhu, T.; Yang, J.; Guo, X.; Gui, L.; et al. Myb10-D confers PHS-3D resistance to pre-harvest sprouting by regulating NCED in ABA biosynthesis pathway of wheat. New Phytol. 2021, 230, 1940–1952. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Sun, M.; He, C.; Yu, K.; Zhao, B.; Li, R.; Li, J.; Yang, Z.; Wang, X.; et al. Multi-Omics Analyses Reveal Systemic Insights into Maize Vivipary. Plants 2021, 10, 2437. [Google Scholar] [CrossRef]
- Xiang, Z.; Zhang, L.; Zhang, M.; Yao, Y.; Qian, Q.; Wei, Z.; Cui, B.; Wang, D.; Quan, C.; Lu, M.; et al. OsNCED5 confers cold stress tolerance through regulating ROS homeostasis in rice. Plant Physiol. Biochem. 2025, 220, 109455. [Google Scholar] [CrossRef]
- Bose, J.; Rodrigo-Moreno, A.; Shabala, S. ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 2014, 65, 1241–1257. [Google Scholar] [CrossRef] [PubMed]
- Avico, E.H.; Acevedo, R.M.; Duarte, M.J.; Rodrigues Salvador, A.; Nunes-Nesi, A.; Ruiz, O.A.; Sansberro, P.A. Integrating Transcriptional, Metabolic, and Physiological Responses to Drought Stress in Ilex paraguariensis Roots. Plants 2023, 12, 2404. [Google Scholar] [CrossRef]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef]
- Nadarajah, K.K. ROS Homeostasis in Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Env. 2012, 35, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Shakar, M.; Yaseen, M.; Mahmood, R.; Ahmad, I. Calcium carbide induced ethylene modulate biochemical profile of Cucumis sativus at seed germination stage to alleviate salt stress. Sci. Hortic. 2016, 213, 179–185. [Google Scholar] [CrossRef]
- Abbasi, A.R.; Hajirezaei, M.; Hofius, D.; Sonnewald, U.; Voll, L.M. Specific Roles of α- and γ-Tocopherol in Abiotic Stress Responses of Transgenic Tobacco. Plant Physiol. 2007, 143, 1720–1738. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, W.C.; Han, C.; Wang, S.; Bai, M.Y.; Song, C.P. Reactive oxygen species: Multidimensional regulators of plant adaptation to abiotic stress and development. J. Integr. Plant Biol. 2024, 66, 330–367. [Google Scholar] [CrossRef]
- Fedoreyeva, L.I. ROS as Signaling Molecules to Initiate the Process of Plant Acclimatization to Abiotic Stress. Int. J. Mol. Sci. 2024, 25, 11820. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, Y.; Lyu, S.; Liu, Y.; Jian, S.; Deng, S. MpNAC1, a transcription factor from the mangrove associate Millettia pinnata, confers salt and drought stress tolerance in transgenic Arabidopsis and rice. Plant Physiol. Biochem. 2024, 211, 108721. [Google Scholar] [CrossRef] [PubMed]
- Kramer, D.M.; Johnson, G.; Kiirats, O.; Edwards, G.E. New Fluorescence Parameters for the Determination of QA Redox State and Excitation Energy Fluxes. Photosynth. Res. 2004, 79, 209. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).