Elucidating the Molecular Mechanisms of Physiological Fruit Abscission in Actinidia arguta Through Comparative Transcriptomics and Transient Genetic Transformation
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Scanning Electron Microscopy (SEM) Analysis
2.3. Quantification of Plant Hormones and Activity Assays of Cell Wall-Modifying Enzymes
2.4. RNA-Seq and Data Analysis and Weighted Gene Co-Expression Network Analysis (WGCNA)
2.5. Quantitative PCR (qPCR)
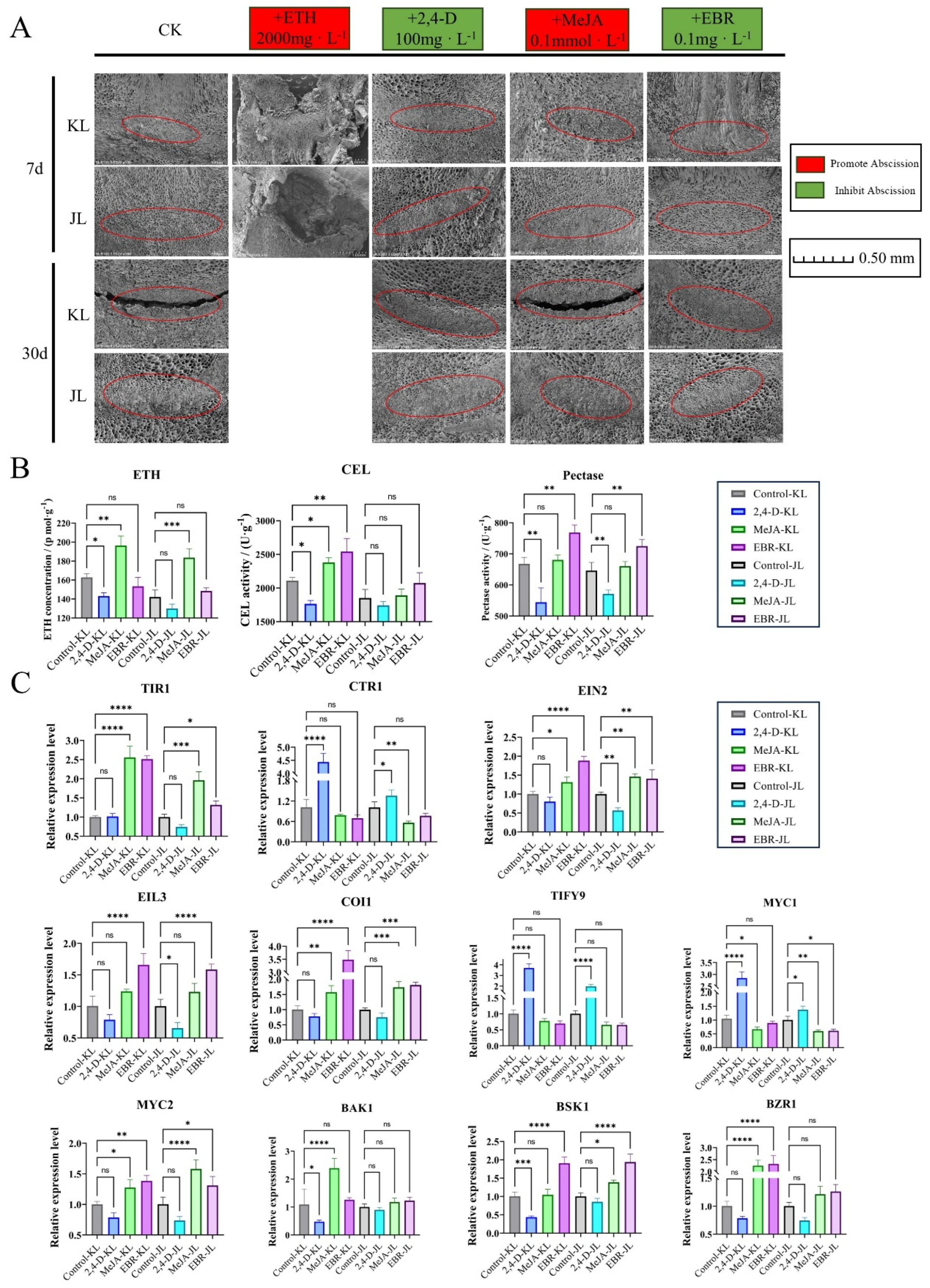
2.6. Treatment with Different Plant Growth Regulators
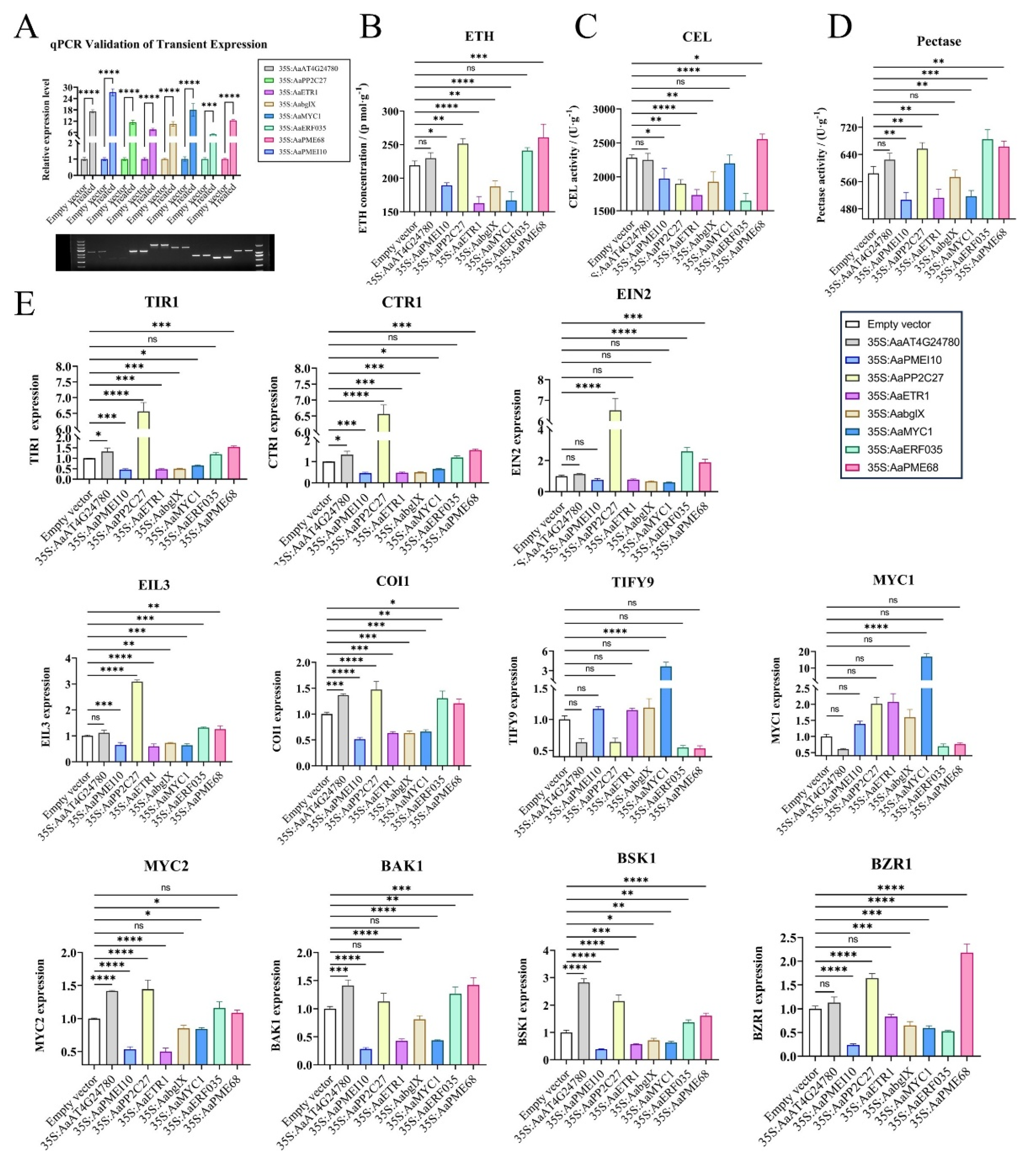
2.7. Overexpression of Candidate Genes
3. Results
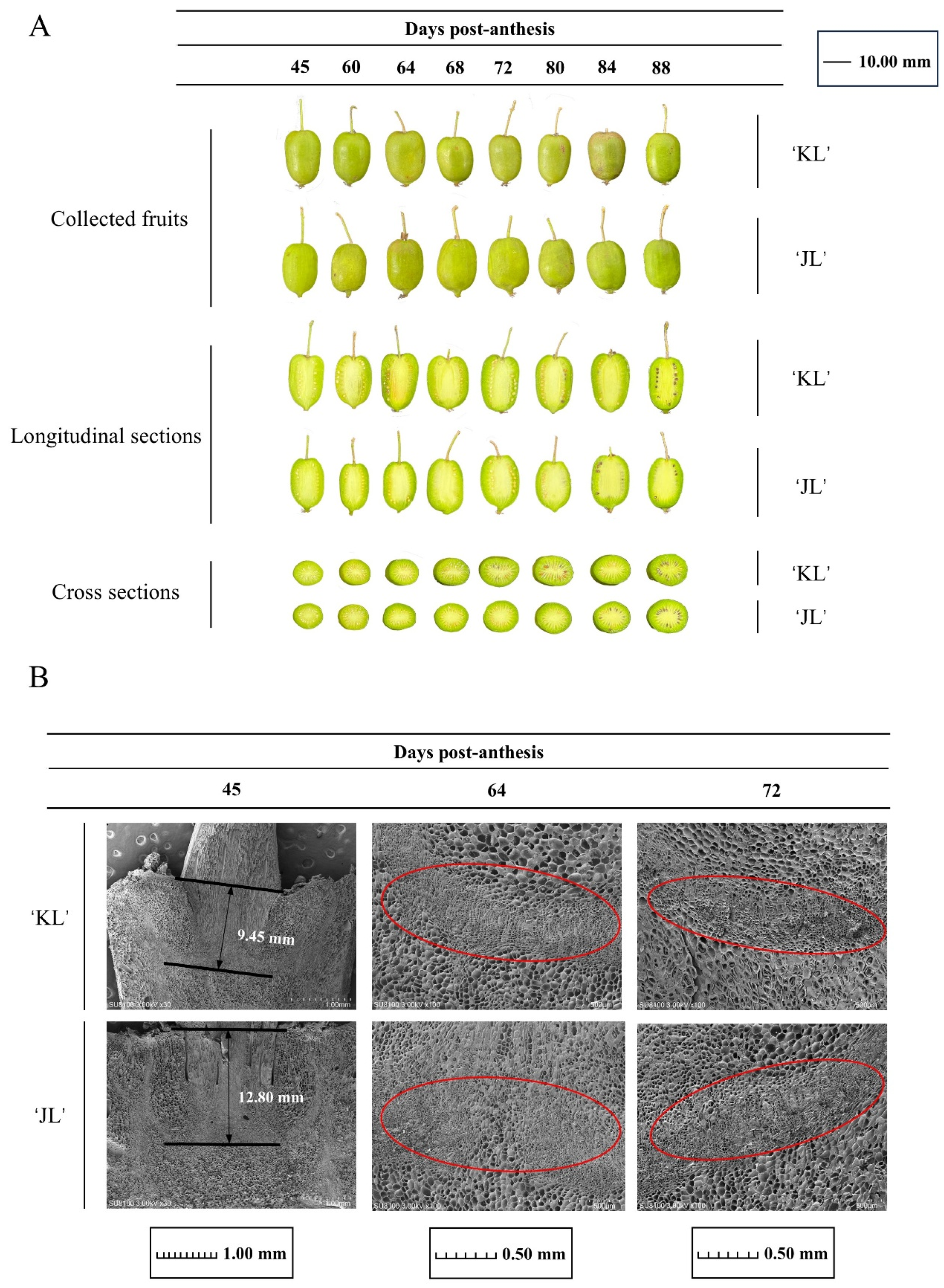
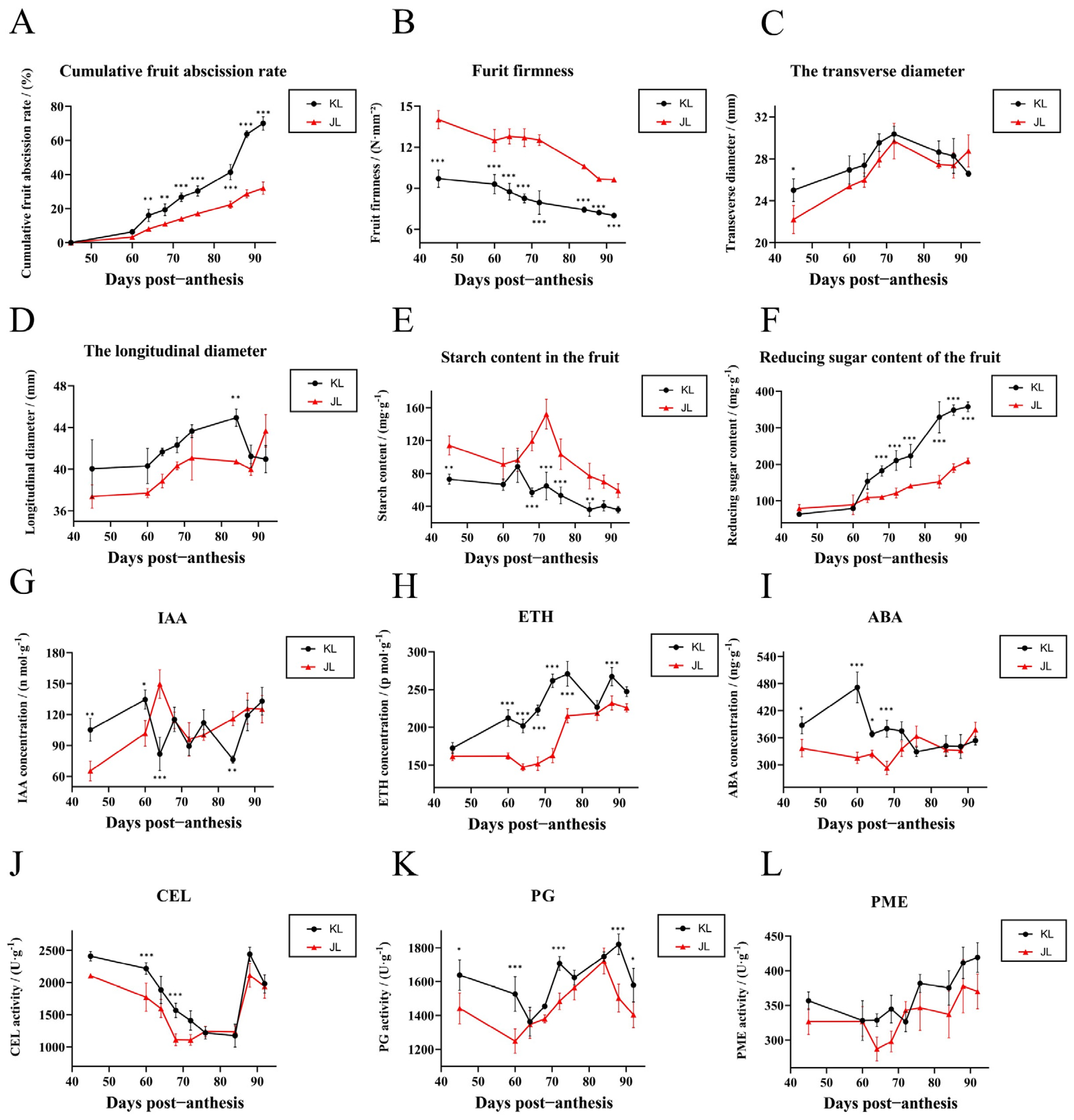
3.1. Physiological Responses of the FAZ During Development in Two A. arguta Varieties
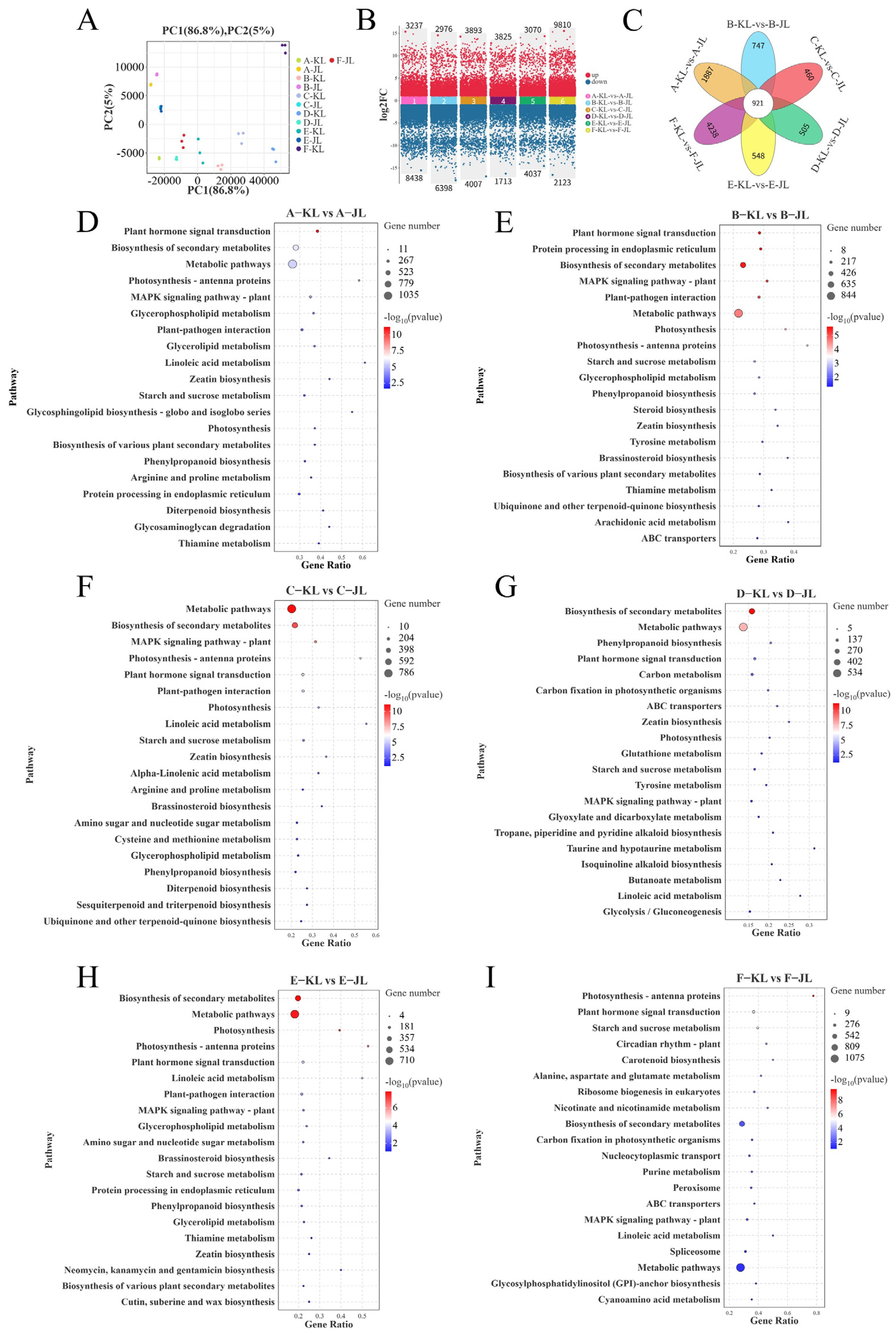
3.2. Comparative Transcriptomic Analysis
3.2.1. Transcriptomic Analysis and Enrichment Analyses of Differentially Expressed Genes (DEGs)
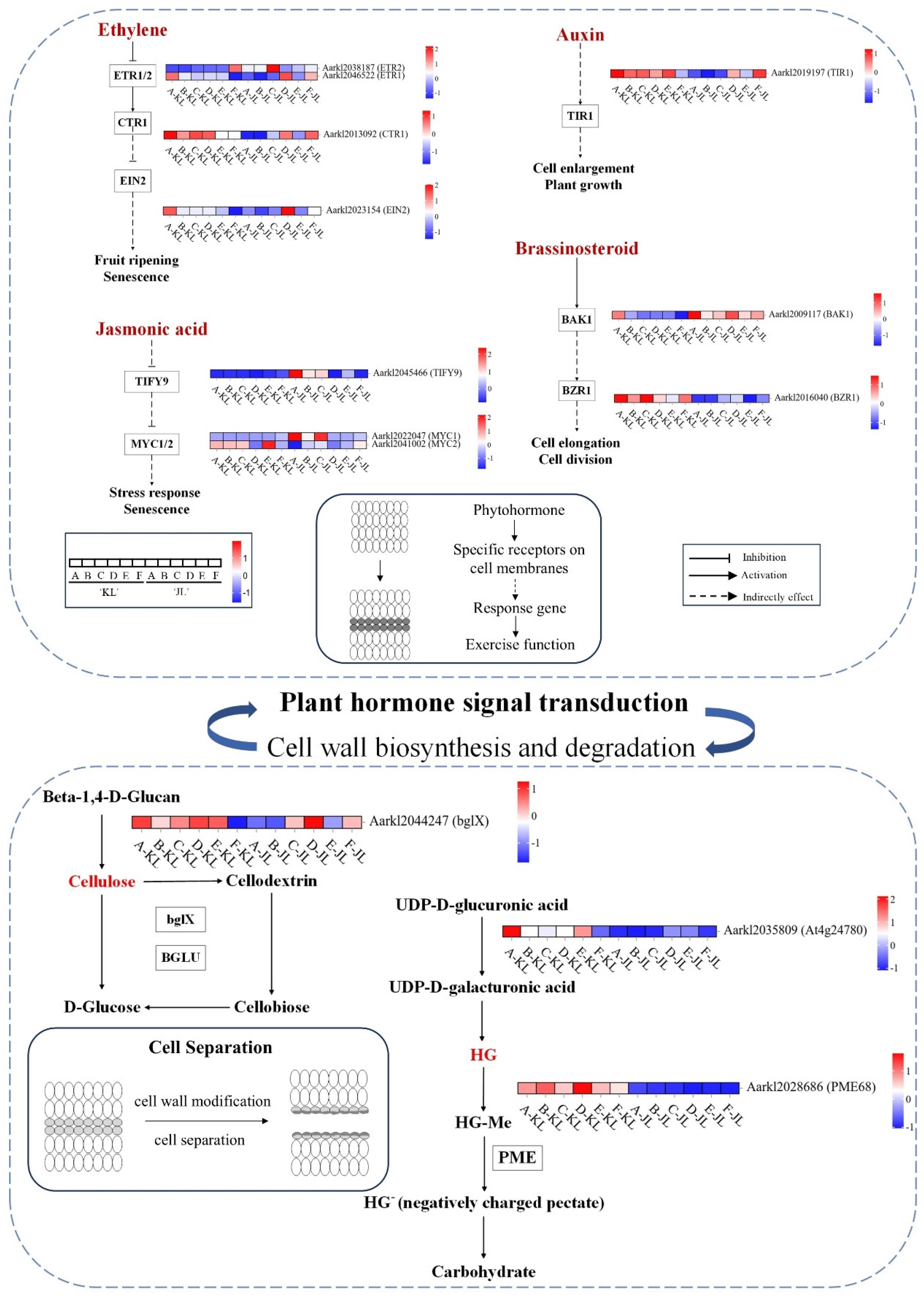
3.2.2. Heatmaps of Gene Expression in Plant Hormone Signal Transduction and Cell Wall Synthesis/Degradation Pathways
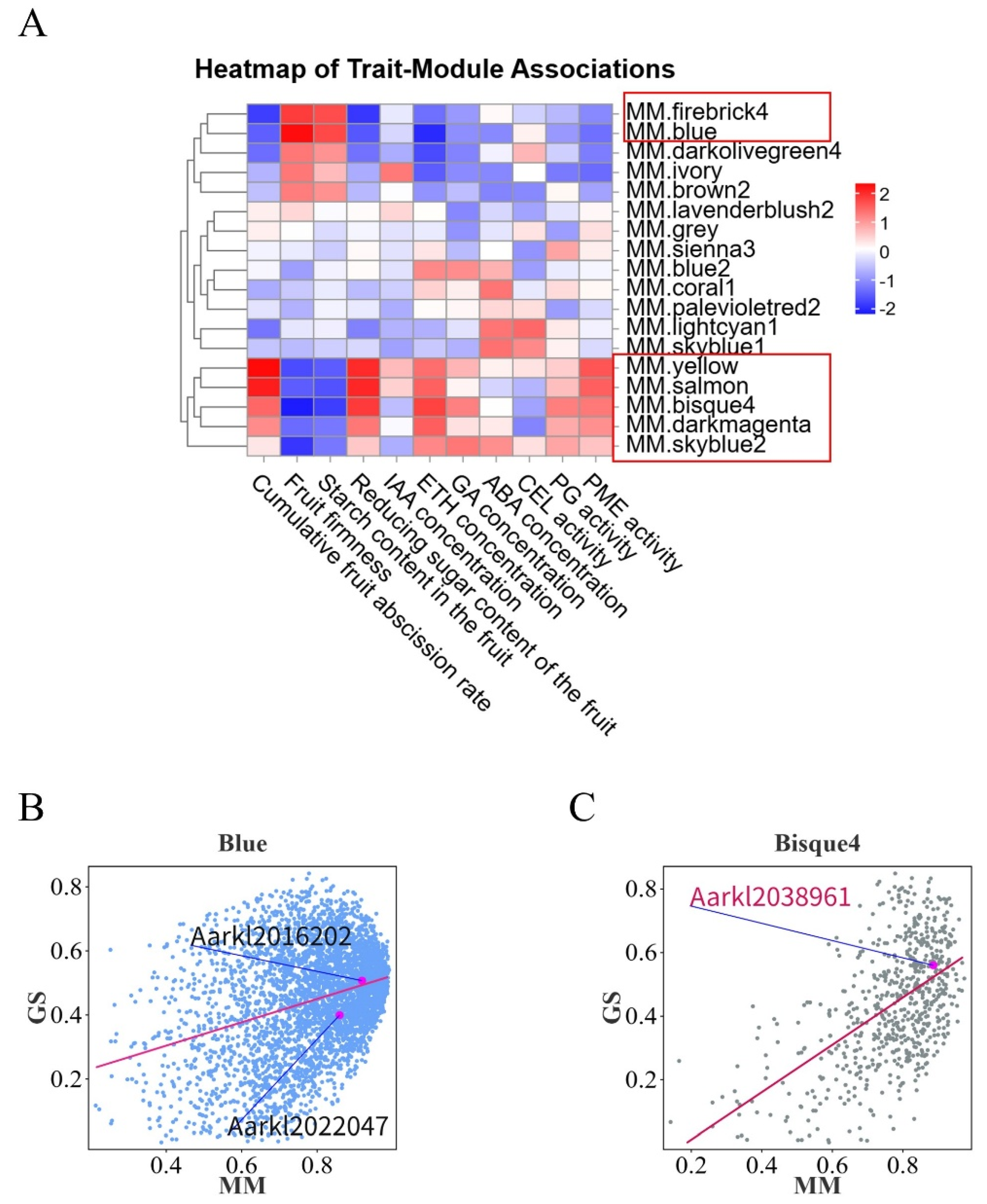
3.2.3. WGCNA Identifies Core Regulatory Genes Governing Abscission
3.3. Validation of Exogenous Plant Growth Regulator Treatments
4. Discussion
4.1. Changes in the AZ During A. arguta Development
4.2. Transcriptomic Profiling and Functional Enrichment of Abscission-Associated Differentially Expressed Genes
4.2.1. Plant Hormone Signal Transduction Plays a Pivotal Role in Regulating A. arguta Abscission Process
4.2.2. Effects of Carbohydrates on the Abscission of A. arguta Fruits
4.2.3. Cell Wall-Modifying Enzymes Are the Executors of Fruit Abscission in A. arguta
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shin, Y.; Chane, A.; Jung, M.; Lee, Y. Recent Advances in Understanding the Roles of Pectin as an Active Participant in Plant Signaling Networks. Plants 2021, 10, 1712. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y. More than cell wall hydrolysis: Orchestration of cellular dynamics for organ separation. Curr. Opin. Plant Biol. 2019, 51, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Olsson, V.; Butenko, M.A. Abscission in plants. Curr. Biol. 2018, 28, R338–R339. [Google Scholar] [CrossRef]
- Daher, F.B.; Braybrook, S.A. How to let go: Pectin and plant cell adhesion. Front. Plant Sci. 2015, 6, 523. [Google Scholar] [CrossRef]
- Chen, J.; Ren, B.; Bian, C.; Qin, D.; Zhang, L.; Li, J.; Wei, J.; Wang, A.; Huo, J.; Gang, H. Transcriptomic and metabolomic analyses reveal molecular mechanisms associated with the natural abscission of blue honeysuckle (Lonicera caerulea L.) ripe fruits. Plant Physiol. Biochem. 2023, 199, 107740. [Google Scholar] [CrossRef]
- Sawicki, M.; Barka, E.A.; Clement, C.; Vaillant-Gaveau, N.; Jacquard, C. Cross-talk between environmental stresses and plant metabolism during reproductive organ abscission. J. Exp. Bot. 2015, 66, 1707–1719. [Google Scholar] [CrossRef]
- Wang, R.; Li, R.; Cheng, L.; Wang, X.; Fu, X.; Dong, X.; Qi, M.; Jiang, C.; Xu, T.; Li, T. SlERF52 regulates SlTIP1;1 expression to accelerate tomato pedicel abscission. Plant Physiol. 2021, 185, 1829–1846. [Google Scholar] [CrossRef]
- Sun, N.; Itamura, H. Ethylene production trend in young ‘Fuyu’ persimmon fruit during attachment to or detachment from the tree. Acta Hortic. 2022, 1338, 135–142. [Google Scholar] [CrossRef]
- Dong, Q.; Gong, G.; Peng, Z.; Li, Y.; Hou, Y.; Hong, Q. Analysis on the relationship between pre-harvest fruit drops and content of endogenous hormone in different parts of fruit in citrus. Plant Physiol. J. 2018, 54, 1569–1575. [Google Scholar]
- Gómez-Cadenas, A.; Mehouachi, J.; Tadeo, F.R.; Primo-Millo, E.; Talon, M. Hormonal regulation of fruitlet abscission induced by carbohydrate shortage in citrus. Planta 2000, 210, 636–643. [Google Scholar] [CrossRef]
- Ma, X.; Li, C.; Huang, X.; Wang, H.; Wu, H.; Zhao, M.; Li, J. Involvement of HD-ZIP I transcription factors LcHB2 and LcHB3 in fruitlet abscission by promoting transcription of genes related to the biosynthesis of ethylene and ABA in litchi. Tree Physiol. 2019, 39, 1600–1613. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.-L.; Wen, Z.; Yang, K.; Tian, T.; Qiao, G.; Hong, Y.; Wen, X.-P. Comparative Proteomics Profiling Illuminates the Fruitlet Abscission Mechanism of Sweet Cherry as Induced by Embryo Abortion. Int. J. Mol. Sci. 2020, 21, 1200. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Do, V.G.; Kim, S.; Kweon, H.; McGhie, T.K. Cold stress triggers premature fruit abscission through ABA-dependent signal transduction in early developing apple. PLoS ONE 2021, 16, e0249975. [Google Scholar] [CrossRef]
- Kucko, A.; Florkiewicz, A.B.; Wolska, M.; Mietki, J.; Kapusta, M.; Domagalski, K.; Wilmowicz, E. Jasmonate-Dependent Response of the Flower Abscission Zone Cells to Drought in Yellow Lupine. Plants 2022, 11, 527. [Google Scholar] [CrossRef]
- Ma, X.; Yuan, Y.; Li, C.; Wu, Q.; He, Z.; Li, J.; Zhao, M. Brassinosteroids suppress ethylene-induced fruitlet abscission through LcBZR1/2-mediated transcriptional repression of LcACS1/4 and LcACO2/3 in litchi. Hortic. Res. 2021, 8, 105. [Google Scholar] [CrossRef]
- Wen, J.; Wang, Y.; Cao, W.; He, Y.; Sun, Y.; Yuan, P.; Sun, B.; Yan, Y.; Qin, H.; Fan, S.; et al. Comprehensive Evaluation of Ten Actinidia arguta Wines Based on Color, Organic Acids, Volatile Compounds, and Quantitative Descriptive Analysis. Foods 2023, 12, 3345. [Google Scholar] [CrossRef]
- Latocha, P.; Lata, B.; Stasiak, A. Phenolics, ascorbate and the antioxidant potential of kiwiberry vs common kiwifruit: The effect of cultivar and tissue type. J. Funct. Foods 2015, 19, 155–163. [Google Scholar] [CrossRef]
- Zhao, S.; Fugui, Y.; Yueshen, M.; Jingcai, Z.; Jinru, Y. Kui Lu—A new cultivar of Actinidia arguta Planch. Acta Hortic. Sin. 1994, 21, 207–208. [Google Scholar]
- Cao, W.; Shu, N.; Wen, J.; Yang, Y.; Wang, Y.; Lu, W. Widely Targeted Metabolomics Was Used to Reveal the Differences between Non-Volatile Compounds in Different Wines and Their Associations with Sensory Properties. Foods 2023, 12, 290. [Google Scholar] [CrossRef]
- Qin, H.; Yang, M.; Ai, J.; Fan, S.; Wang, Z.; Xu, P.; Liu, Y.; Zhao, Y.; Zhang, Q.; Zhang, B.; et al. A new cultivar of Actinidia arguta Planch. ‘Jialu’. J. Fruit Sci. 2015, 32, 733–735. [Google Scholar]
- Tranbarger, T.J.; Domonhedo, H.; Cazemajor, M.; Dubreuil, C.; Fischer, U.; Morcillo, F. The PIP Peptide of INFLORESCENCE DEFICIENT IN ABSCISSION Enhances Populus Leaf and Elaeis guineensis Fruit Abscission. Plants 2019, 8, 143. [Google Scholar] [CrossRef] [PubMed]
- Tranbarger, T.J.; Tadeo, F.R. Diversity and Functional Dynamics of Fleshy Fruit Abscission Zones. Annu. Plant Rev. Online 2020, 3, 151–213. [Google Scholar] [CrossRef]
- Xie, R.; Ge, T.; Zhang, J.; Pan, X.; Ma, Y.; Yi, S.; Zheng, Y. The molecular events of IAA inhibiting citrus fruitlet abscission revealed by digital gene expression profiling. Plant Physiol. Biochem. 2018, 130, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Gong, Z.; Hu, G.; Ma, X.; Bai, R.; Yu, R.; Zhang, Q.; Deng, W.; Li, Z.; Wuriyanghan, H. Tomato SlBL plays an important role in fruit pedicel organogenesis and abscission. Hortic. Res. 2021, 8, 78. [Google Scholar] [CrossRef]
- Wen, Y.; Shao, B.; Hao, Z.; Wang, C.; Sun, T.; Han, Y.; Tian, J.; Zhang, F. Preliminary Study on Programmed Cell Death during Calyx Abscission of Korla Fragrant Pear. Horticulturae 2024, 10, 637. [Google Scholar] [CrossRef]
- Parra, R.; Gomez-Jimenez, M.C. Spatio-temporal immunolocalization of extensin protein and hemicellulose polysaccharides during olive fruit abscission. Planta 2020, 252, 32. [Google Scholar] [CrossRef]
- Shi, Y.; Song, B.; Liang, Q.; Su, D.; Lu, W.; Liu, Y.; Li, Z. Molecular regulatory events of flower and fruit abscission in horticultural plants. Hortic. Plant J. 2023, 9, 867–883. [Google Scholar] [CrossRef]
- Sriskantharajah, K.; El Kayal, W.; Torkamaneh, D.; Ayyanath, M.M.; Saxena, P.K.; Sullivan, A.J.; Paliyath, G.; Subramanian, J. Transcriptomics of Improved Fruit Retention by Hexanal in ‘Honeycrisp’ Reveals Hormonal Crosstalk and Reduced Cell Wall Degradation in the Fruit Abscission Zone. Int. J. Mol. Sci. 2021, 22, 8830. [Google Scholar] [CrossRef]
- Domingos, S.; Fino, J.; Cardoso, V.; Sanchez, C.; Ramalho, J.C.; Larcher, R.; Paulo, O.S.; Oliveira, C.M.; Goulao, L.F. Shared and divergent pathways for flower abscission are triggered by gibberellic acid and carbon starvation in seedless Vitis vinifera L. BMC Plant Biol. 2016, 16, 38. [Google Scholar] [CrossRef]
- Florkiewicz, A.B.; Kucko, A.; Kapusta, M.; Burchardt, S.; Przywieczerski, T.; Czeszewska-Rosiak, G.; Wilmowicz, E. Drought Disrupts Auxin Localization in Abscission Zone and Modifies Cell Wall Structure Leading to Flower Separation in Yellow Lupine. Int. J. Mol. Sci. 2020, 21, 6848. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, L.; Li, R.; Cai, Y.; Wang, X.; Fu, X.; Dong, X.; Qi, M.; Jiang, C.-Z.; Xu, T.; et al. The HD-Zip transcription factor SlHB15A regulates abscission by modulating jasmonoyl-isoleucine biosynthesis. Plant Physiol. 2022, 189, 2396–2412. [Google Scholar] [CrossRef] [PubMed]
- Fidelibus, M.W.; Petracek, P.; McArtney, S. Jasmonic Acid Activates the Fruit-Pedicel Abscission Zone of ‘Thompson Seedless’ Grapes, Especially with Co-Application of 1-Aminocyclopropane-1-carboxylic Acid. Plants 2022, 11, 1245. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chai, L.; Tong, N.; Yu, H.; Jiang, W. Potential Carbohydrate Regulation Mechanism Underlying Starvation-Induced Abscission of Tomato Flower. Int. J. Mol. Sci. 2022, 23, 1952. [Google Scholar] [CrossRef]
- Mesejo, C.; Martinez-Fuentes, A.; Reig, C.; Agusti, M. Ringing branches reduces fruitlet abscission by promoting PIN1 expression in ‘Orri’ mandarin. Sci. Hortic. 2022, 306, 111451. [Google Scholar] [CrossRef]
- Yi, J.-W.; Ge, H.-T.; Abbas, F.; Zhao, J.-T.; Huang, X.-M.; Hu, G.-B.; Wang, H.-C. Function of a non-enzymatic hexokinase LcHXK1 as glucose sensor in regulating litchi fruit abscission. Tree Physiol. 2023, 43, 130–141. [Google Scholar] [CrossRef]
- Zhao, N.; Geng, Z.; Zhao, G.; Liu, J.; An, Z.; Zhang, H.; Ai, P.; Wang, Y. Integrated analysis of the transcriptome and metabolome reveals the molecular mechanism regulating cotton boll abscission under low light intensity. BMC Plant Biol. 2024, 24, 182. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Y.; Liao, F.; Li, T.; Li, X.; Wu, B.; Hong, S.-B.; Xu, K.; Zang, Y.; Zheng, W. Genome-wide identification of GH9 gene family and the assessment of its role during fruit abscission zone formation in Vaccinium ashei. Plant Cell Rep. 2023, 42, 1589–1609. [Google Scholar] [CrossRef]
- Tsuchiya, M.; Satoh, S.; Iwai, H. Distribution of XTH, expansin, and secondary-wall-related CesA in floral and fruit abscission zones during fruit development in tomato (Solanum lycopersicum). Front. Plant Sci. 2015, 6, 323. [Google Scholar] [CrossRef]
- Phetsirikoon, S.; Paull, R.E.; Chen, N.; Ketsa, S.; van Doorn, W.G. Increased hydrolase gene expression and hydrolase activity in the abscission zone involved in chilling-induced abscission of Dendrobium flowers. Postharvest Biol. Technol. 2016, 117, 217–229. [Google Scholar] [CrossRef]
- Paul, P.; Dhatt, B.K.; Miller, M.; Folsom, J.J.; Wang, Z.; Krassovskaya, I.; Liu, K.; Sandhu, J.; Yu, H.; Zhang, C.; et al. MADS78 and MADS79 Are Essential Regulators of Early Seed Development in Rice. Plant Physiol. 2020, 182, 933–948. [Google Scholar] [CrossRef]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, P.; Wang, Y.; Sun, Y.; Liu, G.; Qin, H.; Fan, S.; Yan, Y.; Sun, B.; Lu, W. Elucidating the Molecular Mechanisms of Physiological Fruit Abscission in Actinidia arguta Through Comparative Transcriptomics and Transient Genetic Transformation. Plants 2025, 14, 1645. https://doi.org/10.3390/plants14111645
Yuan P, Wang Y, Sun Y, Liu G, Qin H, Fan S, Yan Y, Sun B, Lu W. Elucidating the Molecular Mechanisms of Physiological Fruit Abscission in Actinidia arguta Through Comparative Transcriptomics and Transient Genetic Transformation. Plants. 2025; 14(11):1645. https://doi.org/10.3390/plants14111645
Chicago/Turabian StyleYuan, Pengqiang, Yanli Wang, Yining Sun, Guoliang Liu, Hongyan Qin, Shutian Fan, Yiping Yan, Bowei Sun, and Wenpeng Lu. 2025. "Elucidating the Molecular Mechanisms of Physiological Fruit Abscission in Actinidia arguta Through Comparative Transcriptomics and Transient Genetic Transformation" Plants 14, no. 11: 1645. https://doi.org/10.3390/plants14111645
APA StyleYuan, P., Wang, Y., Sun, Y., Liu, G., Qin, H., Fan, S., Yan, Y., Sun, B., & Lu, W. (2025). Elucidating the Molecular Mechanisms of Physiological Fruit Abscission in Actinidia arguta Through Comparative Transcriptomics and Transient Genetic Transformation. Plants, 14(11), 1645. https://doi.org/10.3390/plants14111645





