Genome-Wide Identification and Expression Analysis of the LbDof Transcription Factor Family Genes in Lycium barbarum
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Properties of LbDof
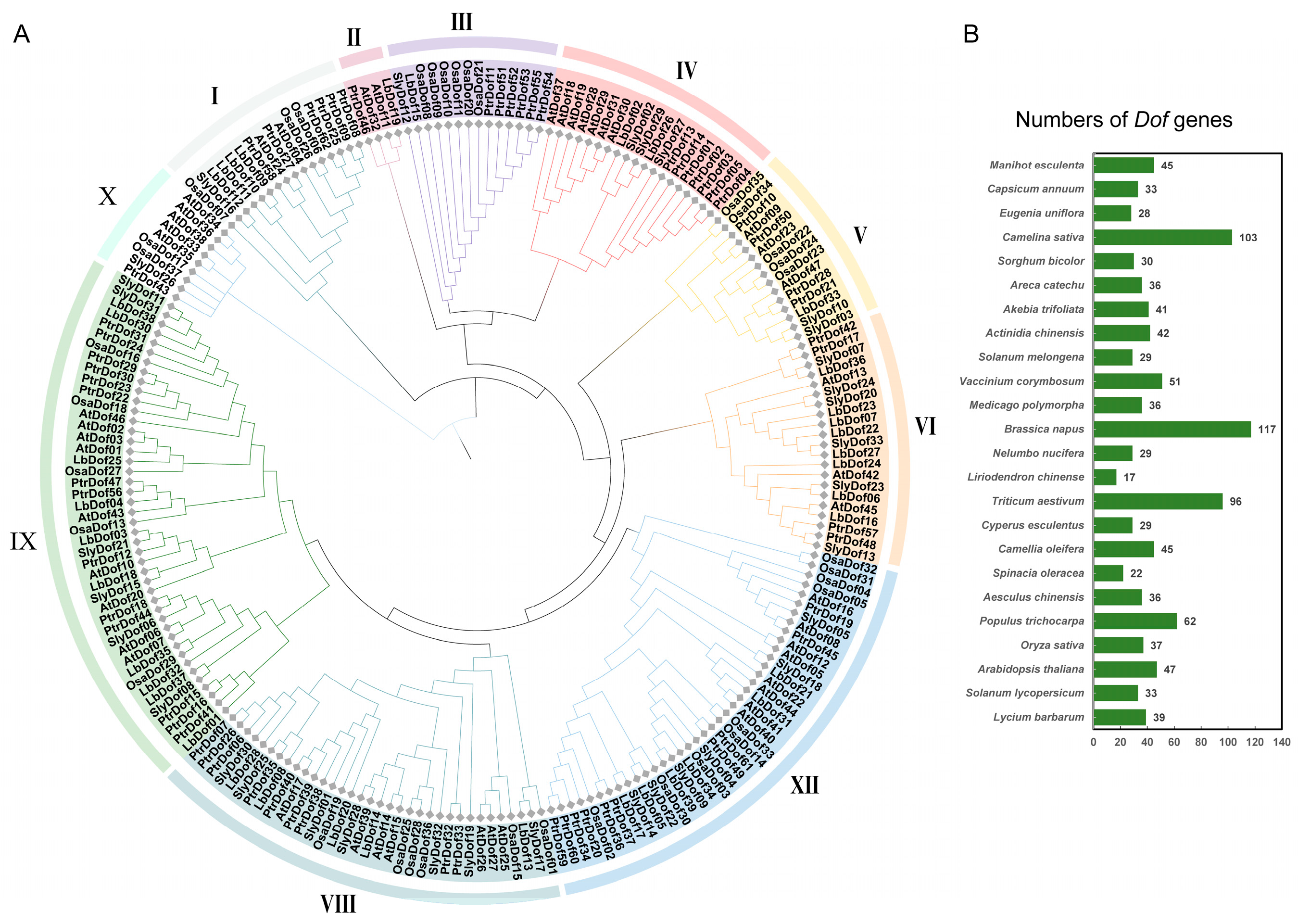
2.2. Phylogenetic Analysis of LbDofs
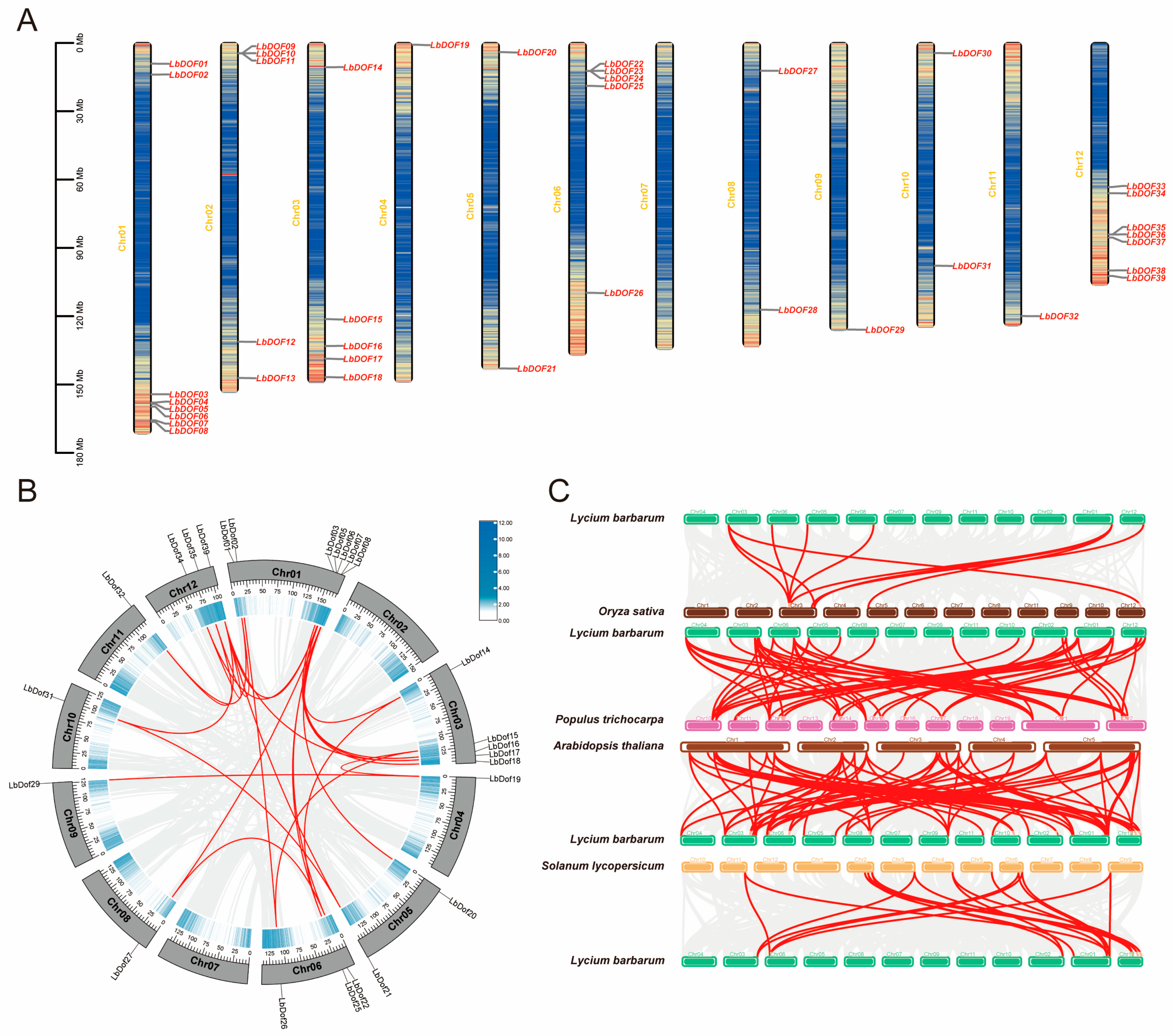
2.3. Chromosomal Location and Collinearity Analysis
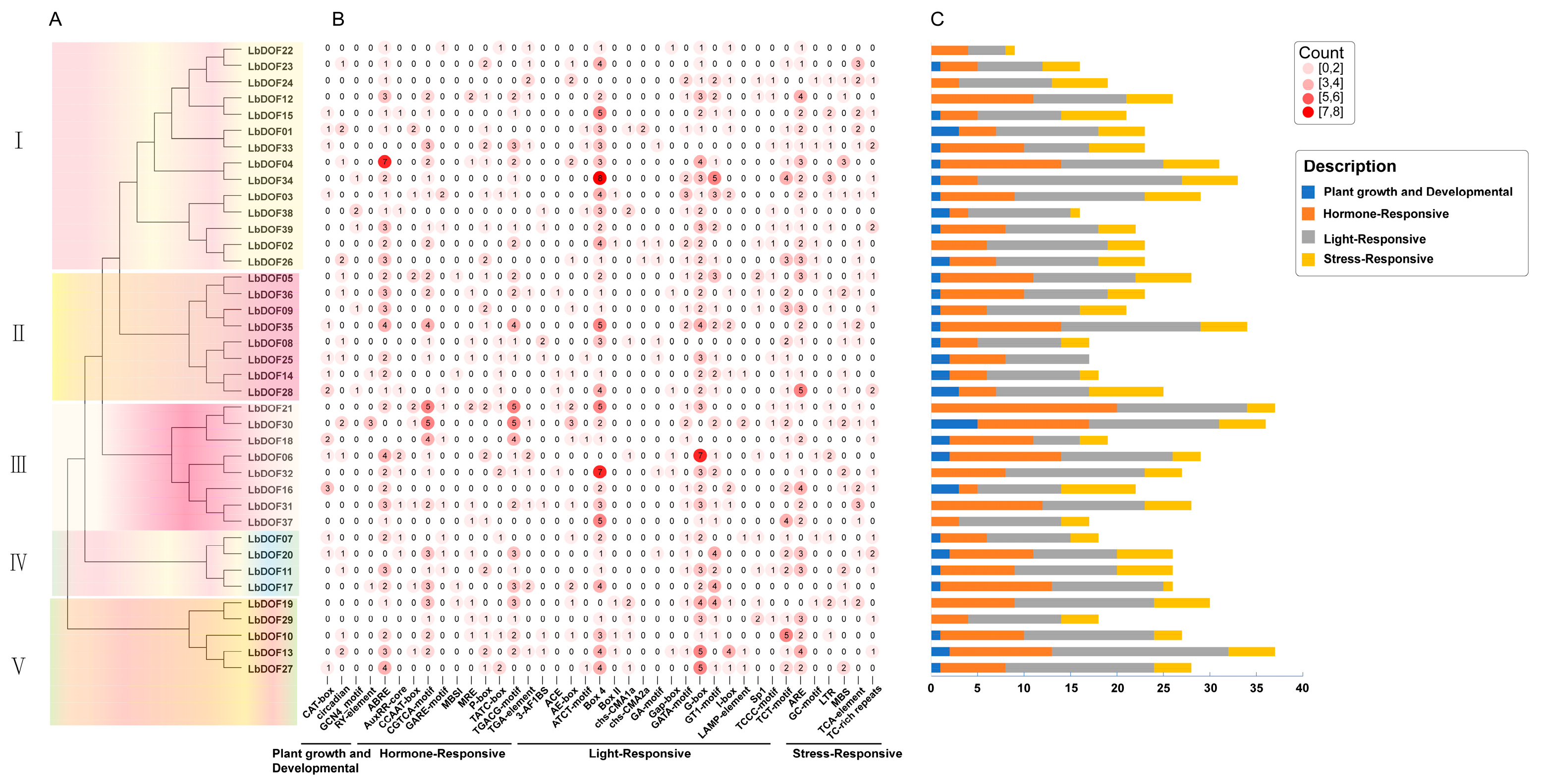
2.4. Analyses of Gene Structures, Motifs, and Domains of LbDofs
2.5. Cis-Regulatory Element Analysis of LbDof Genes
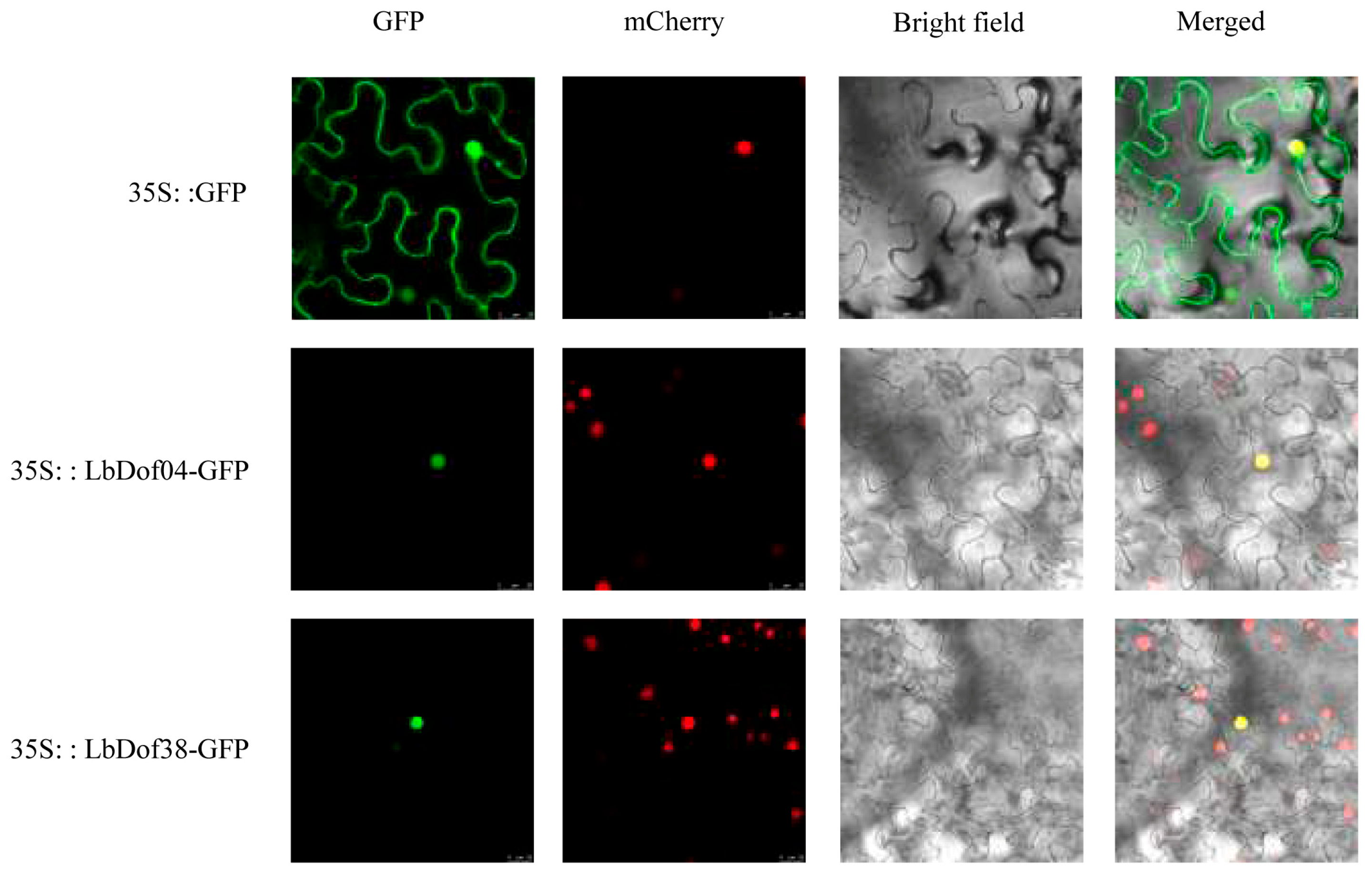
2.6. Subcellular Localization Analysis of LbDof
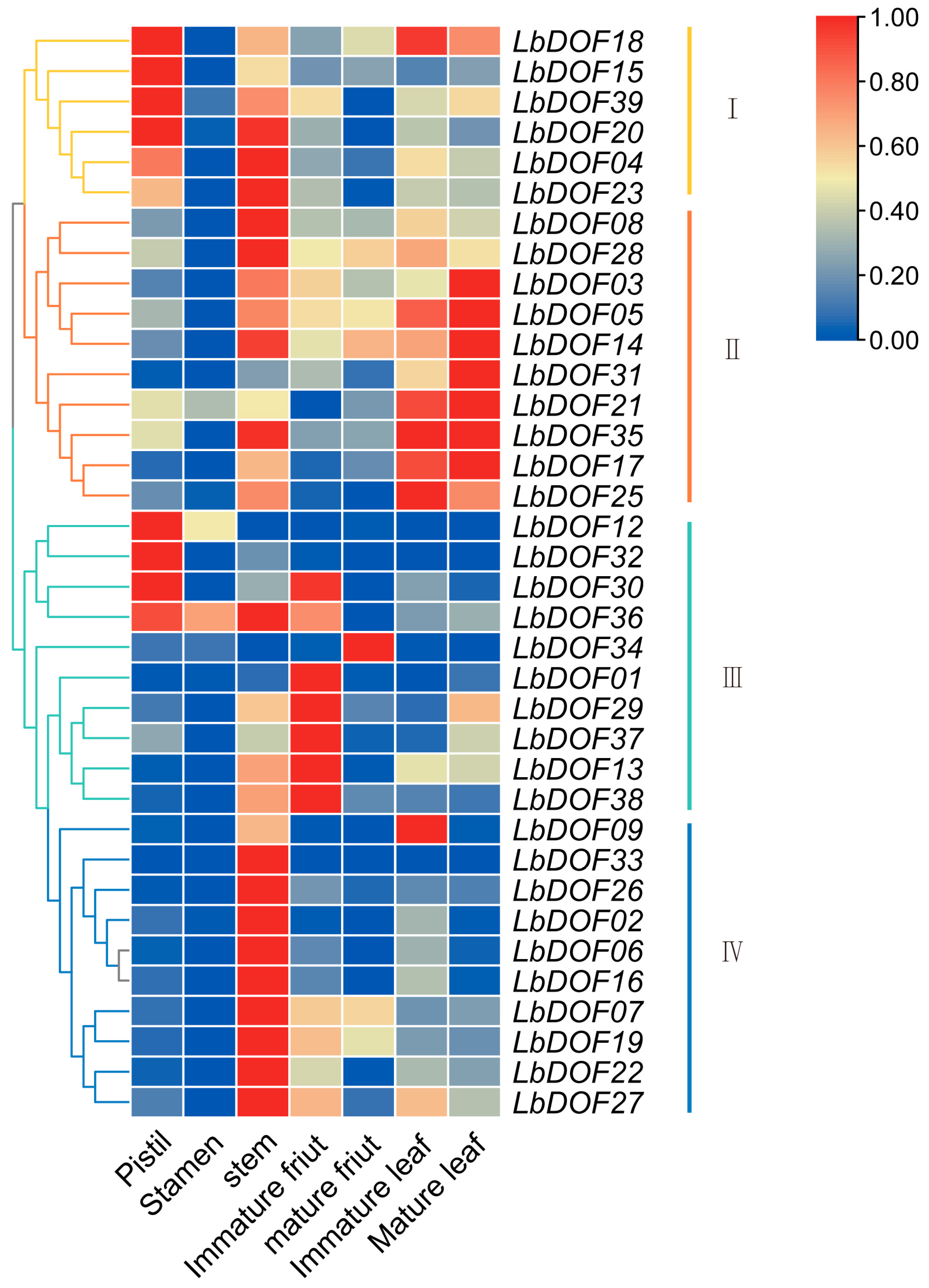
2.7. Expression Pattern of LbDof in L. barbarum Under Different Organs
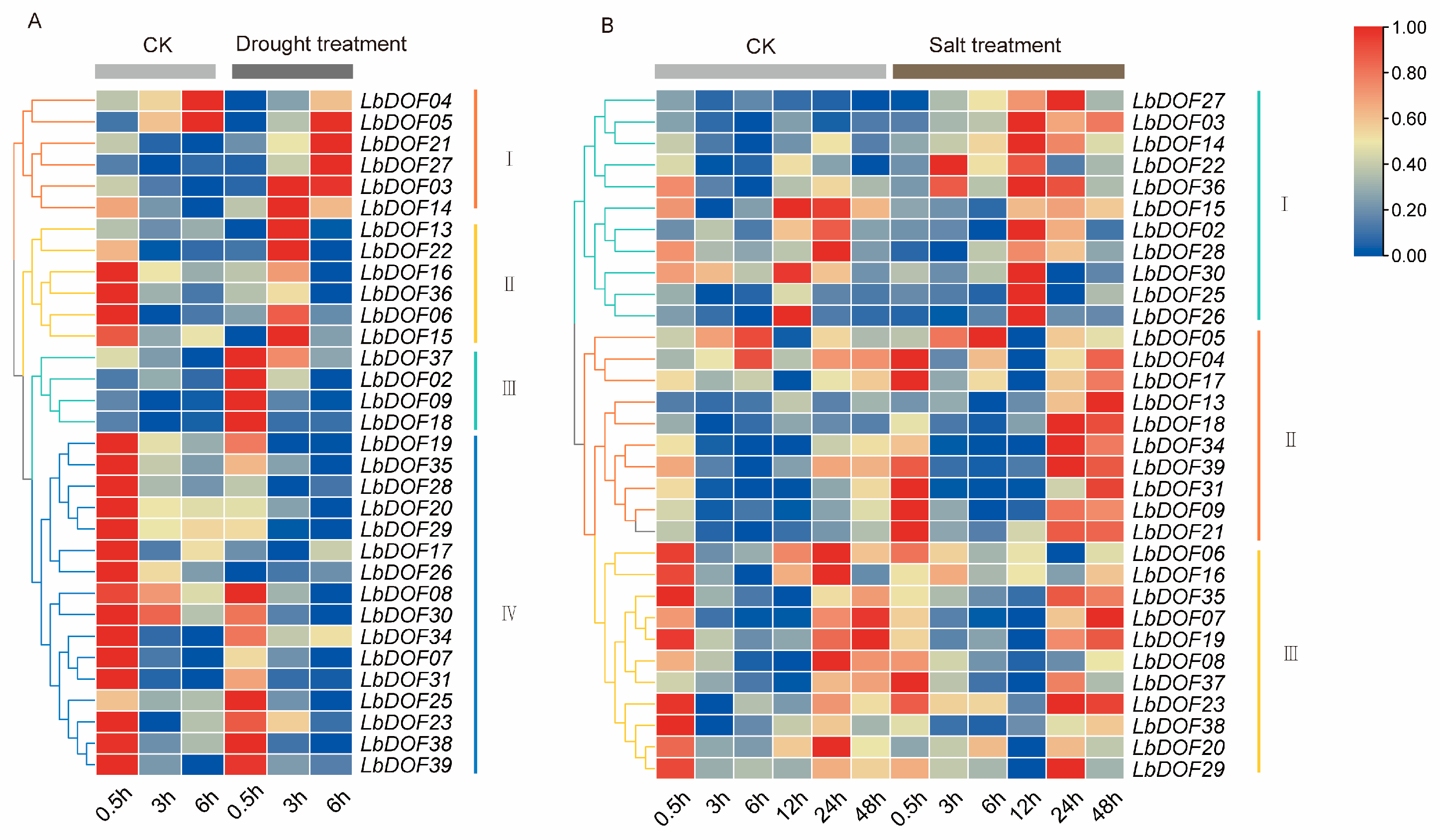
2.8. Expression Pattern of LbDofs Genes in L. barbarum Under Stress
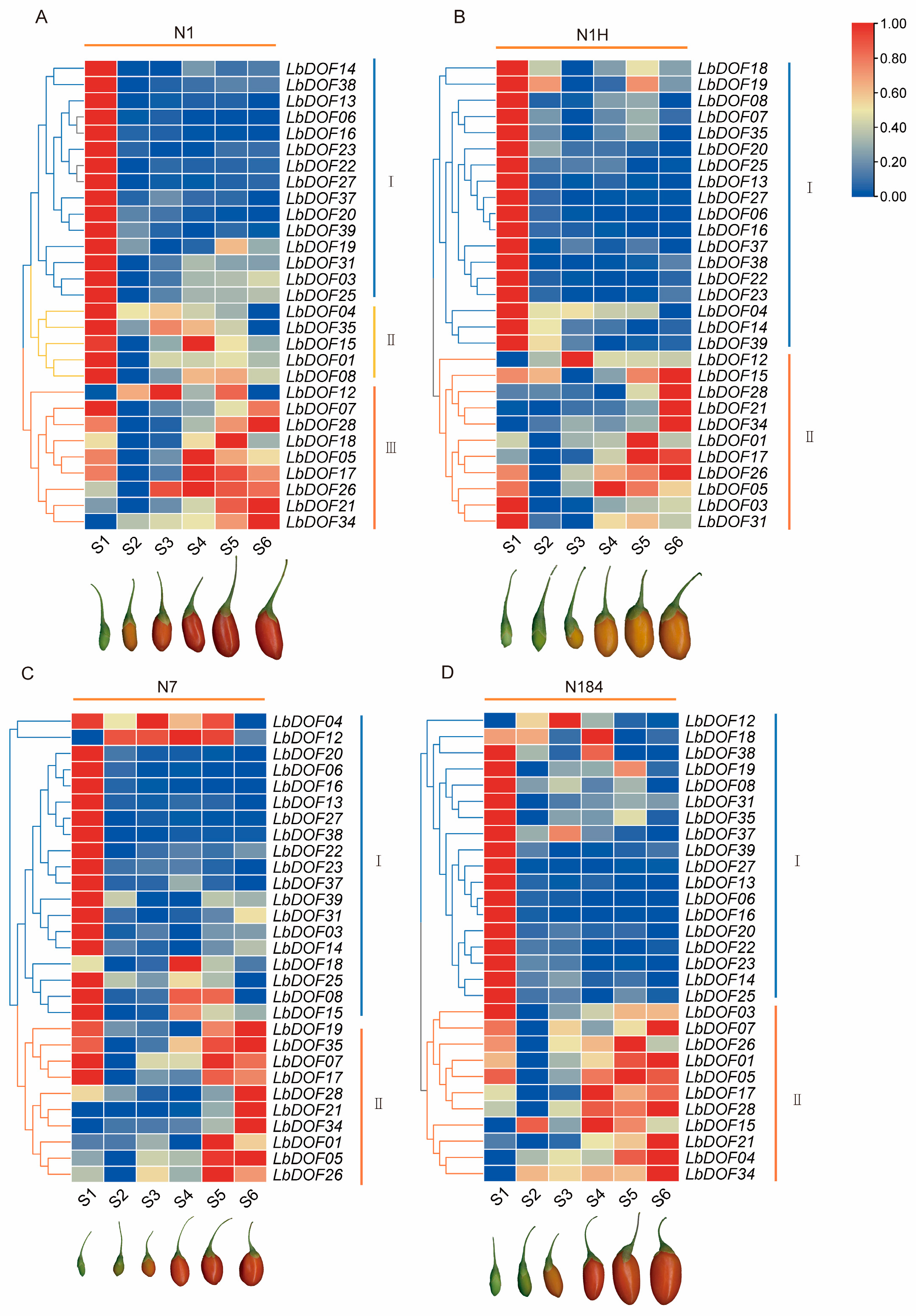
2.9. Expression Pattern of Lbdofs During Fruit Development of Different Cultivars of L. barbarum
3. Discussion
4. Materials and Methods
4.1. Identification of LbDof in L. barbarum
4.2. Gene Structure and Motif Analysis
4.3. Physicochemical Characteristics and Phylogenetic Analyses
4.4. Cis-Regulatory Element Analysis of Lbdof Genes
4.5. Collinearity Analysis and Calculation of Ka/Ks Values
4.6. Plant Materials and Treatments
4.7. Expression Patterns of LbDofs
4.8. Subcellular Localization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Yang, T.; Hu, Y.; Yan, Y.; Zhou, W.; Chen, G.; Zeng, X.; Cao, Y. Characterization and evaluation of antioxidant and anti-inflammatory activities of flavonoids from the fruits of Lycium barbarum. Foods 2022, 11, 306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Rao, S.; Wang, Y.; Qin, Y.; Qin, K.; Chen, J. Chromosome doubling enhances biomass and carotenoid content in Lycium chinense. Plants 2024, 13, 439. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, H.; Dong, X.; Yang, S.; Ma, S.; Ni, J. Quality evaluation of Lycium Barbarum (Wolfberry) from different regions in china based on polysaccharide structure, yield, and bioactivities. Chin. Med. 2019, 14, 49. [Google Scholar] [CrossRef]
- Zamanighomi, M.; Lin, Z.; Wang, Y.; Jiang, R.; Wong, W.H. Predicting transcription factor binding motifs from DNA-binding domains, chromatin accessibility and gene expression data. Nucleic Acids Res. 2017, 45, 5666–5677. [Google Scholar] [CrossRef][Green Version]
- Liu, Y.; Liu, N.; Deng, X.; Liu, D.; Li, M.; Cui, D.; Hu, Y.; Yan, Y. Genome-wide analysis of wheat DNA-binding with one finger (Dof) transcription factor genes: Evolutionary characteristics and diverse abiotic stress responses. BMC Genom. 2020, 21, 276. [Google Scholar] [CrossRef]
- Li, T.; Wang, X.; Elango, D.; Zhang, W.; Li, M.; Zhang, F.; Pan, Q.; Wu, Y. Genome-Wide identification, phylogenetic and expression pattern analysis of Dof transcription factors in blueberry (Vaccinium corymbosum L.). PeerJ 2022, 10, e14087. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, M.; Sakuraba, Y.; Yanagisawa, S. A Jasmonate-Activated MYC2-Dof2.1-MYC2 transcriptional loop promotes leaf senescence in Arabidopsis. Plant Cell 2020, 32, 242–262. [Google Scholar] [CrossRef]
- Gaillochet, C.; Burko, Y.; Platre, M.P.; Zhang, L.; Simura, J.; Willige, B.C.; Kumar, S.V.; Ljung, K.; Chory, J.; Busch, W. HY5 and phytochrome activity modulate shoot-to-root coordination during thermomorphogenesis in Arabidopsis. Development 2020, 147, dev192625. [Google Scholar] [CrossRef]
- Xiao, Q.; Liu, T.; Ling, M.; Ma, Q.; Cao, W.; Xing, F.; Huang, T.; Zhang, Y.; Duan, H.; Liu, Z. Genome-Wide identification of Dof gene family and the mechanism dissection of SbDof21 regulating starch biosynthesis in Sorghum. Int. J. Mol. Sci. 2022, 23, 12152. [Google Scholar] [CrossRef]
- Xu, D.; Li, X.; Wu, X.; Meng, L.; Zou, Z.; Bao, E.; Bian, Z.; Cao, K. Tomato SlCDF3 delays flowering time by regulating different FT-Like genes under long-day and short-day conditions. Front. Plant Sci. 2021, 12, 650068. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Xu, L.; Wang, C.; Luo, Y.; Feng, S.; Yuan, Y.; Yang, Q.; Feng, B. Genome-Wide identification of DNA binding with one finger (Dof) gene family in tartary buckwheat (Fagopyrum tataricum) and analysis of its expression pattern after exogenous hormone stimulation. Biology 2022, 11, 173. [Google Scholar] [CrossRef] [PubMed]
- Corrales, A.R.; Carrillo, L.; Lasierra, P.; Nebauer, S.G.; Dominguez-Figueroa, J.; Renau-Morata, B.; Pollmann, S.; Granell, A.; Molina, R.V.; Vicente-Carbajosa, J.; et al. Multifaceted role of cycling Dof factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764. [Google Scholar] [CrossRef]
- Chen, Y.; Cao, J. Comparative analysis of Dof transcription factor family in maize. Plant Mol. Biol. Rep. 2015, 33, 1245–1258. [Google Scholar] [CrossRef]
- Li, H.; Huang, W.; Liu, Z.W.; Wang, Y.X.; Zhuang, J. Transcriptome-Based analysis of Dof family transcription factors and their responses to abiotic stress in tea plant (Camellia sinensis). Int. J. Genom. 2016, 2016, 5614142. [Google Scholar] [CrossRef]
- Liu, J.; Meng, Q.; Xiang, H.; Shi, F.; Ma, L.; Li, Y.; Liu, C.; Liu, Y.; Su, B. Genome-Wide Analysis of Dof Transcription Factors and Their Response to Cold Stress in Rice (Oryza sativa L.). BMC Genom. 2021, 22, 800. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, P.; Wang, W.; Kong, L.; Tian, S.; Qin, G. Genome-wide binding analysis of the tomato transcription factor SlDof1 reveals its regulatory impacts on fruit ripening. Mol. Hortic. 2021, 1, 9. [Google Scholar] [CrossRef]
- Li, Y.; Tian, M.; Feng, Z.; Zhang, J.; Lu, J.; Fu, X.; Ma, L.; Wei, H.; Wang, H. GhDof1.7, a Dof Transcription factor, plays positive regulatory role under salinity stress in upland cotton. Plants 2023, 12, 3740. [Google Scholar] [CrossRef]
- Zhang, Z.; Yuan, L.; Liu, X.; Chen, X.; Wang, X. Evolution analysis of Dof transcription factor family and their expression in response to multiple abiotic stresses in Malus domestica. Gene 2018, 639, 137–148. [Google Scholar] [CrossRef]
- Corrales, A.-R.; Nebauer, S.G.; Carrillo, L.; Fernández-Nohales, P.; Marqués, J.; Renau-Morata, B.; Granell, A.; Pollmann, S.; Vicente-Carbajosa, J.; Molina, R.-V.; et al. Characterization of tomato cycling Dof factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 2014, 65, 995–1012. [Google Scholar] [CrossRef]
- Xu, J.; Dai, H. Brassica napus Cycling Dof Factor1 (BnCDF1) is involved in flowering time and freezing tolerance. Plant Growth Regul. 2016, 80, 315–322. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, Y.; Zhang, C.; Zhang, T.; Hu, T.; Ye, J.; Zhang, J.; Wang, T.; Li, H.; Ye, Z. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J. Integr. Plant Biol. 2013, 55, 552–566. [Google Scholar] [CrossRef]
- Wang, S.; Wang, R.; Yang, C. Selection and functional identification of Dof genes expressed in response to nitrogen in Populus Simonii × Populus Nigra. Open Life Sci. 2022, 17, 756–780. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, M.Y.; Wang, F.; Tang, J.; Xiong, A.S. Genome-Wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genom. 2015, 16, 33. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Ma, Y.; Lu, Y.; Yue, J.; Ming, R. Expression profiling of the Dof gene family under abiotic stresses in spinach. Sci. Rep. 2021, 11, 14429. [Google Scholar] [CrossRef] [PubMed]
- Skirycz, A.; Jozefczuk, S.; Stobiecki, M.; Muth, D.; Zanor, M.I.; Witt, I.; Mueller-Roeber, B. Transcription factor AtDof4.2 affects phenylpropanoid metabolism in arabidopsis thaliana. New Phytol. 2007, 175, 425–438. [Google Scholar] [CrossRef]
- Song, A.; Gao, T.; Li, P.; Chen, S.; Guan, Z.; Wu, D.; Xin, J.; Fan, Q.; Zhao, K.; Chen, F. Transcriptome-Wide identification and expression profiling of the Dof transcription factor gene family in Chrysanthemum morifolium. Front. Plant Sci. 2016, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Baloglu, M.C.; Eldem, V.; Hajyzadeh, M.; Unver, T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE 2014, 9, e96014. [Google Scholar] [CrossRef]
- Kushwaha, H.; Gupta, S.; Singh, V.K.; Rastogi, S.; Yadav, D. Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol. Biol. Rep. 2011, 38, 5037–5053. [Google Scholar] [CrossRef]
- Yanagisawa, S. Dof domain proteins: Plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 2004, 45, 386–391. [Google Scholar] [CrossRef]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Malviya, N.; Kushwaha, H.; Nasim, J.; Bisht, N.C.; Singh, V.K.; Yadav, D. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 2015, 241, 549–562. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Gómez, C.; Posé, D.; Martín-Pizarro, C. Insights into transcription factors controlling strawberry fruit development and ripening. Front. Plant Sci. 2022, 13, 1022369. [Google Scholar] [CrossRef]
- Feng, B.H.; Han, Y.C.; Xiao, Y.Y.; Kuang, J.F.; Fan, Z.Q.; Chen, J.Y.; Lu, W.J. The banana fruit Dof transcription factor MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes. J. Exp. Bot. 2016, 67, 2263–2275. [Google Scholar] [CrossRef]
- Kloosterman, B.; Abelenda, J.A.; Gomez Mdel, M.; Oortwijn, M.; de Boer, J.M.; Kowitwanich, K.; Horvath, B.M.; van Eck, H.J.; Smaczniak, C.; Prat, S.; et al. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 2013, 495, 246–250. [Google Scholar] [CrossRef]
- Yang, Y.; He, Z.; Bing, Q.; Duan, X.; Chen, S.; Zeng, M.; Liu, X. Two Dof transcription factors promote flavonoid synthesis in kumquat fruit by activating C-glucosyltransferase. Plant Sci. 2022, 318, 111234. [Google Scholar] [CrossRef]
- Zhao, Y.; Su, X.; Wang, X.; Wang, M.; Chi, X.; Aamir Manzoor, M.; Li, G.; Cai, Y. Comparative genomic analysis of TCP genes in six rosaceae species and expression pattern analysis in Pyrus bretschneideri. Front. Genet. 2021, 12, 669959. [Google Scholar] [CrossRef]
- Wei, P.C.; Tan, F.; Gao, X.Q.; Zhang, X.Q.; Wang, G.Q.; Xu, H.; Li, L.J.; Chen, J.; Wang, X.C. Overexpression of AtDof4.7, an Arabidopsis Dof family transcription factor, induces floral organ abscission deficiency in Arabidopsis. Plant Physiol. 2010, 153, 1031–1045. [Google Scholar] [CrossRef] [PubMed]
- Lucas-Reina, E.; Romero-Campero, F.J.; Romero, J.M.; Valverde, F. An evolutionarily conserved DOF-CONSTANS module controls plant photoperiodic signaling. Plant Physiol. 2015, 168, 561–574. [Google Scholar] [CrossRef]
- Gaur, V.S.; Singh, U.S.; Kumar, A. Transcriptional profiling and in silico analysis of Dof transcription factor gene family for understanding their regulation during seed development of rice Oryza sativa L. Mol. Biol. Rep. 2011, 38, 2827–2848. [Google Scholar] [CrossRef]
- Negi, J.; Moriwaki, K.; Konishi, M.; Yokoyama, R.; Nakano, T.; Kusumi, K.; Hashimoto-Sugimoto, M.; Schroeder, J.I.; Nishitani, K.; Yanagisawa, S.; et al. A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr. Biol. 2013, 23, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.F.; Zhang, Y.Q.; Wei, W.; Chen, H.W.; Song, Q.X.; Liu, Y.F.; Zhao, M.Y.; Wang, F.; Zhang, B.C.; Lin, Q.; et al. The transcription factor AtDof4.2 regulates shoot branching and seed coat formation in Arabidopsis. Biochem. J. 2013, 449, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Washio, K. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol. 2003, 133, 850–863. [Google Scholar] [CrossRef]
- Chen, X.; Wang, D.; Liu, C.; Wang, M.; Wang, T.; Zhao, Q.; Yu, J. Maize transcription factor Zmdof1 involves in the regulation of Zm401 gene. Plant Growth Regul. 2012, 66, 271–284. [Google Scholar] [CrossRef]
- Tolosa, L.N.; Zhang, Z. The role of major transcription factors in solanaceous food crops under different stress conditions: Current and future perspectives. Plants 2020, 9, 56. [Google Scholar] [CrossRef]
- Liao, B.; Liang, P.; Tong, L.; Lu, L.; Lu, Y.; Zheng, R.; Zheng, X.; Chen, J.; Hao, Z. The role of liriodendron Dof gene family in abiotic stress response. Plants 2024, 13, 2009. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Wang, J.; Chen, X.; Wang, F.; Peng, P.; Zhou, Y.; Miao, Y.; Zhang, Y.; Gao, Y.; Qi, Y.; et al. Rice OsDof15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol. 2019, 223, 798–813. [Google Scholar] [CrossRef]
- Shangguan, L.; Chen, M.; Fang, X.; Xie, Z.; Zhang, K.; Zheng, T.; Pu, Y.; Fang, J. Comparative study of DAM, Dof, and WRKY gene families in fourteen species and their expression in Vitis vinifera. 3 Biotech 2020, 10, 72. [Google Scholar] [CrossRef]
- Wang, C.-M.; Zeng, Z.-X.; Su, X.-G.; Lakshmanan, P.; Shan, W.; Kuang, J.-F.; Lu, W.-J.; Chen, J.-Y.; Zhao, Y.-T. A transcriptional repressor BrDof2.4 regulates protease genes involved in postharvest leaf senescence in Chinese flowering cabbage. Postharvest Biol. Technol. 2021, 181, 111680. [Google Scholar] [CrossRef]
- Cao, Y.L.; Li, Y.L.; Fan, Y.F.; Li, Z.; Yoshida, K.; Wang, J.Y.; Ma, X.K.; Wang, N.; Mitsuda, N.; Kotake, T.; et al. Wolfberry genomes and the evolution of Lycium (Solanaceae). Commun. Biol. 2021, 4, 671. [Google Scholar] [CrossRef]
- Harrison, P.W.; Amode, M.R.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2024. Nucleic Acids Res. 2023, 52, D891–D899. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2019, 48, D1104–D1113. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2020, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for On”; bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5:an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant Cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- El Baidouri, M.; Murat, F.; Veyssiere, M.; Molinier, M.; Flores, R.; Burlot, L.; Alaux, M.; Quesneville, H.; Pont, C.; Salse, J. Reconciling the evolutionary origin of bread wheat (Triticum aestivum). New Phytol. 2017, 213, 1477–1486. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Li, W.; Che, J.; Lian, Q.; Wang, C.; Dai, G.; Chen, J. Identification and expression pattern of the carotenoid cleavage oxygenase gene family in Lycium suggest CCOs respond to abiotic stress and promote carotenoids degradation. Forests 2023, 14, 983. [Google Scholar] [CrossRef]


| Gene ID | Dof Gene | Map Position (bp) | Length (AA) | PI | MW (kDa) | Num of Exon | Location |
|---|---|---|---|---|---|---|---|
| Lba01g00421 | LbDof01 | Chr01:9119737-9120378 | 213 | 8.14 | 24.31 | 1 | Nuclear |
| Lba01g00560 | LbDof02 | Chr01:13918702-13920360 | 282 | 8.99 | 30.84 | 3 | Nuclear |
| Lba01g01980 | LbDof03 | Chr01:154257739-154263430 | 306 | 5.55 | 33.42 | 2 | Nuclear |
| Lba01g02222 | LbDof04 | Chr01:157935616-157936527 | 303 | 5.87 | 33.58 | 1 | Nuclear |
| Lba01g02268 | LbDof05 | Chr01:158495049-158497125 | 449 | 5.72 | 49.27 | 2 | Nuclear |
| Lba01g02335 | LbDof06 | Chr01:159697135-159699178 | 304 | 6.35 | 33.62 | 2 | Nuclear |
| Lba01g02725 | LbDof07 | Chr01:165684285-165686213 | 297 | 7.18 | 32.91 | 2 | Nuclear |
| Lba01g02777 | LbDof08 | Chr01:166398894-166400062 | 320 | 9.26 | 34.57 | 2 | Nuclear |
| Lba02g00233 | LbDof09 | Chr02:4517154-4541179 | 659 | 8.53 | 73.62 | 4 | Nuclear |
| Lba02g00236 | LbDof10 | Chr02:4602270-4603229 | 319 | 5.92 | 35.64 | 1 | Nuclear |
| Lba02g00237 | LbDof11 | Chr02:4608282-4609223 | 313 | 5.83 | 35.00 | 1 | Nuclear |
| Lba02g01857 | LbDof12 | Chr02:131223420-131224217 | 265 | 4.56 | 29.72 | 1 | Nuclear |
| Lba02g02528 | LbDof13 | Chr02:147141275-147141931 | 218 | 6.12 | 22.66 | 1 | Nuclear |
| Lba03g00541 | LbDof14 | Chr03:10678916-10680566 | 393 | 9.3 | 41.85 | 3 | Nuclear |
| Lba03g01659 | LbDof15 | Chr03:121309151-121310338 | 261 | 8.76 | 28.50 | 2 | Nuclear |
| Lba03g02183 | LbDof16 | Chr03:133102286-133104176 | 300 | 6.66 | 33.36 | 2 | Nuclear |
| Lba03g02450 | LbDof17 | Chr03:138661121-138664401 | 500 | 6.14 | 54.38 | 2 | Nuclear |
| Lba03g02993 | LbDof18 | Chr03:146745940-146746503 | 187 | 9.34 | 20.29 | 1 | Nuclear |
| Lba04g00060 | LbDof19 | Chr04:794018-796171 | 323 | 8.82 | 35.30 | 3 | Nuclear |
| Lba05g00239 | LbDof20 | Chr05:4035257-4036339 | 307 | 9.37 | 34.08 | 2 | Nuclear |
| Lba05g02493 | LbDof21 | Chr05:142942613-142944872 | 401 | 5.96 | 44.32 | 2 | Nuclear |
| Lba06g00549 | LbDof22 | Chr06:12304979-12306315 | 277 | 6.38 | 31.26 | 2 | Nuclear |
| Lba06g00550 | LbDof23 | Chr06:12308649-12310464 | 281 | 8.08 | 31.61 | 2 | Nuclear |
| Lba06g00551 | LbDof24 | Chr06:12350155-12352434 | 252 | 8.83 | 28.82 | 2 | Nuclear |
| Lba06g00700 | LbDof25 | Chr06:18846366-18847463 | 298 | 9.43 | 32.64 | 2 | Nuclear |
| Lba06g01885 | LbDof26 | Chr06:109723965-109725506 | 287 | 9.19 | 30.91 | 3 | Nuclear |
| Lba08g00407 | LbDof27 | Chr08:12244232-12246259 | 310 | 6.19 | 34.88 | 2 | Nuclear |
| Lba08g01387 | LbDof28 | Chr08:117156146-117157413 | 356 | 9.3 | 38.46 | 2 | Nuclear |
| Lba09g02477 | LbDof29 | Chr09:125873081-125874140 | 252 | 7.55 | 28.00 | 2 | Nuclear |
| Lba10g00246 | LbDof30 | Chr10:4459414-4461261 | 356 | 8.33 | 38.40 | 3 | Nuclear |
| Lba10g01565 | LbDof31 | Chr10:97856988-97859756 | 424 | 6.72 | 46.50 | 2 | Nuclear |
| Lba11g02467 | LbDof32 | Chr11:119952692-119957326 | 303 | 9.13 | 34.16 | 3 | Nuclear |
| Lba12g00530 | LbDof33 | Chr12:63267215-63267769 | 184 | 9.73 | 19.88 | 1 | Nuclear |
| Lba12g00627 | LbDof34 | Chr12:66014533-66017295 | 452 | 5.24 | 49.63 | 2 | Nuclear |
| Lba12g01572 | LbDof35 | Chr12:84113552-84114904 | 395 | 7.76 | 43.91 | 2 | Nuclear |
| Lba12g01573 | LbDof36 | Chr12:84129012-84129893 | 293 | 9.81 | 31.83 | 1 | Nuclear |
| Lba12g01646 | LbDof37 | Chr12:85470852-85471736 | 294 | 5.53 | 33.26 | 1 | Nuclear |
| Lba12g02508 | LbDof38 | Chr12:99924661-99925524 | 287 | 9.14 | 31.10 | 1 | Nuclear |
| Lba12g02669 | LbDof39 | Chr12:102326211-102328664 | 446 | 6.03 | 49.84 | 2 | Nuclear |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wang, H.; Li, W.; Dai, G.; Chen, J. Genome-Wide Identification and Expression Analysis of the LbDof Transcription Factor Family Genes in Lycium barbarum. Plants 2025, 14, 1567. https://doi.org/10.3390/plants14111567
Wang Y, Wang H, Li W, Dai G, Chen J. Genome-Wide Identification and Expression Analysis of the LbDof Transcription Factor Family Genes in Lycium barbarum. Plants. 2025; 14(11):1567. https://doi.org/10.3390/plants14111567
Chicago/Turabian StyleWang, Yuchang, Hongrui Wang, Weinan Li, Guoli Dai, and Jinhuan Chen. 2025. "Genome-Wide Identification and Expression Analysis of the LbDof Transcription Factor Family Genes in Lycium barbarum" Plants 14, no. 11: 1567. https://doi.org/10.3390/plants14111567
APA StyleWang, Y., Wang, H., Li, W., Dai, G., & Chen, J. (2025). Genome-Wide Identification and Expression Analysis of the LbDof Transcription Factor Family Genes in Lycium barbarum. Plants, 14(11), 1567. https://doi.org/10.3390/plants14111567






