Identification and Expression Analysis of NAC Transcription Factors Related to Rust Resistance in Foxtail Millet
Abstract
1. Introduction
2. Results
2.1. Selection and Phylogeny Analysis of the SiNAC Genes
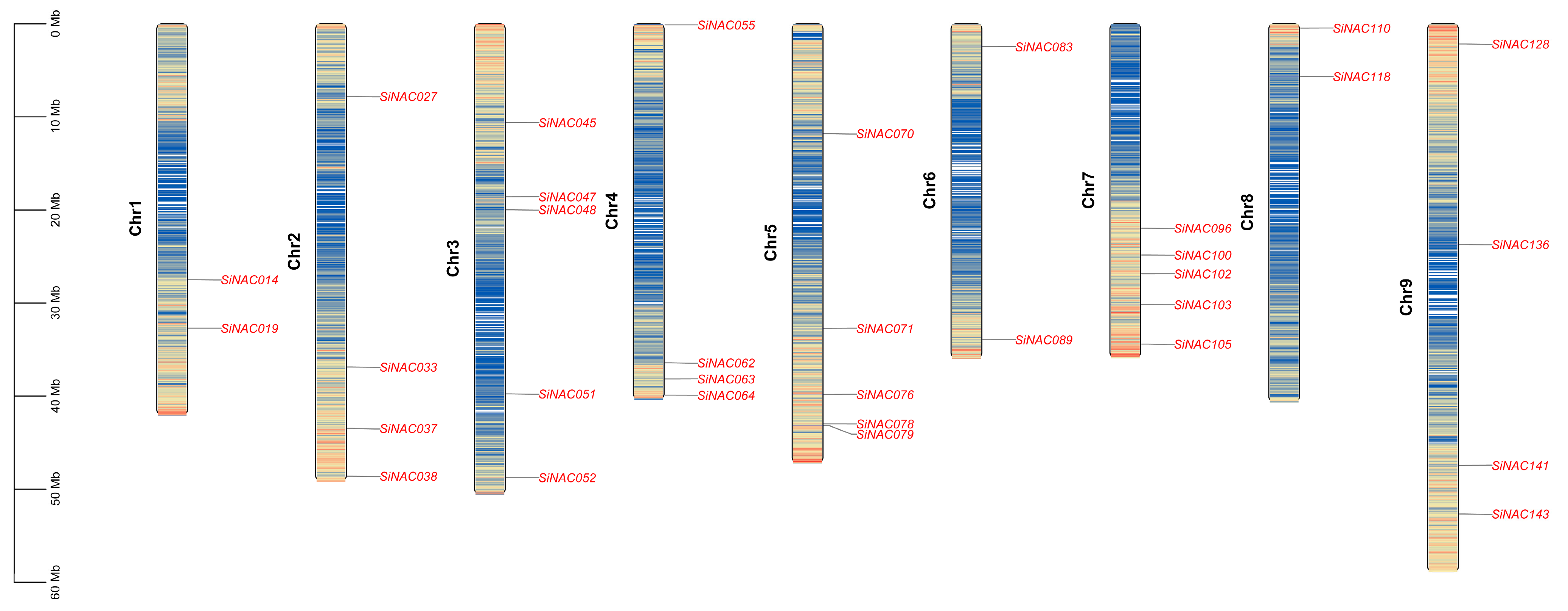
2.2. Chromosomal Distribution of SiNAC Genes
2.3. Cis-Regulatory Element Analysis of SiNAC Gene Promoter
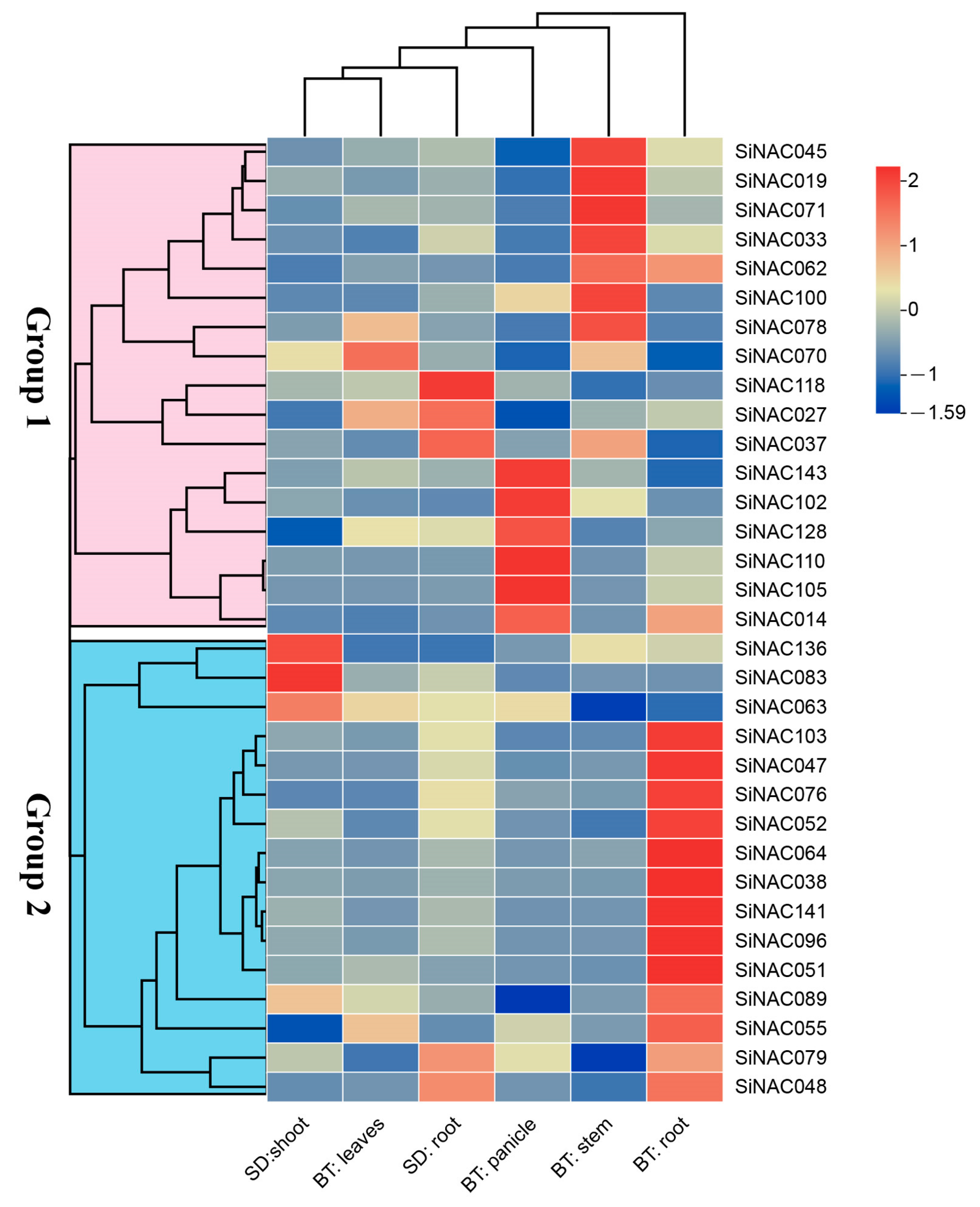
2.4. Expression Patterns of SiNAC Genes Across Developmental Stages and Tissues in Foxtail Millet
2.5. Expression Profiling of SiNAC Genes During Incompatible Interaction Between Foxtail Millet and Usi 93-5
2.6. Comparative Analysis of SiNAC Gene Expression Between Incompatible and Compatible Interactions
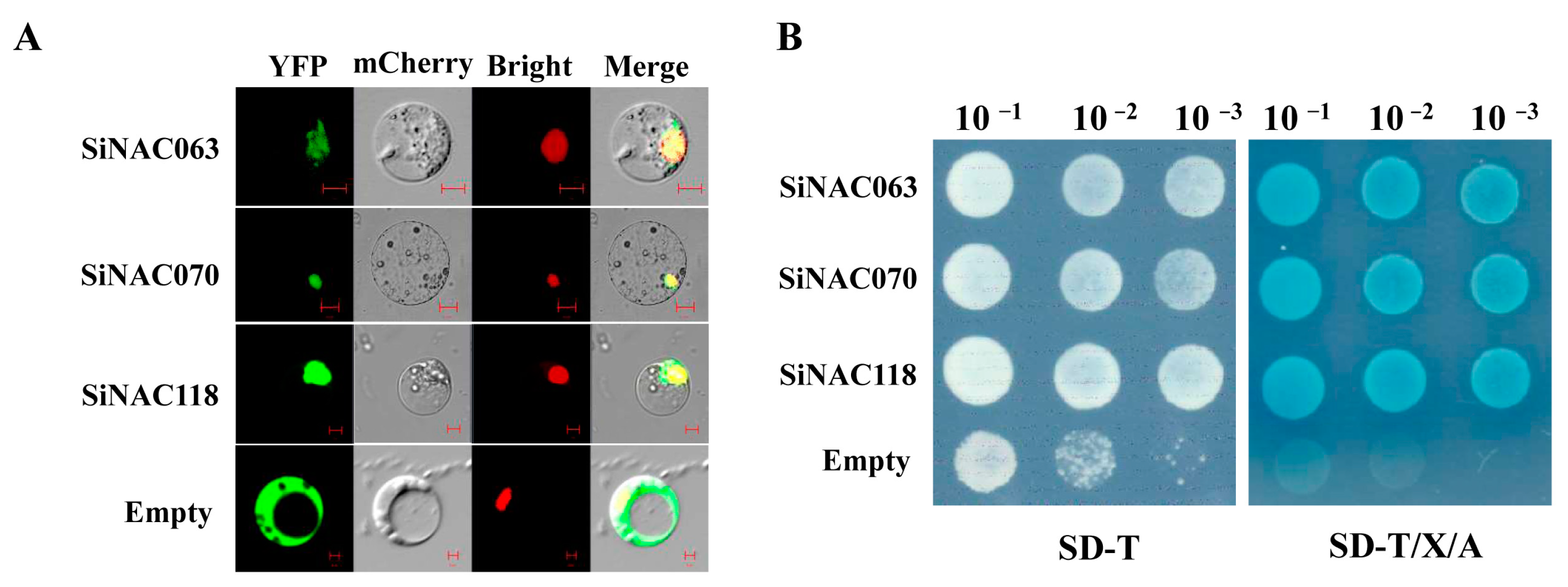
2.7. Functional Characterization of SiNAC Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Inoculation and Sample Collection
4.2. NAC Genes Collection and Phylogenetic Analysis
4.3. Chromosomal Distribution Analysis of the SiNACs
4.4. Analysis of Promoter Cis-Acting Elements
4.5. RNA Extraction and cDNA Synthesis
4.6. qRT-PCR Analysis
4.7. Subcellular Localization of SiNACs Protein
4.8. Transactivation Assay in Yeast Cells
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, Z.Y.; Wang, N.; Dong, L.; Bai, H.; Quan, J.Z.; Liu, L.; Dong, Z.P. Differential gene expression in foxtail millet during incompatible interaction with Uromyces setariae-italicae. PLoS ONE 2015, 10, e0123825. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.S.; Shao, L.X.; Cui, G.X.; Zhang, H.Q. A preliminary study on the epidemic characteristics of millet rust. J. Agric. Univ. Hebei 1997, 20, 30–34. (In Chinese) [Google Scholar]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Shan, W.; Kuang, J.F.; Chen, L.; Xie, H.; Peng, H.H.; Xiao, Y.Y.; Li, X.P.; Chen, W.X.; He, Q.G.; Chen, J.Y.; et al. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J. Exp. Bot. 2012, 63, 5171–5187. [Google Scholar] [CrossRef]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive Analysis of NAC Family Genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Christiansen, M.W.; Holm, P.B.; Gregersen, P.L. Characterization of barley (Hordeum vulgare L.) NAC transcription factors suggests conserved functions compared to both monocots and dicots. BMC Res. Notes 2011, 4, 302–314. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, Y.; Li, B.; Chang, J.; Chen, M.; Li, K.; Yang, G.; He, G. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2015, 15, 268. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Peng, X.; Zhao, Y.; Li, X.; Wu, M.; Chai, W.; Sheng, L.; Wang, Y.; Dong, Q.; Jiang, H.; Cheng, B. Genome-wide identification, classification, and analysis of NAC type gene family in maize. J. Genet. 2015, 94, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Cheng, J.; Hu, H.; Yan, L.; Jia, J.; Wu, B. Genome-wide identification of NAC family genes in oat and functional characterization of AsNAC109 in abiotic stress tolerance. Plants 2024, 13, 1017. [Google Scholar] [CrossRef]
- Wu, M.; Xu, Z.; Fu, C.; Wang, N.; Zhang, R.; Le, Y.; Chen, M.; Yang, N.; Li, Y.; Zhang, X.; et al. NAC Transcription factor GbNTL9 modifies the accumulation and organization of cellulose microfibrils to enhance cotton fiber strength. J. Adv. Res. 2025; in press. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Mandal, S.N.; B, V.S.; Parida, S.K.; Prasad, M. Comprehensive genome-wide survey, genomic constitution, and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.). PLoS ONE 2013, 8, e64594. [Google Scholar] [CrossRef]
- Mao, C.; Ding, J.; Zhang, B.; Xi, D.; Ming, F. OsNAC2 positively affects salt-induced cell death and binds to the OsAP37 and OsCOX11 promoters. Plant J. 2018, 94, 454–468. [Google Scholar] [CrossRef]
- Mao, H.; Li, S.; Chen, B.; Jian, C.; Mei, F.; Zhang, Y.; Li, F.; Chen, N.; Li, T.; Du, L.; et al. Variation in cis-regulation of a NAC transcription factor contributes to drought tolerance in wheat. Mol. Plant 2022, 15, 276–292. [Google Scholar] [CrossRef]
- Yang, C.; Huang, Y.; Lv, P.; Antwi-Boasiako, A.; Begum, N.; Zhao, T.; Zhao, J. NAC transcription factor GmNAC12 improved drought stress tolerance in soybean. Int. J. Mol. Sci. 2022, 23, 12029. [Google Scholar] [CrossRef]
- Sang, R.P.; Kim, H.S.; Lee, K.S.; Hwang, D.J.; Bae, S.C.; Ahn, I.P.; Lee, S.H.; Sun, T.K. Overexpression of rice NAC transcription factor OsNAC58 on increased resistance to bacterial leaf blight. J. Plant Biotechnol. 2017, 44, 149–155. [Google Scholar]
- Zhou, W.; Qian, C.; Li, R.; Zhou, S.; Zhang, R.; Xiao, J.; Wang, X.; Zhang, S.; Xing, L.; Cao, A. TaNAC6s are involved in the basal and broad-spectrum resistance to powdery mildew in wheat. Plant Sci. 2018, 277, 218–228. [Google Scholar] [CrossRef]
- Chen, C.; Jost, M.; Outram, M.A.; Friendship, D.; Chen, J.; Wang, A.; Periyannan, S.; Bartoš, J.; Holušová, K.; Doležel, J.; et al. A pathogen-induced putative NAC transcription factor mediates leaf rust resistance in barley. Nat. Commun. 2023, 14, 5468–5478. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, C.; Shao, X.; Hu, Z.; Li, J.; Wang, P.; Wang, A.; Yu, J.; Shi, K. Transcriptomic and genetic approaches reveal an essential role of the NAC transcription factor SlNAP1 in the growth and defense response of tomato. Hortic. Res. 2020, 7, 209. [Google Scholar] [CrossRef]
- Huang, Y.; Li, T.; Xu, Z.-S.; Wang, F.; Xiong, A.-S. Six NAC transcription factors involved in response to TYLCV infection in resistant and susceptible tomato cultivars. Plant Physiol. Biochem. 2017, 120, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Tsuchida-Mayama, T.; Ichikawa, H.; Mitsuda, N.; Ohme-Takagi, M.; Kaku, H.; Minami, E.; Nishizawa, Y. OsNAC111, a blast disease-responsive transcription factor in rice, positively regulates the expression of defense-related genes. Mol. Plant-Microbe Interact. 2014, 27, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhu, L.; Xu, L.; Guo, W.; Zhang, X. GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks. Plant Cell Rep. 2016, 35, 2167–2179. [Google Scholar] [CrossRef]
- Jia, G.; Thinn, K.S.Z.; Kim, S.H.; Min, J.; Oh, S.K. Capsicum annuum NAC4 (CaNAC4) is a transcription factor with roles in biotic and abiotic stresses. Plant Pathol. J. 2024, 40, 512–524. [Google Scholar] [CrossRef]
- Foresti, C.; Orduña, L.; Matus, J.T.; Vandelle, E.; Danzi, D.; Bellon, O.; Tornielli, G.B.; Amato, A.; Zenoni, S. NAC61 Regulates late- and post-ripening osmotic, oxidative, and biotic stress responses in grapevine. J. Exp. Bot. 2024, 75, 2330–2350. [Google Scholar] [CrossRef]
- Tariq, R.; Hussain, A.; Tariq, A.; Khalid, M.H.B.; Khan, I.; Basim, H.; Ingvarsson, P.K. Genome-wide analyses of the mung bean NAC gene family reveals orthologs, co-expression networking and expression profiling under abiotic and biotic stresses. BMC Plant Biol. 2022, 22, 343. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, S.; Ma, X.; Wang, Y.; Kong, F.; Meng, Q. A stress-associated NAC transcription factor (SlNAC35) from tomato plays a positive role in biotic and abiotic stresses. Physiol. Plant. 2016, 158, 45–64. [Google Scholar] [CrossRef]
- Dong, B.; Liu, Y.; Huang, G.; Song, A.; Chen, S.; Jiang, J.; Chen, F.; Fang, W. Plant NAC transcription factors in the battle against pathogens. BMC Plant Biol. 2024, 24, 958. [Google Scholar] [CrossRef]
- Qiu, J.; Chen, Y.; Liu, Z.; Wen, H.; Jiang, N.; Shi, H.; Kou, Y. The application of zinc oxide nanoparticles: An effective strategy to protect rice from rice blast and abiotic stresses. Environ. Pollut. 2023, 15, 121925. [Google Scholar] [CrossRef]
- Huang, C.; Zhao, J.; Huang, Q.; Peng, L.; Huang, Z.; Li, W.; Sun, S.; He, Y.; Wang, Z. OsNAC3 regulates seed germination involving abscisic acid pathway and cell elongation in rice. New Phytol. 2024, 241, 650–664. [Google Scholar] [CrossRef]
- Puranik, S.; Bahadur, R.P.; Srivastava, P.S.; Prasad, M. Molecular cloning and characterization of a membrane-associated NAC family gene, SiNAC, from foxtail millet [Setaria italica (L.) P. Beauv]. Mol. Biotechnol. 2011, 49, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.H.; Hu, L.Q.; Xue, F.Y.; Li, W.W.; Xu, Z.S.; Li, L.C.; Zhou, Y.B.; Ma, Y.Z.; Diao, X.M.; Jia, G.Q.; et al. Overexpression of millet transcription factor gene SiNAC45 to response of low potassium stress and ABA treatment in transgenic Arabidopsis. Acta Agron. Sin. 2015, 41, 1445–1453. (In Chinese) [Google Scholar] [CrossRef]
- Xie, L.N.; Chen, M.; Min, D.H.; Feng, L.; Xu, Z.S.; Zhou, Y.B.; Xu, D.B.; Li, L.C.; Ma, Y.Z.; Zhang, X.H. The NAC-like transcription factor SiNAC110 in foxtail millet (Setaria italica L.) confers tolerance to drought and high salt stress through an ABA-independent signaling pathway. J. Integr. Agric. 2017, 16, 559–571. [Google Scholar] [CrossRef]
- Feng, H.; Duan, X.; Zhang, Q.; Li, X.; Wang, B.; Huang, L.; Wang, X.; Kang, Z. The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol. Plant Pathol. 2014, 15, 284–296. [Google Scholar] [CrossRef]
- Xia, N.; Zhang, G.; Liu, X.Y.; Deng, L.; Cai, G.L.; Zhang, Y.; Wang, X.J.; Zhao, J.; Huang, L.L.; Kang, Z.S. Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol. Biol. Rep. 2010, 37, 3703–3712. [Google Scholar] [CrossRef]
- Shan, W.; Chen, J.Y.; Kuang, J.F.; Lu, W.J. Banana fruit NAC transcription factor MaNAC5 cooperates with MaWRKYs to enhance the expression of pathogenesis-related genes against Colletotrichum musae. Mol. Plant Pathol. 2016, 17, 330–338. [Google Scholar] [CrossRef]
- Zhao, L.Q.; Pan, W.J.; Ma, J.F.; Weng, Q.Y.; Dong, L.; Quan, J.Z.; Xing, J.H.; Dong, Z.P.; Dong, J.G. Identification of AFLP markers linked to a novel rust resistance gene in foxtail millet. Sci. Agric. Sin. 2010, 43, 4349–4355. (In Chinese) [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, K.; Liu, J.; Zhang, M.; Dong, Z.; Ma, J.; Xuan, P.; Bai, H.; Li, Z. Identification and Expression Analysis of NAC Transcription Factors Related to Rust Resistance in Foxtail Millet. Plants 2025, 14, 1507. https://doi.org/10.3390/plants14101507
Gong K, Liu J, Zhang M, Dong Z, Ma J, Xuan P, Bai H, Li Z. Identification and Expression Analysis of NAC Transcription Factors Related to Rust Resistance in Foxtail Millet. Plants. 2025; 14(10):1507. https://doi.org/10.3390/plants14101507
Chicago/Turabian StyleGong, Keke, Jia Liu, Mengya Zhang, Zhiping Dong, Jifang Ma, Peixue Xuan, Hui Bai, and Zhiyong Li. 2025. "Identification and Expression Analysis of NAC Transcription Factors Related to Rust Resistance in Foxtail Millet" Plants 14, no. 10: 1507. https://doi.org/10.3390/plants14101507
APA StyleGong, K., Liu, J., Zhang, M., Dong, Z., Ma, J., Xuan, P., Bai, H., & Li, Z. (2025). Identification and Expression Analysis of NAC Transcription Factors Related to Rust Resistance in Foxtail Millet. Plants, 14(10), 1507. https://doi.org/10.3390/plants14101507






