Pre-mRNA Splicing Functions in Plant Sexual Reproduction Development
Abstract
1. Introduction
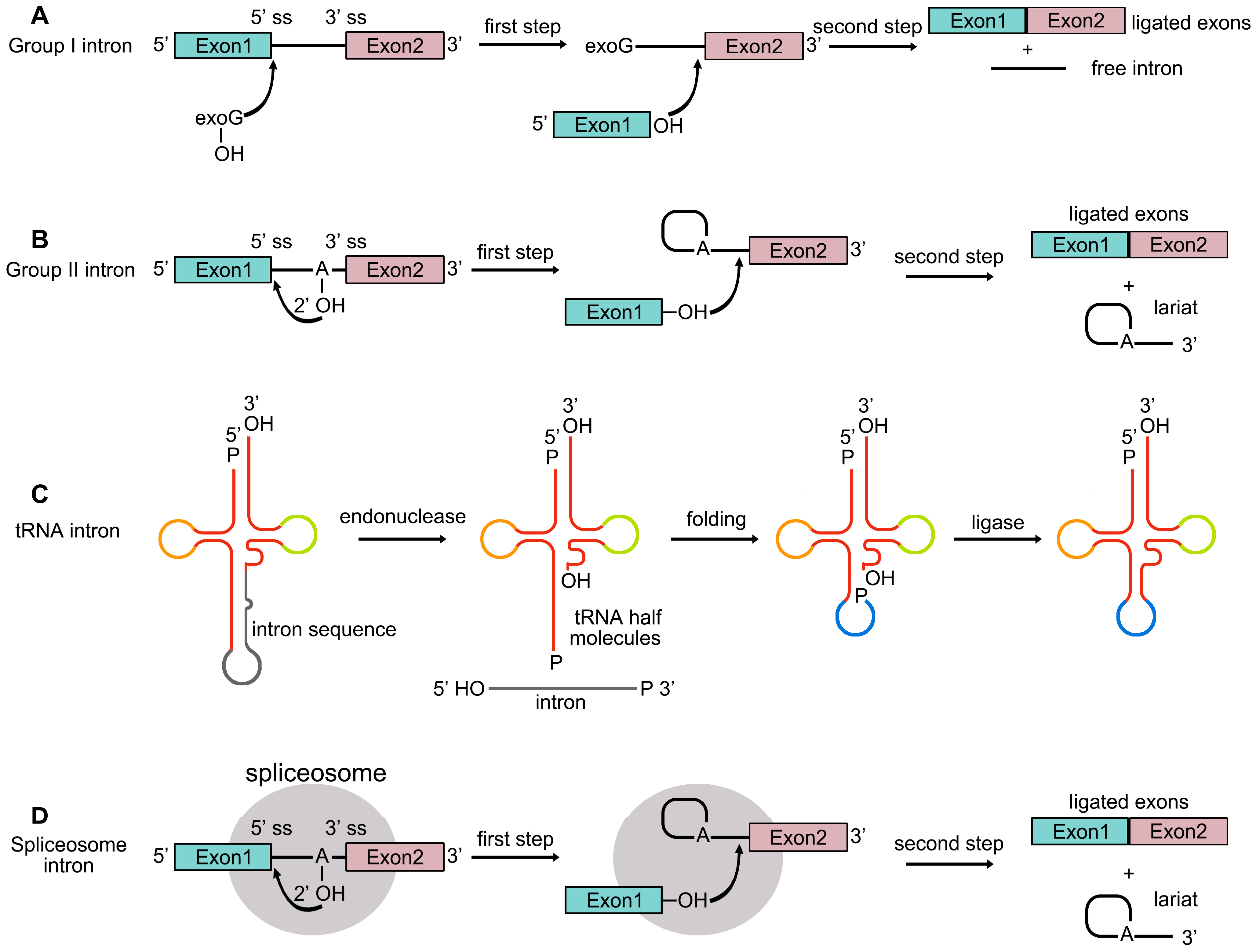
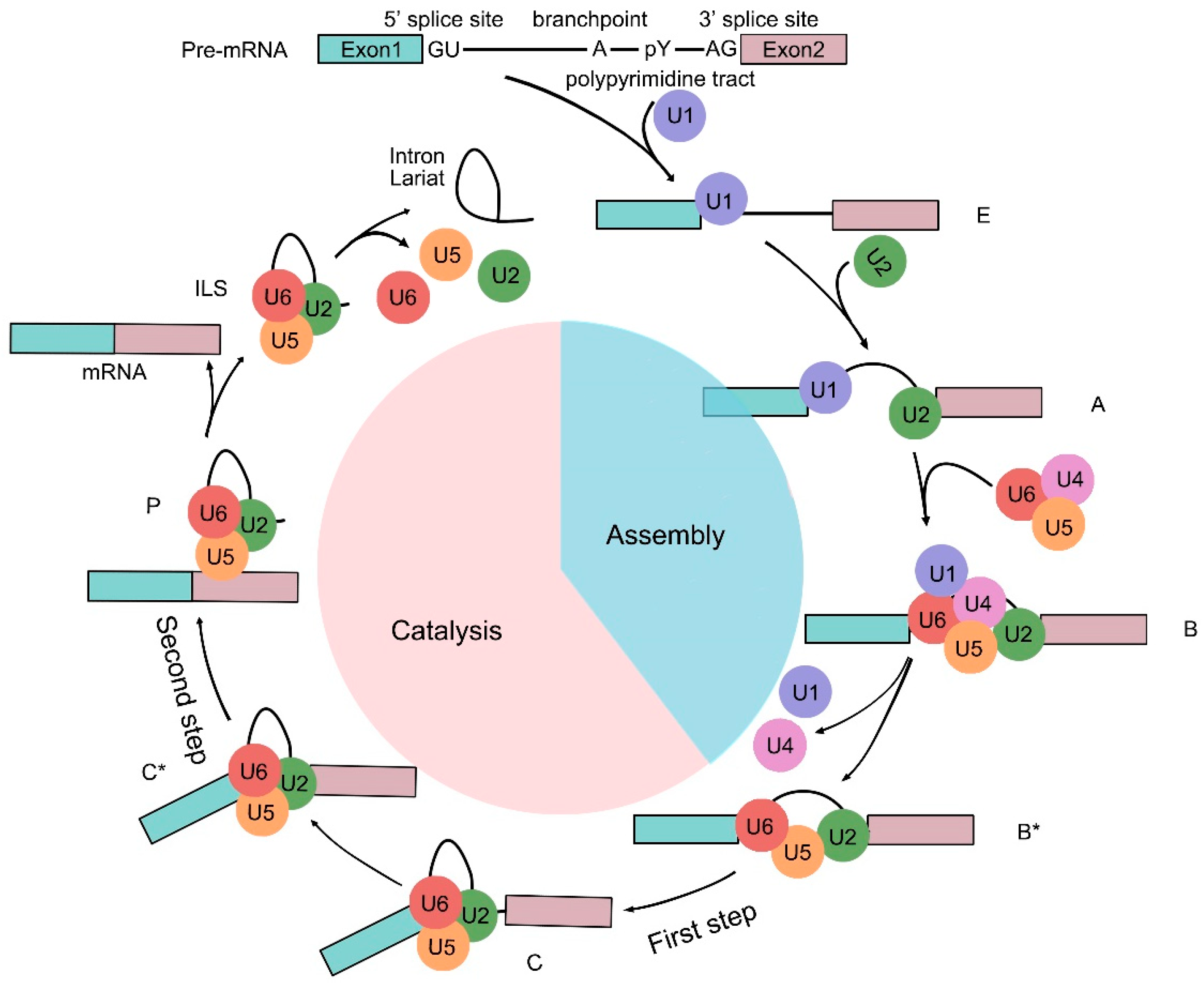
2. Pre-mRNA Splicing
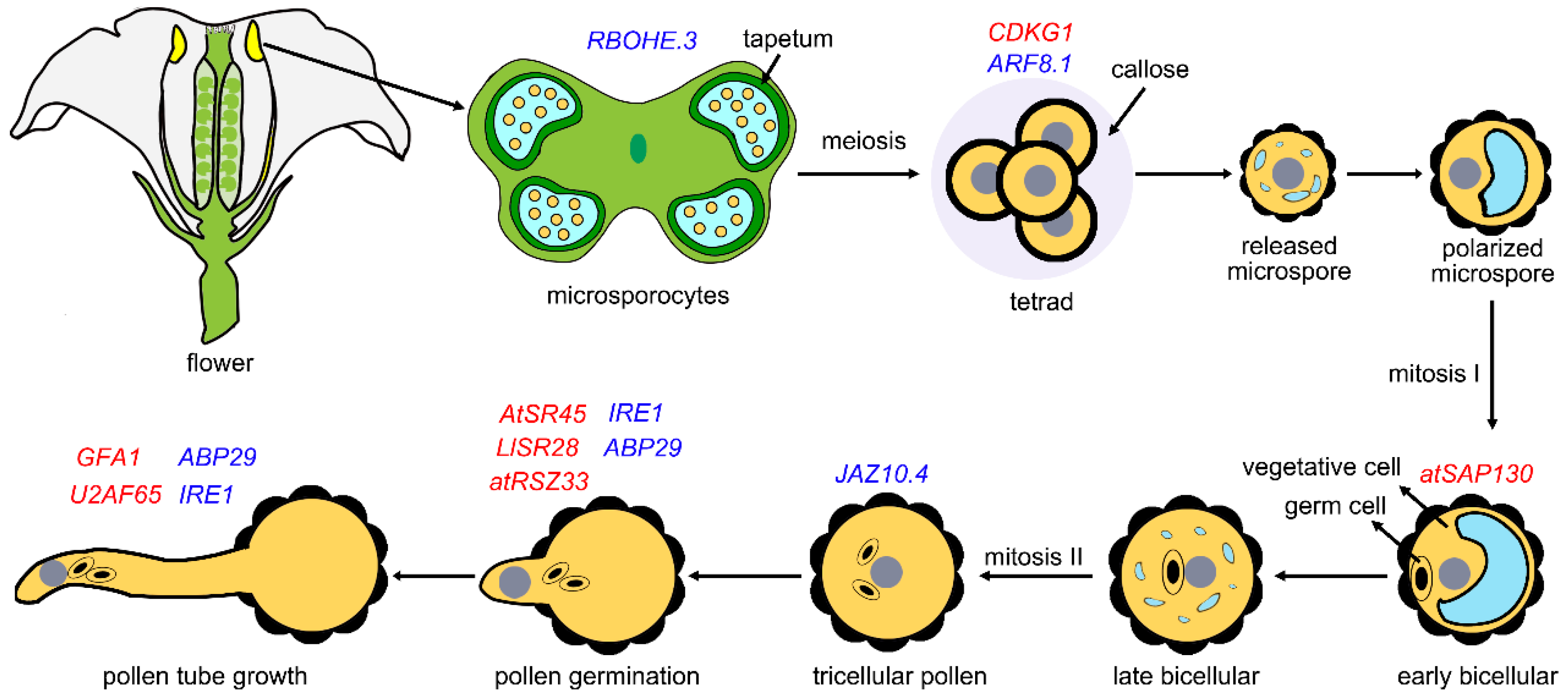
3. Role of Splicing in Pollen Development
| Species | Biological Process | Type of Splicing Factor | Splicing Factor | Targeted Genes | References |
|---|---|---|---|---|---|
| Arabidopsis thaliana | Male gametophyte transmission, pollen tube growth | Non-snRNP, one subunit of U2AF | U2AF65 | Unknown | [32] |
| A. thaliana | Male gametophyte transmission | DEAH-box RNA-dependent ATPase Prp16 | CUV | Genes involved in auxin biosynthesis, polar auxin transport, and auxin perception and signaling | [34] |
| A. thaliana | Pollen tube growth | U5 snRNP component | GFA1 | Unknown | [33,35,36] |
| A. thaliana | Pollen wall formation | Cyclin-dependent protein kinases (SR protein) | CDKG1 | CalS5 | [37] |
| A. thaliana | Male fertility at modestly ET (elevated temperature) | A major component of the UPR signaling pathway | IRE1 | bZIP60 | [38] |
| Lilium longiflorum | Pollen germination | SR protein | LlSR28 | AtVLN1 | [39] |
| A. thaliana | Pollen germination | SR protein | AtSR45 | AtVLN1 | [39] |
| A. thaliana | Pollen germination, pollen tube growth | SR protein | atRSZ33 | Unknown | [40] |
| A. thaliana | Pollen development | A U2 snRNP-associated protein | AtSAP130 | QRT1 and QRT3 | [41] |
| A. thaliana | Male gametophyte transmission | A U4/U6 snRNP associated protein | RDM16 | Unknown | [42] |
| A. thaliana | Male gametophyte development | U5 snRNP component | PRP8A, PRP8B | Unknown | [43] |
4. Splicing and Female Gametogenesis
5. Splicing-Regulated Embryo Development
6. Conclusions and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Ning, M.; Li, Q.; Wang, Y.; Li, Q.; Tao, Y.; Zhang, F.; Hu, F.; Huang, L. Alternative splicing drives the functional diversification of a bHLH transcription factor in the control of growth and drought tolerance in rice. Sci. Bull. 2024, 70, 153–156. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Du, Y.; Huai, J.; Jing, Y.; Lin, R. The RNA helicase UAP56 and the E3 ubiquitin ligase COP1 coordinately regulate alternative splicing to repress photomorphogenesis in Arabidopsis. Plant Cell 2022, 34, 4191–4212. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Xiong, F.; Ren, Q.P.; Wang, X.L.; Wellmer, F. Regulation of flowering transition by alternative splicing: The role of the U2 auxiliary factor. J. Exp. Bot. 2020, 71, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, S.; Xu, X.; Zhou, W.; Wu, L. Alternative splicing: An efficient regulatory approach towards plant developmental plasticity. WIREs RNA 2023, 14, e1758. [Google Scholar] [CrossRef]
- Liu, Y.; Do, S.; Huynh, H.; Li, J.X.; Liu, Y.G.; Du, Z.Y.; Chen, M.X. Importance of pre-mRNA splicing and its study tools in plants. Adv. Biotechnol. 2024, 2, 4. [Google Scholar] [CrossRef]
- Deng, X.; Cao, X. Roles of pre-mRNA splicing and polyadenylation in plant development. Curr. Opin. Plant Biol. 2017, 35, 45–53. [Google Scholar] [CrossRef]
- Haugen, P.; Simon, D.M.; Bhattacharya, D. The natural history of group I introns. Trends Genet. 2005, 21, 111–119. [Google Scholar] [CrossRef]
- Wilkinson, M.E.; Charenton, C.; Nagai, K. RNA splicing by the spliceosome. Annu. Rev. Biochem. 2020, 89, 359–388. [Google Scholar] [CrossRef]
- Pyle, A.M. Group II intron self-splicing. Annu. Rev. Biophys. 2016, 45, 183–205. [Google Scholar] [CrossRef]
- Schmidt, C.A.; Matera, A.G. tRNA introns: Presence, processing, and purpose. WIREs RNA 2019, 11, e1583. [Google Scholar] [CrossRef]
- Wahl, M.C.; Will, C.L.; Luhrmann, R. The spliceosome: Design principles of a dynamic RNP machine. Cell 2009, 136, 701–718. [Google Scholar] [CrossRef] [PubMed]
- Burge, P.A.; Burge, C.B. Classification of introns: U2-type or U12-type. Cell 1997, 91, 875–879. [Google Scholar] [CrossRef]
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Will, C.L.; Luhrmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3, a003707. [Google Scholar] [CrossRef]
- Shi, Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017, 18, 655–670. [Google Scholar] [CrossRef]
- Thatcher, S.R.; Zhou, W.; Leonard, A.; Wang, B.B.; Beatty, M.; Zastrow-Hayes, G.; Zhao, X.; Baumgarten, A.; Li, B. Genome-wide analysis of alternative splicing in zea mays: Landscape and genetic regulation. Plant Cell 2014, 26, 3472–3487. [Google Scholar] [CrossRef]
- Lee, B.M.C. A genomic view of alternative splicing. Nat. Genet. 2002, 30, 13–19. [Google Scholar] [CrossRef]
- Reddy, A.S.N.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013, 25, 3657–3683. [Google Scholar] [CrossRef]
- Shang, X.; Cao, Y.; Ma, L. Alternative splicing in plant genes: A means of regulating the environmental fitness of plants. Int. J. Mol. Sci. 2017, 18, 432. [Google Scholar] [CrossRef]
- Ghelli, R.; Brunetti, P.; Marzi, D.; Cecchetti, V.; Costantini, M.; Lanzoni-Rossi, M.; Scaglia Linhares, F.; Costantino, P.; Cardarelli, M. The full-length Auxin Response Factor 8 isoform ARF8.1 controls pollen cell wall formation and directly regulates TDF1, AMS and MS188 expression. Plant J. 2023, 113, 851–865. [Google Scholar] [CrossRef]
- Ghelli, R.; Brunetti, P.; Napoli, N.; De Paolis, A.; Cecchetti, V.; Tsuge, T.; Serino, G.; Matsui, M.; Mele, G.; Rinaldi, G.; et al. A newly identified flower-specific splice variant of AUXIN RESPONSE FACTOR8 regulates stamen elongation and endothecium lignification in Arabidopsis. Plant Cell 2018, 30, 620–637. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, J. Fresh as an exitron: A flower-specific splice variant of AUXIN RESPONSE FACTOR8 helps shape the stamen. Plant Cell 2018, 30, 518–519. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Huang, X.; Wang, T.; Zhang, Y.; Liu, Q.; Hussey, P.J.; Ren, H. ACTIN BINDING PROTEIN29 from Lilium pollen plays an important role in dynamic actin remodeling. Plant Cell 2007, 19, 1930–1946. [Google Scholar] [CrossRef]
- Pei, Y.X.; Zengjie, Q.; Xuejun, C.; Zhujun, C.; Jiashu, C.; Xin, Y.; Lihua, M.; Xiaohui, L. T1243, an alternative transcript of the mitochondrial T gene in Brassica juncea var. tumida, causes pollen abortion in Arabidopsis thaliana. Plant Sci. 2008, 175, 793–798. [Google Scholar] [CrossRef]
- Ju, L.; Jing, Y.; Shi, P.; Liu, J.; Chen, J.; Yan, J.; Chu, J.; Chen, K.M.; Sun, J. JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol. 2019, 223, 246–260. [Google Scholar] [CrossRef]
- Chung, H.S.; Howe, G.A. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 2009, 21, 131–145. [Google Scholar] [CrossRef]
- Gao, X.Q.; Liu, C.Z.; Li, D.D.; Zhao, T.T.; Li, F.; Jia, X.N.; Zhao, X.Y.; Zhang, X.S. The Arabidopsis KINβγ subunit of the SNRK1 complex regulates pollen hydration on the stigma by mediating the level of reactive oxygen species in pollen. PLoS Genet. 2016, 12, e1006228. [Google Scholar] [CrossRef]
- Zhang, M.J.; Zhang, X.S.; Gao, X.Q. ROS in the male-female interactions during pollination: Function and regulation. Front. Plant Sci. 2020, 11, 177. [Google Scholar] [CrossRef]
- Xie, H.T.; Wan, Z.Y.; Li, S.; Zhang, Y. Spatiotemporal production of reactive oxygen species by NADPH oxidase is critical for tapetal programmed cell death and pollen development in Arabidopsis. Plant Cell 2014, 26, 2007–2023. [Google Scholar] [CrossRef]
- Shao, C.; Yang, B.; Wu, T.; Huang, J.; Tang, P.; Zhou, Y.; Zhou, J.; Qiu, J.; Jiang, L.; Li, H.; et al. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat. Struct. Mol. Biol. 2014, 21, 997–1005. [Google Scholar] [CrossRef]
- Xiong, F.; Ren, J.J.; Yu, Q.; Wang, Y.Y.; Lu, C.C.; Kong, L.J.; Otegui, M.S.; Wang, X.L. AtU2AF65b functions in abscisic acid mediated flowering via regulating the precursor messenger RNA splicing of ABI5 and FLC in Arabidopsis. New Phytol. 2019, 223, 277–292. [Google Scholar] [CrossRef] [PubMed]
- Park, H.Y.; Lee, H.T.; Lee, J.H.; Kim, J.K. Arabidopsis U2AF65 regulates flowering time and the growth of pollen tubes. Front. Plant Sci. 2019, 10, 569. [Google Scholar] [CrossRef] [PubMed]
- Coury, D.A.; Zhang, C.; Ko, A.; Skaggs, M.I.; Christensen, C.A.; Drews, G.N.; Feldmann, K.A.; Yadegari, R. Segregation distortion in Arabidopsis gametophytic factor 1 (gfa1) mutants is caused by a deficiency of an essential RNA splicing factor. Sex. Plant Reprod. 2007, 20, 87–97. [Google Scholar] [CrossRef]
- Tsugeki, R.; Tanaka-Sato, N.; Maruyama, N.; Terada, S.; Kojima, M.; Sakakibara, H.; Okada, K. CLUMSY VEIN, the Arabidopsis DEAH-box Prp16 ortholog, is required for auxin-mediated development. Plant J. 2015, 81, 183–197. [Google Scholar] [CrossRef]
- Liu, M.; Yuan, L.; Liu, N.Y.; Shi, D.Q.; Liu, J.; Yang, W.C. GAMETOPHYTIC FACTOR 1, involved in pre-mRNA splicing, is essential for megagametogenesis and embryogenesis in Arabidopsis. J. Integr. Plant Biol. 2009, 51, 261–271. [Google Scholar] [CrossRef]
- Zhu, D.Z.; Zhao, X.F.; Liu, C.Z.; Ma, F.F.; Wang, F.; Gao, X.Q.; Zhang, X.S. Interaction between RNA helicase ROOT INITIATION DEFECTIVE 1 and GAMETOPHYTIC FACTOR 1 is involved in female gametophyte development in Arabidopsis. J. Exp. Bot. 2016, 67, 5757–5768. [Google Scholar] [CrossRef]
- Huang, X.Y.; Niu, J.; Sun, M.X.; Zhu, J.; Gao, J.F.; Yang, J.; Zhou, Q.; Yang, Z.N. CYCLIN-DEPENDENT KINASE G1 is associated with the spliceosome to regulate CALLOSE SYNTHASE5 splicing and pollen wall formation in Arabidopsis. Plant Cell 2013, 25, 637–648. [Google Scholar] [CrossRef]
- Deng, Y.; Srivastava, R.; Quilichini, T.D.; Dong, H.; Bao, Y.; Horner, H.T.; Howell, S.H. IRE1, a component of the unfolded protein response signaling pathway, protects pollen development in Arabidopsis from heat stress. Plant J. 2016, 88, 193–204. [Google Scholar] [CrossRef]
- Cao, L.J.; Zhao, M.M.; Liu, C.; Dong, H.J.; Li, W.C.; Ren, H.Y. LlSR28 is involved in pollen germination by affecting filamentous actin dynamics. Mol. Plant 2013, 6, 1163–1175. [Google Scholar] [CrossRef]
- Kalyna, M.; Lopato, S.; Barta, A. Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development. Mol. Biol. Cell 2003, 14, 3565–3577. [Google Scholar] [CrossRef]
- Aki, S.; Nakai, H.; Aoyama, T.; Oka, A.; Tsuge, T. AtSAP130/AtSF3b-3 function is required for reproduction in Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 1330–1339. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Miki, D.; Tang, K.; Zhou, H.R.; Zheng, Z.; Chen, W.; Ma, Z.Y.; Yang, L.; Zhang, H.; Liu, R.; et al. A pre-mRNA-splicing factor is required for RNA-directed DNA methylation in Arabidopsis. PLoS Genet. 2013, 9, e1003779. [Google Scholar] [CrossRef] [PubMed]
- Kulichová, K.; Kumar, V.; Steinbachová, L.; Klodová, B.; Timofejeva, L.; Juříček, M.; Honys, D.; Hafidh, S.S. PRP8A and PRP8B spliceosome subunits act coordinately to control pollen tube attraction in Arabidopsis thaliana. Development 2020, 147, dev186742. [Google Scholar] [CrossRef]
- Dong, X.; Hong, Z.; Sivaramakrishnan, M.; Mahfouz, M.; Verma, D.P.S. Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 2005, 42, 315–328. [Google Scholar] [CrossRef]
- Zheng, T.; Nibau, C.; Phillips, D.W.; Jenkins, G.; Armstrong, S.J.; Doonan, J.H. CDKG1 protein kinase is essential for synapsis and male meiosis at high ambient temperature in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2014, 111, 2182–2187. [Google Scholar] [CrossRef]
- Nibau, C.; Dadarou, D.; Kargios, N.; Mallioura, A.; Fernandez-Fuentes, N.; Cavallari, N.; Doonan, J.H. A functional kinase is necessary for Cyclin-Dependent Kinase G1 (CDKG1) to maintain fertility at high ambient temperature in Arabidopsis. Front. Plant Sci. 2020, 11, 586870. [Google Scholar] [CrossRef]
- Ali, G.S.; Palusa, S.G.; Golovkin, M.; Prasad, J.; Manley, J.L.; Reddy, A.S.N. Regulation of plant developmental processes by a novel splicing factor. PLoS ONE 2007, 2, e471. [Google Scholar] [CrossRef]
- Ohtani, M.; Demura, T.; Sugiyama, M. Arabidopsis ROOT INITIATION DEFECTIVE1, a DEAH-box RNA helicase involved in pre-mRNA splicing, is essential for plant development. Plant Cell 2013, 25, 2056–2069. [Google Scholar] [CrossRef]
- Moll, C.; Von Lyncker, L.; Zimmermann, S.; Kägi, C.; Baumann, N.; Twell, D.; Grossniklaus, U.; Groß-Hardt, R. CLO/GFA1 and ATO are novel regulators of gametic cell fate in plants. Plant J. 2008, 56, 913–921. [Google Scholar] [CrossRef]
- Groß-Hardt, R.; Carrington, J.C.; Kägi, C.; Baumann, N.; Moore, J.M.; Baskar, R.; Gagliano, W.B.; Jürgens, G.; Grossniklaus, U. LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis. PLoS Biol. 2007, 5, e47. [Google Scholar] [CrossRef]
- Völz, R.; von Lyncker, L.; Baumann, N.; Dresselhaus, T.; Sprunck, S.; Groß-Hardt, R. LACHESIS-dependent egg-cell signaling regulates the development of female gametophytic cells. Development 2012, 139, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Slane, D.; Lee, C.H.; Kolb, M.; Dent, C.; Miao, Y.; Franz-Wachtel, M.; Lau, S.; Macek, B.; Balasubramanian, S.; Bayer, M.; et al. The integral spliceosomal component CWC15 is required for development in Arabidopsis. Sci. Rep. 2020, 10, 13336. [Google Scholar] [CrossRef] [PubMed]
- Shikata, H.; Shibata, M.; Ushijima, T.; Nakashima, M.; Kong, S.G.; Matsuoka, K.; Lin, C.; Matsushita, T. The RS domain of Arabidopsis splicing factor RRC1 is required for phytochrome B signal transduction. Plant J. 2012, 70, 727–738. [Google Scholar] [CrossRef]
- DaS, B.K.; Xia, L.; Palandjian, L.; Palandjian, O.; Chyung, Y.; Reed, R. Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol. Cell Biol. 1999, 19, 6796–6802. [Google Scholar] [CrossRef]
- Aki, S.S.; Yura, K.; Aoyama, T.; Tsuge, T. SAP130 and CSN1 interact and regulate male gametogenesis in Arabidopsis thaliana. J. Plant Res. 2021, 134, 279–289. [Google Scholar] [CrossRef]
- Maeder, C.; Kutach, A.K.; Guthrie, C. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat. Struct. Mol. Biol. 2008, 16, 42–48. [Google Scholar] [CrossRef]
- Wang, H.; Hill, K.; Perry, S.E. An Arabidopsis RNA lariat debranching enzyme is essential for embryogenesis. J. Biol. Chem. 2004, 279, 1468–1473. [Google Scholar] [CrossRef]
- Xiong, F.; Ren, J.J.; Yu, Q.; Wang, Y.Y.; Kong, L.J.; Otegui, M.S.; Wang, X.L. AtBUD13 affects pre-mRNA splicing and is essential for embryo development in Arabidopsis. Plant J. 2019, 98, 714–726. [Google Scholar] [CrossRef]
- Xiong, F.; Ren, J.J.; Wang, Y.Y.; Zhou, Z.; Qi, H.D.; Otegui, M.S.; Wang, X.L. An Arabidopsis Retention and Splicing complex regulates root and embryo development through pre-mRNA splicing. Plant Physiol. 2022, 190, 621–639. [Google Scholar] [CrossRef]
- Fouquet, R.; Martin, F.; Fajardo, D.S.; Gault, C.M.; Gómez, E.; Tseung, C.W.; Policht, T.; Hueros, G.; Settles, A.M. Maize Rough Endosperm 3 encodes an RNA splicing factor required for endosperm cell differentiation and has a nonautonomous effect on embryo development. Plant Cell 2011, 23, 4280–4297. [Google Scholar] [CrossRef]
- Sasaki, T.; Kanno, T.; Liang, S.C.; Chen, P.Y.; Liao, W.W.; Lin, W.D.; Matzke, A.J.M.; Matzke, M. An Rtf2 domain-containing protein influences Pre-mRNA splicing and is essential for embryonic development in Arabidopsis thaliana. Genetics 2015, 200, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Daras, G.; Rigas, S.; Alatzas, A.; Samiotaki, M.; Chatzopoulos, D.; Tsitsekian, D.; Papadaki, V.; Templalexis, D.; Banilas, G.; Athanasiadou, A.M.; et al. LEFKOTHEA regulates nuclear and chloroplast mRNA splicing in plants. Dev. Cell 2019, 50, 767–779.e767. [Google Scholar] [CrossRef] [PubMed]
- Shao, D.J.; Wei, Y.M.; Yu, Z.Q.; Dai, X.; Gao, X.Q. Arabidopsis AtPRP17 functions in embryo development by regulating embryonic patterning. Planta 2021, 254, 58. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.; Liu, H.H.; Duan, C.Y.; Zhang, B.K.; Wei, G.; Zhang, Y.; Li, S. Arabidopsis JANUS regulates embryonic pattern formation through Pol II-mediated transcription of WOX2 and PIN7. iScience 2019, 19, 1179–1188. [Google Scholar] [CrossRef]
- Xiong, F.; Li, S. Spliceosome component JANUS fulfills a role of mediator in transcriptional regulation during Arabidopsis development. Plant Signal Behav. 2021, 16, 1841974. [Google Scholar] [CrossRef]
- Perea-Resa, C.; Hernández-Verdeja, T.; López-Cobollo, R.; Castellano, M.d.M.; Salinas, J. LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell 2012, 24, 4930–4947. [Google Scholar] [CrossRef]
- Swaraz, A.M.; Park, Y.-D.; Hur, Y. Knock-out mutations of Arabidopsis SmD3-b induce pleotropic phenotypes through altered transcript splicing. Plant Sci. 2011, 180, 661–671. [Google Scholar] [CrossRef]
- Schwartz, B.W.; Yeung, E.C.; Meinke, D.W. Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development 1994, 120, 3235–3245. [Google Scholar] [CrossRef]
- Ren, R.C.; Wang, L.L.; Zhang, L.; Zhao, Y.J.; Wu, J.W.; Wei, Y.M.; Zhang, X.S.; Zhao, X.Y. DEK43 is a p-type pentatricopeptide repeat (PPR) protein responsible for the cis-splicing of nad4 in maize mitochondria. J. Integr. Plant Biol. 2020, 62, 299–313. [Google Scholar] [CrossRef]
- Zhu, C.; Jin, G.; Fang, P.; Zhang, Y.; Feng, X.; Tang, Y.; Qi, W.; Song, R. Maize pentatricopeptide repeat protein DEK41 affects cis-splicing of mitochondrial nad4 intron 3 and is required for normal seed development. J. Exp. Bot. 2019, 70, 3795–3808. [Google Scholar] [CrossRef]
- Sun, F.; Zhang, X.; Shen, Y.; Wang, H.; Liu, R.; Wang, X.; Gao, D.; Yang, Y.Z.; Liu, Y.; Tan, B.C. The pentatricopeptide repeat protein EMPTY PERICARP8 is required for the splicing of three mitochondrial introns and seed development in maize. Plant J. 2018, 95, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Feng, F.; Qi, W.; Xu, L.; Yao, D.; Wang, Q.; Song, R. Dek35 encodes a PPR protein that affects cis-splicing of mitochondrial nad4 intron 1 and seed development in maize. Mol. Plant 2017, 10, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Wang, F.; Li, N.; Shi, D.Q.; Yang, W.C. Pentatricopeptide repeat protein MID1 modulates nad2 intron 1 splicing and Arabidopsis development. Sci. Rep. 2020, 10, 2008. [Google Scholar] [CrossRef]
- de Longevialle, A.o.F.; Meyer, E.H.; Andrés, C.; Taylor, N.L.; Lurin, C.; Millar, A.H.; Small, I.D. The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 2007, 19, 3256–3265. [Google Scholar] [CrossRef]
- Hsieh, W.Y.; Liao, J.C.; Chang, C.Y.; Harrison, T.; Boucher, C.; Hsieh, M.H. The SLOW GROWTH3 pentatricopeptide repeat protein is required for the splicing of mitochondrial NADH dehydrogenase subunit7 intron 2 in Arabidopsis. Plant Physiol. 2015, 168, 490–501. [Google Scholar] [CrossRef]
- Wu, M.W.; Zhao, H.; Zhang, J.D.; Guo, L.; Liu, C.M. RADICLELESS 1 (RL1)-mediated nad4 intron 1 splicing is crucial for embryo and endosperm development in rice (Oryza sativa L.). Biochem. Biophys. Res. Commun. 2020, 523, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Liu, L.; Yu, Y.; Zhu, J.; Gao, H.; Wang, Y.; Wan, J. Lose-of-function of a rice nucleolus-localized pentatricopeptide repeat protein is responsible for the floury endosperm14 mutant phenotypes. Rice 2019, 12, 100. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide pepeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, C. The enigmatic roles of PPR-SMR proteins in plants. Adv. Sci. 2019, 6, 1900361. [Google Scholar] [CrossRef]
- Wang, Y.; Tan, B.C. Pentatricopeptide repeat proteins in plants: Cellular functions, action mechanisms, and potential applications. Plant Commun. 2025, 6, 101203. [Google Scholar] [CrossRef]
- Zheng, P.; Liu, Y.; Liu, X.; Huang, Y.; Sun, F.; Wang, W.; Chen, H.; Jan, M.; Zhang, C.; Yuan, Y.; et al. OsPPR939, a nad5 splicing factor, is essential for plant growth and pollen development in rice. Theor. Appl. Genet. 2021, 134, 923–940. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Du, M.; Zhu, T.; Li, G.; Ding, Y.; Zhang, Q. PPR proteins in plants: Roles, mechanisms, and prospects for rice research. Front. Plant Sci. 2024, 15. [Google Scholar] [CrossRef]
- Lorković, Z.J.; Wieczorek Kirk, D.A.; Lambermon, M.H.; Filipowicz, W. Pre-mRNA splicing in higher plants. Trends Plant Sci. 2000, 5, 160–167. [Google Scholar] [CrossRef]
- Maurel, M.; Chevet, E.; Tavernier, J.; Gerlo, S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 2014, 39, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, Y.; Mishiba, K.I.; Suzuki, E.; Shimada, Y.; Iwata, Y.; Koizumi, N. Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci. Rep. 2011, 1, 29. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, M.; Demura, T.; Sugiyama, M. Differential requirement for the function of SRD2, an snRNA transcription activator, in various stages of plant development. Plant Mol. Biol. 2007, 66, 303–314. [Google Scholar] [CrossRef]
- Ohtani, M.; Sugiyama, M. Involvement of SRD2-mediated activation of snRNA transcription in the control of cell proliferation competence in Arabidopsis. Plant J. 2005, 43, 479–490. [Google Scholar] [CrossRef]
- Ohtani, M.; Demura, T.; Sugiyama, M. Particular significance of SRD2-dependent snRNA accumulation in polarized pattern generation during lateral root development of Arabidopsis. Plant Cell Physiol. 2010, 51, 2002–2012. [Google Scholar] [CrossRef]
- Liu, H.; Ma, X.; Han, H.N.; Hao, Y.J.; Zhang, X.S. AtPRMT5 Regulates Shoot Regeneration through Mediating Histone H4R3 Dimethylation on KRPs and Pre-mRNA splicing of RKP in Arabidopsis. Mol. Plant 2016, 9, 1634–1646. [Google Scholar] [CrossRef]
- El Arbi, N.; Nardeli, S.M.; Šimura, J.; Ljung, K.; Schmid, M. The Arabidopsis splicing factor PORCUPINE/SmE1 orchestrates temperature-dependent root development via auxin homeostasis maintenance. New Phytol. 2024, 244, 1408–1421. [Google Scholar] [CrossRef]
- Chong, G.L.; Foo, M.H.; Lin, W.D.; Wong, M.M.; Verslues, P.E. Highly ABA-Induced 1 (HAI1)-Interacting protein HIN1 and drought acclimation-enhanced splicing efficiency at intron retention sites. Proc. Natl. Acad. Sci. USA 2019, 116, 22376–22385. [Google Scholar] [CrossRef]
- Butt, H.; Bazin, J.; Prasad, K.V.S.K.; Awad, N.; Crespi, M.; Reddy, A.S.N.; Mahfouz, M.M. The rice serine/arginine splicing factor RS33 regulates Pre-mRNA splicing during abiotic stress responses. Cells 2022, 11, 1796. [Google Scholar] [CrossRef]
- Zhao, J.; Yao, B.; Peng, Z.; Yang, X.; Li, K.; Zhang, X. Splicing defect of StDRO2 intron 1 promotes potato root growth by disturbing auxin transport to adapt to drought stress. Hortic. Plant J. 2025, 11, 706–720. [Google Scholar] [CrossRef]
- Fu, D.; Song, Y.; Wu, S.; Peng, Y.; Ming, Y.; Li, Z.; Zhang, X.; Song, W.; Su, Z.; Gong, Z.; et al. Regulation of alternative splicing by CBF-mediated protein condensation in plant response to cold stress. Nat. Plants 2025, 11, 505–517. [Google Scholar] [CrossRef]
- Lev Maor, G.; Yearim, A.; Ast, G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015, 31, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Huppertz, I.; Attig, J.; D’Ambrogio, A.; Easton, L.E.; Sibley, C.R.; Sugimoto, Y.; Tajnik, M.; König, J.; Ule, J. iCLIP: Protein–RNA interactions at nucleotide resolution. Methods 2014, 65, 274–287. [Google Scholar] [CrossRef]
- Ohtani, M. Expanding the plant non-coding RNA world. J. Plant Res. 2016, 130, 3–5. [Google Scholar] [CrossRef]
- Han, X.; Lin, Y.; Gao, X.Q.; Wang, X.L. Non-canonical functions of splicing factors in RNA metabolism. Crit. Rev. Plant Sci. 2020, 39, 493–513. [Google Scholar] [CrossRef]
- Waititu, J.K.; Zhang, C.; Liu, J.; Wang, H. Plant non-coding RNAs: Origin, biogenesis, mode of action and their roles in abiotic stress. Int. J. Mol. Sci. 2020, 21, 8401. [Google Scholar] [CrossRef]
- Li, S.; Liu, K.; Zhou, B.; Li, M.; Zhang, S.; Zeng, L.; Zhang, C.; Yu, B. MAC3A and MAC3B, two core subunits of the MOS4-associated complex, positively influence miRNA biogenesis. Plant Cell 2018, 30, 481–494. [Google Scholar] [CrossRef]
- Zhang, S.; Dou, Y.; Li, S.; Ren, G.; Chevalier, D.; Zhang, C.; Yu, B. DAWDLE Interacts with DICER-LIKE proteins to mediate small RNA biogenesis. Plant Physiol. 2018, 177, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Liu, P.; Wu, C.A.; Yang, G.D.; Xu, R.; Guo, Q.H.; Huang, J.G.; Zheng, C.C. Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell 2012, 48, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef]
- Gai, Y.P.; Yuan, S.S.; Zhao, Y.N.; Zhao, H.N.; Zhang, H.L.; Ji, X.L. A novel lncRNA, MuLnc1, associated with environmental stress in mulberry (Morus multicaulis). Front. Plant Sci. 2018, 9, 669. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Zhang, M.; Xu, X.; Li, X.; Li, C.; Ding, Z. System analysis of microRNAs in the development and aluminium stress responses of the maize root system. Plant Biotechnol. J. 2014, 12, 1108–1121. [Google Scholar] [CrossRef]
- Liu, F.; Xu, Y.; Chang, K.; Li, S.; Liu, Z.; Qi, S.; Jia, J.; Zhang, M.; Crawford, N.M.; Wang, Y. The long noncoding RNA T5120 regulates nitrate response and assimilation in Arabidopsis. New Phytol. 2019, 224, 117–131. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, C.; Baulcombe, D.C.; Chen, Z.J. Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 2012, 109, 5529–5534. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, X.; Sun, F.; Hu, J.; Zha, X.; Su, W.; Yang, J. Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice. Nature Commun. 2018, 9, 3516. [Google Scholar] [CrossRef]
- Qin, T.; Zhao, H.; Cui, P.; Albesher, N.; Xiong, L. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 2017, 175, 1321–1336. [Google Scholar] [CrossRef]
- Vashisht, I.; Dhaka, N.; Jain, R.; Sood, A.; Sharma, N.; Sharma, M.K.; Sharma, R. Non-coding RNAs-mediated environmental surveillance determines male fertility in plants. Plant Physiol. Biochem. 2023, 203, 108030. [Google Scholar] [CrossRef]
- Sharif, R.; Zhu, Y.; Huang, Y.; Sohail, H.; Li, S.; Chen, X.; Qi, X. MicroRNA regulates cytokinin induced parthenocarpy in cucumber (Cucumis sativus L.). Plant Physiol. Biochem. 2024, 212, 108681. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Yang, L.; Zhuang, M.; Lv, H.; Wang, Y.; Zhang, Y.; Ji, J. Plant non-coding RNAs: The new frontier for the regulation of plant development and adaptation to stress. Plant Physiol. Biochem. 2024, 208, 108435. [Google Scholar] [CrossRef] [PubMed]

| Species | Biological Process | Type of Splicing Factor | Splicing Factor | Targeted Genes | References |
|---|---|---|---|---|---|
| A. thaliana | Female gametophyte development | DEAH-box RNA-dependent ATPase Prp22 | RID1 | EDA26, EDA4, FGPS3, GASA4 | [36,48] |
| A. thaliana | Megagametogenesis | U5 snRNP component | GFA1 | EDA26, EDA4, FGPS3, GASA4 | [33,35,36,49] |
| A. thaliana | Female gametophyte development | U2 snRNP-associated protein | ATO | Unknown | [49] |
| A. thaliana | Supernumerary egg cells, female gametophytes development | U4/U6 snRNP-associated protein | LIS | Unknown | [50,51] |
| A. thaliana | Female gametophyte development | A U2 snRNP-associated protein | AtSAP130 | Unknown | [41] |
| A. thaliana | Female gametophyte transmission | A U4/U6 snRNP associated protein | RDM16 | Unknown | [42] |
| A. thaliana | A functional redundancy between PRP8A and PRP8B in female gametophyte development | U5 snRNP component | PRP8A; PRP8B | EDA9, EDA30, EDA35, MEE29 | [43] |
| A. thaliana | Female gametophyte transmission | Potential component of the NTC | CWC15 | Unknown | [52] |
| A. thaliana | Female gametophyte transmission | RS domain (SR-like) protein | RRC1 | Unknown | [53] |
| Species | Biological Process | Type of Splicing Factor | Splicing Factor | Targeted Genes | References |
|---|---|---|---|---|---|
| Zea mays | Kernel development | RNA lariat debranching enzyme | AtDBR1 | Unknown | [57] |
| A. thaliana | Embryogenesis | Nucleus-encoded RNA-binding protein | LEFKO | Unknown | [62] |
| A. thaliana | Early embryogenesis | One composition of the RES complex | AtBUD13 | Early embryo developmental genes, e.g., ZYG1/APC11, PFI and ATML1 | [58] |
| A. thaliana | Early embryogenesis | One composition of the RES complex | DDL | Genes associated with cell proliferation and early embryo development e.g., AGO10/ZLL, ATML1 | [59] |
| A. thaliana | Early embryogenesis | One composition of the RES complex | GDS1 | Genes associated with cell proliferation and early embryo development e.g., AGO10/ZLL, ATML1 | [59] |
| A. thaliana | Embryogenesis | U5 snRNP component | PRP8A | Unknown | [61,68] |
| Z. mays | Kernel development | U2AF35-related protein | Rgh3 | Unknown | [60] |
| Z. mays | Kernel development | Mitochondrion-targeted P-type PRP protein | DEK43/DEK41 | Mitochondrial nad4 introns 1 and 3 | [69,70] |
| Z. mays | Kernel development | Mitochondria-targeted P-type PPR protein | EMP8 | nad1 intron 4, nad4 intron 1 and nad2 intron 1 | [71] |
| Z. mays | Kernel development | Mitochondria-targeted P-type PPR protein | DEK35 | Mitochondrial nad4 intron 1 | [72] |
| A. thaliana | Embryogenesis | Mitochondria-targeted P-type PPR protein | MID1 | Mitochondrial nad2 intron 1 | [73] |
| A. thaliana | Seed development | Mitochondria-targeted P-type PPR protein | OTP43 | Mitochondrial nad1 Intron 1 | [74] |
| A. thaliana | Seed development | Mitochondria-targeted P-type PPR protein | SLO3 | Mitochondrial nad7 Intron 2 | [75] |
| A. thaliana | Embryogenesis | Rtf2-domain splicing-related protein | AtRTF2 | Unknown | [61] |
| Oryza sativa | Grain development | Mitochondrion-targeted P-type PRP protein | RL1 | Mitochondrial nad4 intron 1 | [76] |
| O. sativa | Grain development | Nucleolus-localized PPR protein | FLO14/OsNPPR3 | nad 1–2 and nad 2 | [77] |
| A. thaliana | Embryogenesis | DEAH-box RNA-dependent ATPase Prp16 | CUV | Genes involved in auxin biosynthesis, polar auxin transport, and auxin perception and signaling | [34] |
| A. thaliana | Embryogenesis | U5 snRNP component | GFA1 | Unknown | [35] |
| A. thaliana | Embryogenesis | SR protein | AtRSZ33 | Unknown | [40] |
| A. thaliana | Embryogenesis | A homolog of the yeast splicing factor PRP17 | AtPRP17 | Unknown | [63] |
| A. thaliana | Embryonic pattern formation | U2 snRNP assembly factor | JANUS | Unknown | [64,65] |
| A. thaliana | Embryogenesis | U6 SnRNP component (SM-like protein) | LSM | Genes involved in embryo development e.g., EMB2785, EMB2016 | [66] |
| A. thaliana | Embryogenesis | RS domain (SR-like) protein | RRC1 | Unknown | [53] |
| A. thaliana | Embryogenesis | SnRNP core protein | SmD3 | Unknown | [67] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shao, D.; Gao, X.; Wei, Y. Pre-mRNA Splicing Functions in Plant Sexual Reproduction Development. Plants 2025, 14, 1472. https://doi.org/10.3390/plants14101472
Shao D, Gao X, Wei Y. Pre-mRNA Splicing Functions in Plant Sexual Reproduction Development. Plants. 2025; 14(10):1472. https://doi.org/10.3390/plants14101472
Chicago/Turabian StyleShao, Dongjie, Xinqi Gao, and Yiming Wei. 2025. "Pre-mRNA Splicing Functions in Plant Sexual Reproduction Development" Plants 14, no. 10: 1472. https://doi.org/10.3390/plants14101472
APA StyleShao, D., Gao, X., & Wei, Y. (2025). Pre-mRNA Splicing Functions in Plant Sexual Reproduction Development. Plants, 14(10), 1472. https://doi.org/10.3390/plants14101472






