Pantoea stewartii subsp. stewartii an Inter-Laboratory Comparative Study of Molecular Tests and Comparative Genome Analysis of Italian Strains
Abstract
1. Introduction
- (i)
- Comparison of different molecular tests to detect Pss: real-time PCR Scala et al., (2023) [7], real-time PCR Tambong et al., (2008) [9] and real-time PCR Pal et al., (2019) [10] (hereafter named Scala, Tambong and Pal, respectively) through interlaboratory comparisons (ILC). ILC combines the results obtained from a Proficiency Test (PT) and a Test Performance Study (TPS) conducted on the same set of samples;
- (ii)
- Complete sequencing of five bacterial isolates collected in Italy between 2015 and 2022, utilizing the Oxford Nanopore MinION technology alongside the Illumina methodology. The study concerned several key objectives: (a) sequencing and assembling Pss strains; (b) conducting genomic characterization studies to compare the isolates with existing data; and (c) performing a comparative analysis of the sequenced genomes.
2. Results
2.1. Proficiency Test (PT), Test Performance Study (TPS) and ILC (Interlaboratory Comparison)
2.1.1. Sample Homogeneity and Stability Test
2.1.2. Proficiency Test (PT)
2.1.3. Test Performance Study (TPS)
2.1.4. Interlaboratory Comparison (ICL)
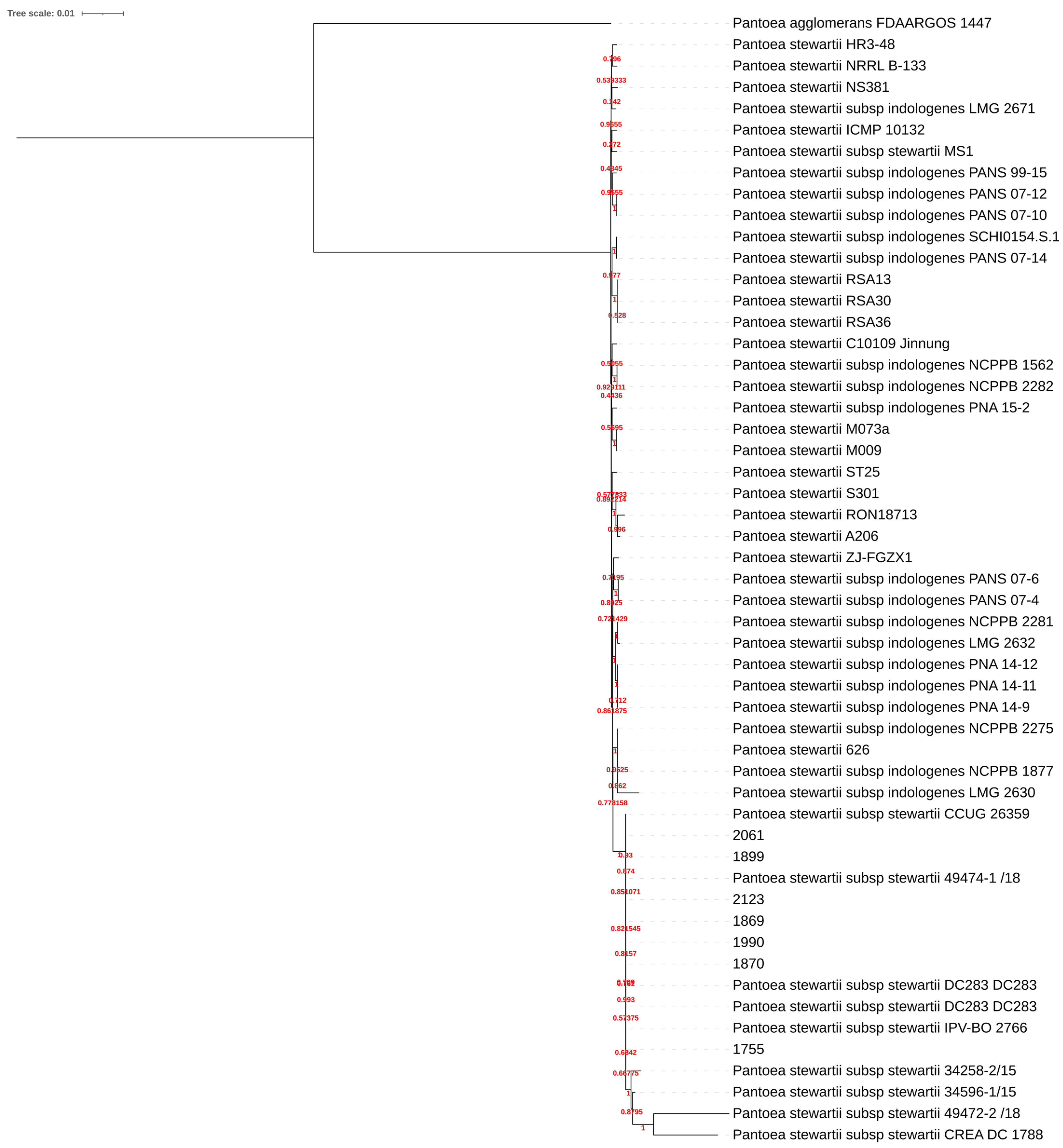
2.2. De Novo Genome Assembly
3. Discussion
4. Materials and Methods
4.1. Proficiency Test (PT), Test Performance Study (TPS) and ICL (Interlaboratory Comparison)
4.2. Plant Material, Bacterial Strains and Sample Preparation
4.3. DNA Extraction
4.4. Homogeneity and Stability
4.5. Performance Criteria Evaluation and Outliers
4.6. Genome Analysis
4.7. Genome Annotation, Pangenome and Phylogenomics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-Based Phylogeny and Taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales Ord. Nov. Divided into the Families Enterobacteriaceae, Erwiniaceae Fam. Nov., Pectobacteriaceae Fam. Nov., Yersiniaceae Fam. Nov., Hafniaceae Fam. Nov., Morganellaceae Fam. Nov., and Budviciaceae Fam. Nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef] [PubMed]
- Genomics-Informed Multiplex PCR Scheme for Rapid Identification of Rice-Associated Bacteria of the Genus Pantoea. Available online: https://apsjournals.apsnet.org/doi/epdf/10.1094/PDIS-07-20-1474-RE (accessed on 10 March 2025).
- PM 7/60 (2) Pantoea stewartii subsp. stewartii. EPPO Bull. 2016, 46, 226–236. [CrossRef]
- Coplin, D.L.; Majerczak, D.R.; Zhang, Y.; Kim, W.-S.; Jock, S.; Geider, K. Identification of Pantoea stewartii subsp. stewartii by PCR and Strain Differentiation by PFGE. Plant Dis. 2002, 86, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Roper, M.C. Pantoea stewartii subsp. stewartii: Lessons Learned from a Xylem-Dwelling Pathogen of Sweet Corn. Mol. Plant Pathol. 2011, 12, 628–637. [Google Scholar] [CrossRef]
- Pantoea stewartii subsp. stewartii (ERWIST) [World Distribution]|EPPO Global Database. Available online: https://gd.eppo.int/taxon/ERWIST/distribution (accessed on 24 February 2025).
- Scala, V.; Faino, L.; Costantini, F.; Crosara, V.; Albanese, A.; Pucci, N.; Reverberi, M.; Loreti, S. Analysis of Italian Isolates of Pantoea stewartii subsp. stewartii and Development of a Real-Time PCR-Based Diagnostic Method. Front. Microbiol. 2023, 14, 1129229. [Google Scholar] [CrossRef]
- Pest Categorisation of Pantoea stewartii subsp. stewartii-2018-EFSA Journal-Wiley Online Library. Available online: https://efsa.onlinelibrary.wiley.com/doi/full/10.2903/j.efsa.2018.5356 (accessed on 10 March 2025).
- Tambong, J.T.; Mwange, K.N.; Bergeron, M.; Ding, T.; Mandy, F.; Reid, L.M.; Zhu, X. Rapid Detection and Identification of the Bacterium Pantoea stewartii in Maize by TaqMan® Real-time PCR Assay Targeting the cpsD Gene. J. Appl. Microbiol. 2008, 104, 1525–1537. [Google Scholar] [CrossRef]
- Pal, N.; Block, C.C.; Gardner, C.A.C. A Real-Time PCR Differentiating Pantoea stewartii subsp. stewartii from P. stewartii subsp. indologenes in Corn Seed. Plant Dis. 2019, 103, 1474–1486. [Google Scholar] [CrossRef]
- Dreo, T. Report on the Results of the Test Performance Study for Molecular Detection of Pantoea stewartii subsp. stewartii in Asymptomatic Plant Material (Maize Seeds); v1.0, Valitest, GA n°773139; National Institute of Biology: Ljubljana, Slovenia, 2020. [Google Scholar]
- Thapa, S.P.; Park, D.H.; Wilson, C.; Hur, J.H.; Lim, C.K. Multiplex PCR Assay for the Detection of Pantoea stewartii subsp. stewartii Using Species-Specific Genetic Markers. Australas. Plant Pathol. 2012, 41, 559–564. [Google Scholar] [CrossRef]
- McDonald, B.A.; Stukenbrock, E.H. Rapid Emergence of Pathogens in Agro-Ecosystems: Global Threats to Agricultural Sustainability and Food Security. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 20160026. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The Global Burden of Pathogens and Pests on Major Food Crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef]
- EFSA Panel on Plant Health (PLH); Bragard, C.; Baptista, P.; Chatzivassiliou, E.; Di Serio, F.; Gonthier, P.; Jaques Miret, J.A.; Justesen, A.F.; Magnusson, C.S.; Milonas, P.; et al. Pest Categorisation of Euzophera Semifuneralis. EFSA J. 2023, 21, e08120. [Google Scholar] [CrossRef] [PubMed]
- Gehring, I.; Wensing, A.; Gernold, M.; Wiedemann, W.; Coplin, D.L.; Geider, K. Molecular Differentiation of Pantoea stewartii subsp. indologenes from Subspecies Stewartii and Identification of New Isolates from Maize Seeds. J. Appl. Microbiol. 2014, 116, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Goss, E.M. Genome-Enabled Analysis of Plant-Pathogen Migration. Annu. Rev. Phytopathol. 2015, 53, 121–135. [Google Scholar] [CrossRef]
- Xu, J.; Wang, N. Where Are We Going with Genomics in Plant Pathogenic Bacteria? Genomics 2019, 111, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Straub, C.; Colombi, E.; McCann, H.C. Population Genomics of Bacterial Plant Pathogens. Phytopathology® 2021, 111, 23–31. [Google Scholar] [CrossRef] [PubMed]
- De Maayer, P.; Aliyu, H.; Vikram, S.; Blom, J.; Duffy, B.; Cowan, D.A.; Smits, T.H.M.; Venter, S.N.; Coutinho, T.A. Phylogenomic, Pan-Genomic, Pathogenomic and Evolutionary Genomic Insights into the Agronomically Relevant Enterobacteria Pantoea ananatis and Pantoea stewartii. Front. Microbiol. 2017, 8, 01755. [Google Scholar] [CrossRef]
- Walterson, A.M.; Stavrinides, J. Pantoea: Insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 2015, 39, 968–984. [Google Scholar] [CrossRef]
- Agarwal, G.; Gitaitis, R.D.; Dutta, B. Pan-Genome of Novel Pantoea stewartii subsp. indologenes Reveals Genes Involved in Onion Pathogenicity and Evidence of Lateral Gene Transfer. Microorganisms 2021, 9, 1761. [Google Scholar] [CrossRef] [PubMed]
- Documento Tecnico Ufficiale del Servizio Fitosanitario Nazionale n. 27. Protezione Delle Piante. 2022. Available online: https://www.protezionedellepiante.it/attivita-divulgativa-3-aprile-2024-corso-better-training-for-safer-food-btsf-plant-disease-outbreaks-contingency-planning-for-priority-pests/ (accessed on 14 March 2025).
- PM 7/122 (2) Guidelines for the Organization of Interlaboratory Comparisons by Plant Pest Diagnostic Laboratories. EPPO Bull. 2022, 52, 604–618. [CrossRef]
- Tian, Q.; Zhou, H.; Zhao, Z.; Zhang, Y.; Zhao, W.; Cai, L.; Guo, T. Single and dual RPA-CRISPR/Cas assays for point-of-need detection of Stewart’s wilt pathogen (Pantoea stewartii subsp. stewartii) of corn and Maize dwarf mosaic virus. Pest Manag. Sci. 2025, 81, 1988–1999. [Google Scholar] [CrossRef]
- Kristensen, T.; Sørensen, L.H.; Pedersen, S.K.; Jensen, J.D.; Mordhorst, H.; Lacy-Roberts, N.; Lukjancenko, O.; Luo, Y.; Hoffmann, M.; Hendriksen, R.S. Results of the 2020 Genomic Proficiency Test for the network of European Union Reference Laboratory for Antimicrobial Resistance assessing whole-genome-sequencing capacities. Microb. Genom. 2023, 9, mgen001076. [Google Scholar] [CrossRef] [PubMed]
- Petter, F.; Trontin, C.; Anthoine, G.; Ravnikar, M.; Dreo, T.; Lukežič, T.; Vučurović, A.; Mehle, N. Introduction to Interlaboratory Comparisons. In Critical Points for the Organisation of Test Performance Studies in Microbiology; Plant Pathology in the 21st Century; Vučurović, A., Mehle, N., Anthoine, G., Dreo, T., Ravnikar, M., Eds.; Springer: Cham, Switzerland, 2022; Volume 12. [Google Scholar] [CrossRef]
- Estoup, A.; Guillemaud, T. Reconstructing Routes of Invasion Using Genetic Data: Why, How and so What? Mol. Ecol. 2010, 19, 4113–4130. [Google Scholar] [CrossRef] [PubMed]
- Mack, R.N.; Simberloff, D.; Mark Lonsdale, W.; Evans, H.; Clout, M.; Bazzaz, F.A. Biotic Invasions: Causes, Epidemiology, Global Consequences, and Control. Ecol. Appl. 2000, 10, 689–710. [Google Scholar] [CrossRef]



| Real-Time PCR | DSE% | DSP% | ACC% |
|---|---|---|---|
| Scala App | 100 | 100 | 100 |
| Scala Prom | 100 | 100 | 100 |
| Pal | 100 | 100 | 100 |
| Tambong | 100 | 83.3 | 100 |
| Sample | Raw | Trimmed |
|---|---|---|
| Pss_1990 | 4,970,917 | 4,901,306 |
| Pss_1870 | 3,853,357 | 3,812,740 |
| Pss_1869 | 4,877,111 | 4,849,311 |
| Pss_2061 | 5,057,085 | 5,033,398 |
| Pss_2123 | 4,793,392 | 4,764,577 |
| Sample | Contigs | Dimension bp | Completeness | Contamination |
|---|---|---|---|---|
| Pss_1990 | 27 | 5,442,216 | 99.34 | 0.74 |
| Pss_1870 | 21 | 5,403,016 | 99.34 | 0.74 |
| Pss_1869 | 58 | 5,362,941 | 99.34 | 0.74 |
| Pss_2061 | 538 | 5,171,712 | 99.34 | 0.43 |
| Pss_2123 | 487 | 5,171,801 | 99.34 | 0.25 |
| Sample | Genes |
|---|---|
| Pss_1990 | 5669 |
| Pss_1870 | 5607 |
| Pss_1869 | 5553 |
| Pss_2061 | 5255 |
| Pss_2123 | 5295 |
| Pss_1755 | 5884 |
| Pss_1899 | 5751 |
| Sample ID | Sample Type | Phytosanitary Status | Host |
|---|---|---|---|
| S1 | Healthy SE | Negative | Maize seed (Zea mays) |
| S2 | SE spiked with 104 cfu/mL of Pss | Positive | Maize seed (Zea mays) |
| S3 | Bacterial suspension 106 cfu/mL of Psi | Negative | Bacterial strain |
| S4 | SE | Negative | Maize seed (Zea mays) |
| S5 | SE spiked with 105 cfu/mL of Pss | Positive | Maize seed (Zea mays) |
| S6 | SE spiked with 104 cfu/mL of Pss | Positive | Maize seed (Zea mays) |
| S7 | Bacterial suspension 106 cfu/mL of Pan | Negative | Bacterial strain |
| S8 | SE | Negative | Maize seed (Zea mays) |
| S9 | SE spiked with 105 cfu/mL of Pss | Positive | Maize seed (Zea mays) |
| S10 | SE spiked with 105 cfu/mL of Pss | Positive | Maize seed (Zea mays) |
| S11 | SE | Negative | Maize seed (Zea mays) |
| S12 | SE spiked with 106 cfu/mL of Pss | Positive | Maize seed (Zea mays) |
| PAC1 | Bacterial DNA (106 cfu/mL of Pss) | Positive | Bacterial strain |
| NAC1 | Bacterial DNA (106 cfu/mL of Psi) | Negative | Bacterial strain |
| NAC2 | Water DEPC (Diethyl pyrocarbonate) | Negative |
| Official Laboratories (OLs) |
|---|
| Agenzia Agris, Ussana (SU) |
| Agenzia Lucana di Sviluppo e di Innovazione in Agricoltura—Centro ricerche metapontum Agrobios ALSIA-CRMA, Metaponto (MT) |
| Agenzia Settore Agroalimentare delle Marche—SFR Regione Marche, Osimo (AN) |
| ARSAC, San Marco Argentano (CS) |
| Centro di Sperimentazione Laimburg (BZ) |
| CREA-DC Centro difesa e certificazione—Roma |
| CRSFA, Centro di Ricerca, Sperimentazione e Formazione in Agricoltura “Basile Caramia”, Locorotondo (BA) |
| DAFNE, Università degli Studi della Tuscia, Dipartimento di Scienze agrarie e forestali, Viterbo (VT) |
| ERSA, Laboratorio di Fitopatologia e Biotecnologie, Udine (UD) |
| Fondazione Edmund Mach, San Michele all’Adige (TN) |
| Laboratorio Fitopatologico Regione Campania, Napoli (NA) |
| Laboratorio Fitopatologico Regione Emilia-Romagna, Bologna (BO) |
| Laboratorio fitosanitario e settore fitosanitario e servizi tecnico-scientifici, Direzione Agricoltura e cibo, Regione Piemonte, Torino (TO) |
| Laboratorio SFR Regione Lombardia, Vertemate con Minoprio (MI) |
| Laboratorio Fitopatologico Regione Liguria, Genova (GE) |
| Regione Toscana SFR e di vigilanza del controllo agroforestale—Laboratorio Fitopatologico Regionale, Pistoia (PT) |
| Regione Veneto Unità organizzativa fitosanitario, Buttapietra (VR) |
| Species Name | Year | Location | Sample | Assembly |
|---|---|---|---|---|
| Pantoea stewartii subsp. stewartii | 2015 | Italy, Emilia Romagna | Pss_1869 | This study |
| Pantoea stewartii subsp. stewartii | 2015 | Italy, Emilia Romagna | Pss_1870 | This study |
| Pantoea stewartii subsp. stewartii | 2018 | Italy, Emilia Romagna | Pss_1990 | This study |
| Pantoea stewartii subsp. stewartii | 2021 | Italy, Emilia Romagna | Pss_2061 | This study |
| Pantoea stewartii subsp. stewartii | 2022 | Italy, Emilia Romagna | Pss_2123 | This study |
| Code | Assembly Name | Organisms | Strain | Date of Submission |
|---|---|---|---|---|
| GCF_011044475.1 | ASM1104447v1 | Pantoea stewartii | ZJ-FGZX1 | 1 March 2020 |
| GCF_025765915.1 | ASM2576591v1 | Pantoea stewartii | HR3-48 | 20 October 2022 |
| GCF_002082215.1 | ASM208221v1 | Pantoea stewartii subsp. stewartii | DC283 | 10 April 2017 |
| GCF_000757405.2 | LMG2632assem4 | Pantoea stewartii subsp. indologenes | LMG 2632 | 23 September 2014 |
| GCF_008801695.1 | ASM880169v1 | Pantoea stewartii subsp. stewartii | CCUG 26359 | 1 October 2019 |
| GCF_029433915.1 | ASM2943391v1 | Pantoea stewartii | ICMP 10132 | 27 March 2023 |
| GCF_001310285.1 | Pantoea_stewartii_A206_1.0 | Pantoea stewartii | A206 | 13 October 2015 |
| GCF_013277595.1 | ASM1327759v1 | Pantoea stewartii | 626 | 4 June 2020 |
| GCF_025599245.1 | ASM2559924v1 | Pantoea stewartii | ST25 | 5 October 2022 |
| GCF_014218605.1 | ASM1421860v1 | Pantoea stewartii | NRRL B-133 | 17 August 2020 |
| GCF_000803205.1 | ASM80320v1 | Pantoea stewartii | M073a | 17 December 2014 |
| GCF_029991035.1 | ASM2999103v1 | Pantoea stewartii | C10109_Jinnung | 12 May 2023 |
| GCF_001310295.1 | Pantoea_stewartii_S301_1.0 | Pantoea stewartii | S301 | 13 October 2015 |
| GCF_001476375.1 | ASM147637v1 | Pantoea stewartii | RSA36 | 22 December 2015 |
| GCF_001476355.1 | ASM147635v1 | Pantoea stewartii | NS381 | 22 December 2015 |
| GCF_000786255.1 | ASM78625v1 | Pantoea stewartii | M009 | 23 November 2014 |
| GCF_001476795.1 | ASM147679v1 | Pantoea stewartii | RSA30 | 22 December 2015 |
| GCF_001477215.1 | ASM147721v1 | Pantoea stewartii | RSA13 | 22 December 2015 |
| GCF_000248395.1 | ASM24839v2 | Pantoea stewartii subsp. stewartii | DC283 | 22 February 2012 |
| GCF_030370575.1 | ASM3037057v1 | Pantoea stewartii subsp. indologenes | LMG 2671 | 28 June 2023 |
| GCF_017052135.1 | ASM1705213v1 | Pantoea stewartii subsp. indologenes | PNA 14-12 | 27 February 2021 |
| GCF_017052195.1 | ASM1705219v1 | Pantoea stewartii subsp. indologenes | PNA 14-11 | 27 February 2021 |
| GCF_017051895.1 | ASM1705189v1 | Pantoea stewartii subsp. indologenes | NCPPB 2275 | 27 February 2021 |
| GCF_017051975.1 | ASM1705197v1 | Pantoea stewartii subsp. indologenes | PANS 07-10 | 27 February 2021 |
| GCF_017052015.1 | ASM1705201v1 | Pantoea stewartii subsp. indologenes | PANS 07-12 | 27 February 2021 |
| GCF_017052375.1 | ASM1705237v1 | Pantoea stewartii subsp. indologenes | PNA 14-9 | 27 February 2021 |
| GCF_017052175.1 | ASM1705217v1 | Pantoea stewartii subsp. indologenes | PNA 15-2 | 27 February 2021 |
| GCF_017051945.1 | ASM1705194v1 | Pantoea stewartii subsp. indologenes | PANS 99-15 | 27 February 2021 |
| GCF_017051805.1 | ASM1705180v1 | Pantoea stewartii subsp. indologenes | NCPPB 2281 | 27 February 2021 |
| GCF_017051935.1 | ASM1705193v1 | Pantoea stewartii subsp. indologenes | PANS 07-14 | 27 February 2021 |
| GCF_017052095.1 | ASM1705209v1 | Pantoea stewartii subsp. indologenes | PANS 07-4 | 27 February 2021 |
| GCF_017052115.1 | ASM1705211v1 | Pantoea stewartii subsp. indologenes | PANS 07-6 | 27 February 2021 |
| GCF_017051815.1 | ASM1705181v1 | Pantoea stewartii subsp. indologenes | NCPPB 2282 | 27 February 2021 |
| GCF_017051845.1 | ASM1705184v1 | Pantoea stewartii subsp. indologenes | NCPPB 1562 | 27 February 2021 |
| GCF_030144305.1 | ASM3014430v1 | Pantoea stewartii subsp. indologenes | SCHI0154.S.1 | 31 May 2023 |
| GCF_017051875.1 | ASM1705187v1 | Pantoea stewartii subsp. indologenes | NCPPB 1877 | 27 February 2021 |
| GCF_030370585.1 | ASM3037058v1 | Pantoea stewartii subsp. stewartii | IPV-BO 2766 | 28 June 2023 |
| GCF_030370545.1 | ASM3037054v1 | Pantoea stewartii subsp. stewartii | 49474.1 /18 | 28 June 2023 |
| GCF_010273335.1 | ASM1027333v1 | Pantoea stewartii subsp. stewartii | MS1 | 6 February 2020 |
| GCF_030064655.1 | ASM3006465v1 | Pantoea stewartii | 23 May 2023 | |
| GCA_030336395.1 | ASM3033639v1 | Pantoea stewartii subsp. indologenes | LMG 2630 | 22 June 2023 |
| GCA_030336485.1 | ASM3033648v1 | Pantoea stewartii subsp. stewartii | 34258.2/15 | 22 June 2023 |
| GCA_030370555.1 | ASM3037055v1 | Pantoea stewartii subsp. stewartii | 49472.2 /18 | 28 June 2023 |
| GCA_030336515.1 | ASM3033651v1 | Pantoea stewartii subsp. stewartii | 34596.1/15 | 22 June 2023 |
| GCA_030370565.1 | ASM3037056v1 | Pantoea stewartii subsp. stewartii | CREA_DC_1788 | 28 June 2023 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scala, V.; Pucci, N.; Fiorani, R.; L’Aurora, A.; Polito, A.; Marsico, M.D.; Aiese Cigliano, R.; Barra, E.; Ciarroni, S.; De Amicis, F.; et al. Pantoea stewartii subsp. stewartii an Inter-Laboratory Comparative Study of Molecular Tests and Comparative Genome Analysis of Italian Strains. Plants 2025, 14, 1470. https://doi.org/10.3390/plants14101470
Scala V, Pucci N, Fiorani R, L’Aurora A, Polito A, Marsico MD, Aiese Cigliano R, Barra E, Ciarroni S, De Amicis F, et al. Pantoea stewartii subsp. stewartii an Inter-Laboratory Comparative Study of Molecular Tests and Comparative Genome Analysis of Italian Strains. Plants. 2025; 14(10):1470. https://doi.org/10.3390/plants14101470
Chicago/Turabian StyleScala, Valeria, Nicoletta Pucci, Riccardo Fiorani, Alessia L’Aurora, Alessandro Polito, Marco Di Marsico, Riccardo Aiese Cigliano, Eleonora Barra, Serena Ciarroni, Francesca De Amicis, and et al. 2025. "Pantoea stewartii subsp. stewartii an Inter-Laboratory Comparative Study of Molecular Tests and Comparative Genome Analysis of Italian Strains" Plants 14, no. 10: 1470. https://doi.org/10.3390/plants14101470
APA StyleScala, V., Pucci, N., Fiorani, R., L’Aurora, A., Polito, A., Marsico, M. D., Aiese Cigliano, R., Barra, E., Ciarroni, S., De Amicis, F., Fascella, S., Gaffuri, F., Gallmetzer, A., Giacobbi, F., Grieco, P. D., Gualandri, V., Mason, G., Pasqua di Bisceglie, D., Rizzo, D., ... Loreti, S. (2025). Pantoea stewartii subsp. stewartii an Inter-Laboratory Comparative Study of Molecular Tests and Comparative Genome Analysis of Italian Strains. Plants, 14(10), 1470. https://doi.org/10.3390/plants14101470






