Genetic Diversity and Conservation of Bomarea ovallei (Phil.) Ravenna: Microsatellite Markers Reveal Population Vulnerability in the Atacama Desert
Abstract
1. Introduction
2. Results
3. Discussion
3.1. Novel SSR Marker for B. ovallei Using NGS
3.2. Genetic Diversity Assessment of B. ovallei
3.3. Genetic Relationships and Population Structure
3.4. Practical Implications
4. Materials and Methods
4.1. Materials
4.2. DNA Extraction and NGS Technology
4.3. SSR Locus Search
4.4. Evaluation of New SSR Markers
4.5. Data Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clarke, J.D.A. Antiquity of aridity in the Chilean Atacama Desert. Geomorphology 2006, 73, 101–114. [Google Scholar] [CrossRef]
- Contreras, R.; van den Brink, L.; Sepúlveda, B.; Aguayo, F.; Porcile, V. Phylogenetic relationships of plant species from the flowering desert of the Atacama Region. Bol. Latinoam. Caribe Plantas Med. Aromát. 2020, 19, 300–313. [Google Scholar]
- Gutiérrez, J. El desierto Florido en la Región de Atacama. In Libro Rojo de la Flora Nativa y de los Sitios Prioritarios Para su Conservación: Región de Atacama; Squeo, F., Arancio, G., Gutiérrez, J., Eds.; Universidad de La Serena: La Serena, Chile, 2008; pp. 285–291. [Google Scholar]
- Squeo, F.; Arancio, G.; Gutiérrez, J.R.; Letelier, L.; Arroyo, M.T.K.; León-Lobos, P.; Rentería-Arrieta, L. Flora Amenazada de la Región de Atacama y Estrategias Para Su Conservación; Universidad de La Serena: La Serena, Chile, 2008. [Google Scholar]
- Contreras-Díaz, R.; Huanca-Mamani, W.; van den Brink, L.; Fuentes, M.; Arias-Aburto, M. The complete chloroplast genome of the endangered species garra de león [Bomarea ovallei (Phil.) Ravenna] from Chile. Indian J. Genet. Plant Breed. 2022, 82, 365–368. [Google Scholar] [CrossRef]
- Chacón, J.; de Assis, M.C.; Meerow, A.W.; Renner, S.S. From East Gondwana to Central America: Historical biogeography of the Alstroemeriaceae. J. Biogeogr. 2012, 39, 1806–1818. [Google Scholar] [CrossRef]
- Hofreiter, A. Leontochir: A Synonym of Bomarea (Alstroemeriaceae)? Harv. Pap. Bot. 2006, 11, 53–60. [Google Scholar] [CrossRef]
- Tribble, C.M.; Alzate-Guarín, F.; Gándara, E.; Vartoumian, A.; Burleigh, J.B.; Zenil-Ferguson, R.; Specht, C.D.; Rothfels, C.J. The rapid radiation of Bomarea (Alstroemeriaceae: Liliales), driven by the rise of the Andes. Evolution 2024, 78, 221–236. [Google Scholar] [CrossRef]
- Hofreiter, A. The Genus Bomarea (Alstroemeriaceae) in Bolivia and Southern South America. Harv. Pap. Bot. 2005, 9, 343–374. [Google Scholar]
- Finot, V.; Baeza, C.; Muñoz-Schick, M.; Ruiz, E.; Espejo, J.; Alarcón, D.; Carrasco, P.; Eyzaguirre, P.N.y.M.T. Guía de Campo Alstromerias Chilenas. CORMA. 2018. Available online: www.corma.cl (accessed on 10 March 2025).
- Gostel, M.R.; Kress, W.J. The Expanding Role of DNA Barcodes: Indispensable Tools for Ecology, Evolution, and Conservation. Diversity 2022, 14, 213. [Google Scholar] [CrossRef]
- Amiteye, S. Basic Concepts and Methodologies of DNA Marker Systems in Plant Molecular Breeding. Heliyon 2021, 7, e08093. [Google Scholar] [CrossRef]
- Aburto, M.A.; Medina, D.R.; Díaz, R.C. Molecular taxonomic analysis of individuals of the genus lathyrus found in the northern extreme of Chile, using DNA barcoding. Bol. Soc. Argent. Bot. 2020, 55, 479–492. [Google Scholar] [CrossRef]
- Wu, Y.; Huang, L.; Zhou, D.; Fu, X.; Li, C.; Wei, S.; Peng, J.; Kuang, M. Development and application of perfect SSR markers in cotton. J. Cott. Res. 2020, 3, 21. [Google Scholar] [CrossRef]
- Alves, S.I.A.; Dantas, C.W.D.; Macedo, D.B.; Ramos, R.T.J. What are microsatellites and how to choose the best tool: A user-friendly review of SSR and 74 SSR mining tools. Front. Genet. 2021, 15, 1474611. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.d.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Lyu, D.; Sun, S.; Shan, X.; Wang, W. Inbreeding evaluation using microsatellite confirmed inbreeding depression in growth in the Fenneropenaeus chinensis natural population. Front. Genet. 2023, 14, 1077814. [Google Scholar] [CrossRef] [PubMed]
- Tambarussi, E.V.; Boshier, D.H.; Vencovsky, R.; Freitas, M.L.M.; Di-Dio, O.J.; Sebbenn, A.M. Several Small: How Inbreeding Affects Conservation of Cariniana legalis Mart. Kuntze (Lecythidaceae) the Brazilian Atlantic Forest’s Largest Tree. Int. For. Rev. 2016, 18, 502–510. [Google Scholar] [CrossRef]
- Heinken, T.; Weber, E. Consequences of habitat fragmentation for plant species: Do we know enough? Perspect. Plant Ecol. Evol. Syst. 2013, 15, 205–216. [Google Scholar] [CrossRef]
- Contreras, R.; Carevic, F.; Aburto, M.A.; Huanca-Mamani, W.; Díaz-Martin, B. Development of new microsatelite loci for Geoffroea decorticans (Gillies ex Hook. & Arn.) Burkart. Bol. Soc. Argent. Bot. 2021, 56, 533–545. [Google Scholar] [CrossRef]
- Chun, S.W.; Lee, J.W.; Ahn, J.Y. Development and characterization of novel microsatellite markers in Tilia amurensis Rupr. using next-generation sequencing. Mol. Biol. Rep. 2022, 49, 1637–1641. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, J.; Jiang, M.; Wang, W.; Jiang, J.; Li, Y.; Yang, L.; Zhou, X. Development of genome-wide SSR markers in rapeseed by next generation sequencing. Gene 2021, 798, 145798. [Google Scholar] [CrossRef]
- Contreras, R.; Carevic, F.S.; Porcile, V.; Arias, M. Development of SSR loci in Prosopis tamarugo Phillipi and assessment of their transferability to species of the strombocarpa section. For. Syst. 2020, 29, e012. [Google Scholar] [CrossRef]
- Anastassopoulos, E.; Keil, M. Assessment of natural and induced genetic variation in Alstroemeria using random amplified polymorphic DNA (RAPD) markers. Euphytica 1996, 90, 235–244. [Google Scholar] [CrossRef]
- Aros, D.; Meneses, C.; Infante, R. Genetic diversity of wild species and cultivated varieties of alstroemeria estimated through morphological descriptors and RAPD markers. Sci. Hortic. 2006, 108, 86–90. [Google Scholar] [CrossRef]
- Han, T.H.; De Jeu, M.; Van Eck, H.; Jacobsen, E. Genetic diversity of Chilean and Brazilian Alstroemeria species assessed by AFLP analysis. Heredity 2000, 84, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Sakazono, S.; Hiramatsu, M.; Watanabe, M.; Okubo, H. Development and characterization of microsatellite markers for Lilium longiflorum (Liliaceae). Appl. Plant Sci. 2013, 1, 1300014. [Google Scholar] [CrossRef]
- Nakamasu, R.; Sakaguchi, S.; Setoguchi, H. Development and characterization of est-ssr markers for Tricyrtis sect. Tricyrtis (liliaceae). Acta Phytotaxon. Geobot. 2020, 71, 73–76. [Google Scholar] [CrossRef]
- Sakazono, S.; Hiramatsu, M.; Huang, K.L.; Huang, C.L.; Okubo, H. Phylogenetic relationship between degree of self-compatibility and floral traits in Lilium longiflorum thunb. (Liliaceae). J. Japanese Soc. Hortic. Sci. 2012, 81, 80–90. [Google Scholar] [CrossRef][Green Version]
- Souto, C.P.; Aizen, M.A.; Premoli, A.C. Effects of crossing distance and genetic relatedness on pollen performance in Alstroemeria aurea (Alstroemeriaceae). Am. J. Bot. 2002, 89, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Aizen, M.A.; Basilio, A. Within and among flower sex-phase distribution in Alstroemeria aurea (Alstroemeriaceae). Can. J. Bot. 1995, 73, 1986–1994. [Google Scholar] [CrossRef]
- Tambarussi, E.V.; Boshier, D.; Vencovsky, R.; Freitas, M.L.M.; Sebbenn, A.M. Inbreeding depression from selfing and mating between relatives in the Neotropical tree Cariniana legalis Mart. Kuntze. Conserv. Genet. 2017, 18, 225–234. [Google Scholar] [CrossRef]
- Wu, X.; Duan, L.; Chen, Q.; Zhang, D. Genetic diversity, population structure, and evolutionary relationships within a taxonomically complex group revealed by AFLP markers: A case study on Fritillaria cirrhosa D. Don and closely related species. Glob. Ecol. Conserv. 2020, 24, e01323. [Google Scholar] [CrossRef]
- Beavis, W.; Campbell, A.; Edwards, J.; Fei, S.-Z.; Lübberstedt, T.; Merrick, L.; Muenchrath, D. Crop Genetics. In Crop Genetics; Suza, W.P., Lamkey, K.R., Eds.; Iowa State University Digital Press: Ames, IA, USA, 2023. [Google Scholar] [CrossRef]
- Eshel, G.; Araus, V.; Undurraga, S.; Soto, D.C.; Moraga, C.; Montecinos, A.; Moyano, T.; Maldonado, J.; Díaz, F.P.; Varala, K.; et al. Plant ecological genomics at the limits of life in the Atacama Desert. Proc. Natl. Acad. Sci. USA 2021, 118, 1–11. [Google Scholar] [CrossRef]
- Vargas, M.; Jofré, E.; Navarrete, C.; Bravo, J.; Jamett, F.; Inostroza-Blancheteau, C.; Ibáñez, C. Sexual and asexual reproductive aspects of Leontochir ovallei, a rare and endangered geophyte of the Atacama Desert. Rev. Chil. Hist. Nat. 2018, 91, 5. [Google Scholar] [CrossRef]
- Ravenna, P. New or interesting Alstroemeriaceae. Onira 2000, 5, 35–41. [Google Scholar]
- Contreras-Díaz, R.; Carevic, F.S.; van den Brink, L. Comparative analysis of the complete mitogenome of Geoffroea decorticans: A native tree surviving in the Atacama Desert. Front. Genet. 2023, 14, 1226052. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Mokhtar, M.M.; Alsamman, A.M.; El Allali, A. MegaSSR: A web server for large scale microsatellite identification, classification, and marker development. Front. Plant Sci. 2023, 14, 1219055. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenALEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Brookfield, J.F.Y. A simple new method for estimating null allele frequency from heterozygote deficiency. Mol. Ecol. 1996, 5, 453–455. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.; Skolnick, M.; Davis, R. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Hammer, Ø.; Harper, A.T.D.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Paleontol. Electrón. 2001, 4, 9. Available online: https://palaeo-electronica.org/2001_1/past/past.pdf (accessed on 6 March 2025).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Li, Y.L.; Liu, J.X. StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef]

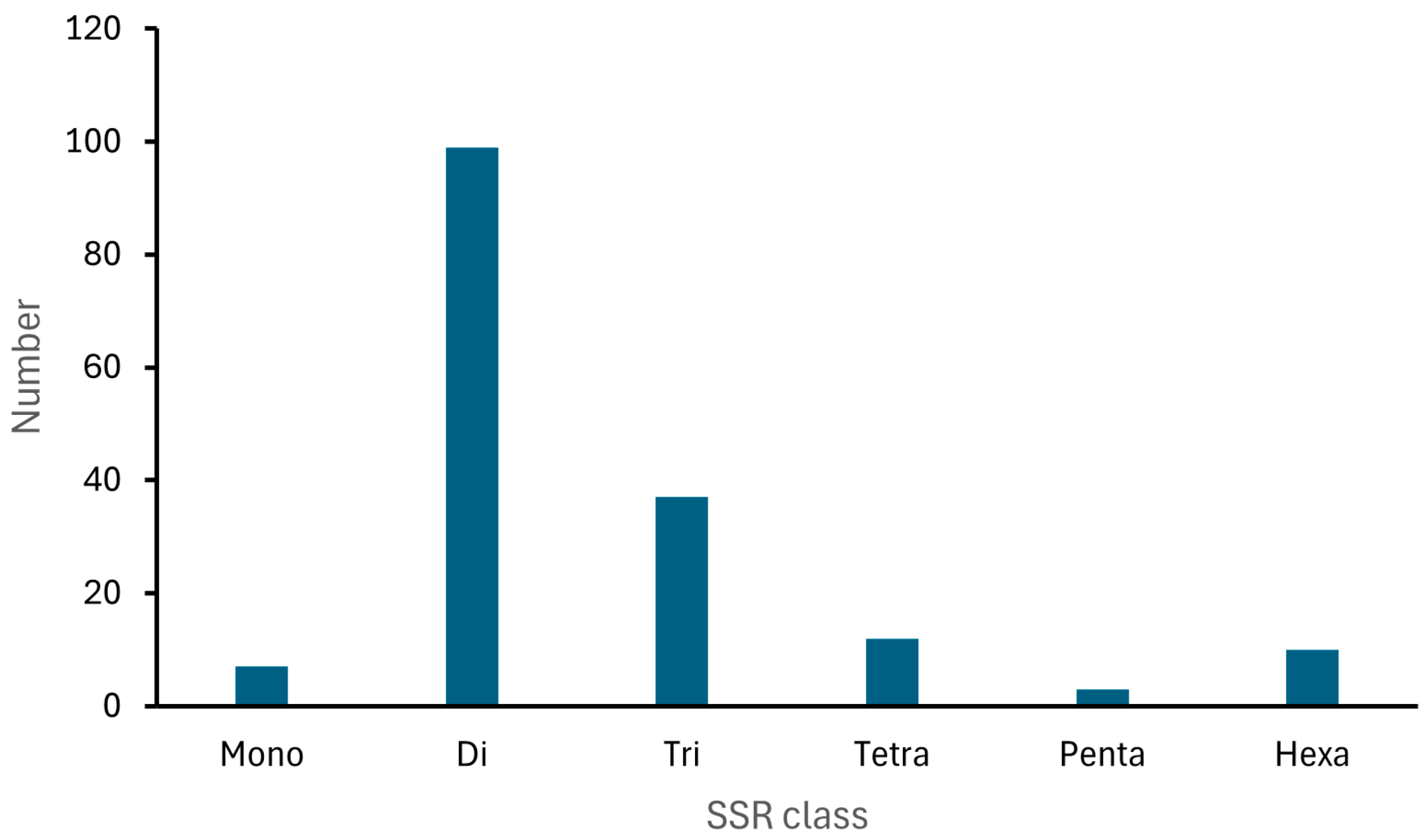
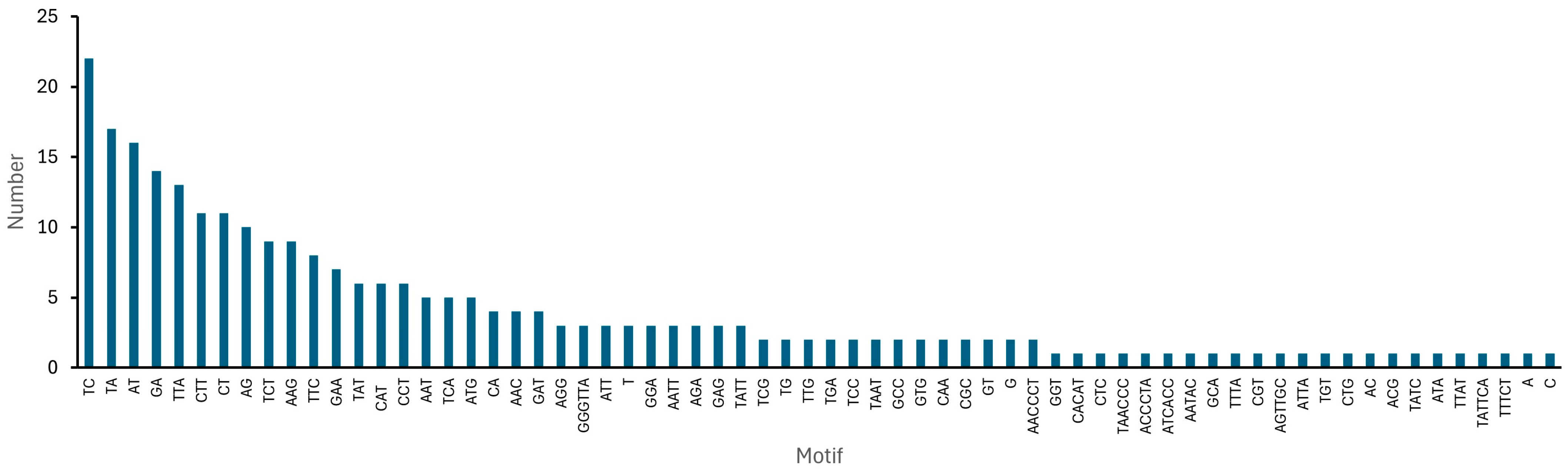

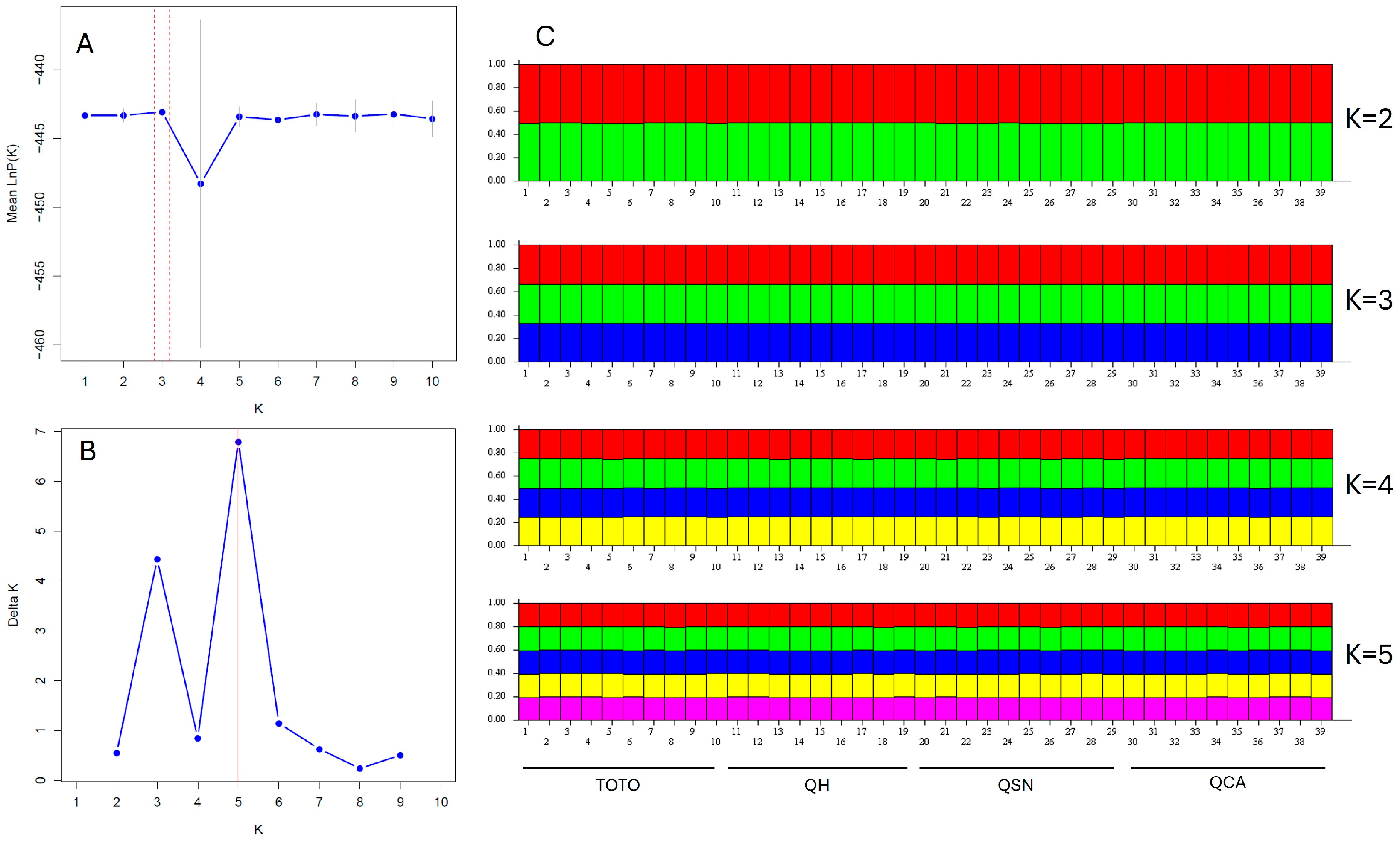
| Category | Total Number |
|---|---|
| Total number of sequences examined | 18,709 |
| Total size examined sequences (bp) | 6,252,133 |
| Total number of identified SSRs | 268 |
| Number of SSR-containing sequences | 254 |
| Number of sequences containing more than one SSR | 13 |
| Number of SSRs present in compound formation | 9 |
| Locus | 5′-Sequence-3′ | Accession | Motif | Size (bp) | T°a (°C) |
|---|---|---|---|---|---|
| SSRBO8819 | F-AGTCGAGTTTCTCTGATCTTAAA R-ACAGAATCCAAAATAAAACTGGG | PV126292 | (TTA)9 | 153 | 59 |
| SSRBO16864 | F-ATCATCAAAGATAGTAGCGGAAT R-CTGATCTTTGTCTCTCTCCG | PV126301 | (TCA)9 | 169 | 59 |
| SSRBO9629 | F-CCCTTTTATGGTTTGTTGTAGCA R-AGCCAAGAACAAACCGTAAAG | PV126315 | (CT)21 | 195 | 59 |
| SSRBO14256 | F-GGTCTTAGGGTGTGGGCTAA R-GTGGCATGGAAAGGAACACA | PV126317 | (CT)14 | 169 | 59 |
| SSRBO29 | F-TGTTCAATCAAGTCATGGGCA R-CTGGTGTAGTATCATGCATGCA | PV126319 | (TGA)7 | 204 | 59 |
| SSRBO3955 | F-TCGCACTTTCTCTCTGTCCT R-AAACTCTATCTAAACATCCGCCA | PV126320 | (TGA)6 | 232 | 59 |
| SSRBO1064 | F-TGAGTTGCTGTGGTGGTATG R-TCACCAAACAACCATAACACCA | PV126321 | (TTG)11 | 105 | 57 |
| SSRBO20205 | F-TCAGGCGATTGGTTGGAAAG R-GTTTGGGACGCGGTTTCTTT | PV126324 | (TTC)7 | 118 | 59 |
| Locus | Allele Size (bp) | Na | Ne | I | Ho | He | Null Allele | PIC |
|---|---|---|---|---|---|---|---|---|
| SSRBO8819 | 144, 147, 150, 156 | 4 | 2.54 (0.21) | 0.991 (0.11) | 1.000 (0.000) | 0.598 (0.035) | −0.251 | 0.70 |
| SSRBO16864 | 154 | 1 | 1.00 | 0 | 0 | 0 | - | 0.00 |
| SSRBO9629 | 197, 199, 201, 203 | 4 | 2.58 (0.24) | 1.075 (0.07) | 0.519 (0.089) | 0.603 (0.036) | 0.052 | 0.69 |
| SSRBO14256 | 167, 169, 171, 173, 181, 183 | 6 | 1.80 (0.10) | 0.715 (0.08) | 0.589 (0.042) | 0.439 (0.032) | −0.104 | 0.48 |
| SSRBO29 | 195 | 1 | 1.00 | 0 | 0 | 0 | - | 0.00 |
| SSRBO3955 | 229, 235, 241, 247 | 4 | 1.92 (0.28) | 0.691 (0.11) | 0.711 (0.167) | 0.448 (0.071) | −0.181 | 0.53 |
| SSRBO1064 | 105 | 1 | 1.00 | 0 | 0 | 0 | - | 0.00 |
| SSRBO20205 | 109, 112, 115 | 3 | 2.29 (0.09) | 0.884 (0.06) | 1.000 (0.000) | 0.562 (0.021) | −0.280 | 0.56 |
| Average | 3 | 1.76 (0.12) | 0.545 (0.08) | 0.477 (0.075) | 0.331 (0.048) | 0.37 |
| Population | N | Na | Ne | I | Ho | He | Fis |
|---|---|---|---|---|---|---|---|
| TOTO | 10 | 1.875 (0.29) | 1.696 (0.23) | 0.489 (0.11) | 0.488 (0.164) | 0.321 (0.097) | −0.528 (0.190) |
| QH | 9 | 2.125 (0.39) | 1.752 (0.26) | 0.531 (0.17) | 0.472 (0.154) | 0.324 (0.102) | −0.441 (0.073) |
| QSN | 10 | 2.250 (0.41) | 1.736 (0.22) | 0.554 (0.16) | 0.500 (0.165) | 0.334 (0.099) | −0.486 (0.169) |
| QCA | 10 | 2.375 (0.46) | 1.880 (0.31) | 0.603 (0.19) | 0.450 (0.151) | 0.346 (0.109) | −0.313 (0.131) |
| TOTO | QH | QSN | QCA | |
|---|---|---|---|---|
| TOTO | 0.000 | |||
| QH | 0.112 | 0.000 | ||
| QSN | 0.063 | 0.103 | 0.000 | |
| QCA | 0.059 | 0.078 | 0.092 | 0.000 |
| N° | Latitude S | Longitude W | Altitude (Masl) | Populations |
|---|---|---|---|---|
| 1–10 | 27°53′45.09″ S | 70°59′17.25″ W | 153–162 | TOTO |
| 11–19 | 27°59′25.39″ S | 71°7′49.86″ W | 83–93 | QH |
| 20–29 | 28°01′39.4″ S | 71°07′51.2″ W | 84–91 | QSN |
| 30–39 | 28°6′27.64″ S | 71°6′23.81″ W | 39–41 | QCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rozas-Lazcano, V.; Mamani-Gómez, M.; Rojas-Jopia, I.; Arias-Aburto, M.; Contreras-Díaz, R. Genetic Diversity and Conservation of Bomarea ovallei (Phil.) Ravenna: Microsatellite Markers Reveal Population Vulnerability in the Atacama Desert. Plants 2025, 14, 1468. https://doi.org/10.3390/plants14101468
Rozas-Lazcano V, Mamani-Gómez M, Rojas-Jopia I, Arias-Aburto M, Contreras-Díaz R. Genetic Diversity and Conservation of Bomarea ovallei (Phil.) Ravenna: Microsatellite Markers Reveal Population Vulnerability in the Atacama Desert. Plants. 2025; 14(10):1468. https://doi.org/10.3390/plants14101468
Chicago/Turabian StyleRozas-Lazcano, Valeska, Mariel Mamani-Gómez, Irina Rojas-Jopia, Mariana Arias-Aburto, and Roberto Contreras-Díaz. 2025. "Genetic Diversity and Conservation of Bomarea ovallei (Phil.) Ravenna: Microsatellite Markers Reveal Population Vulnerability in the Atacama Desert" Plants 14, no. 10: 1468. https://doi.org/10.3390/plants14101468
APA StyleRozas-Lazcano, V., Mamani-Gómez, M., Rojas-Jopia, I., Arias-Aburto, M., & Contreras-Díaz, R. (2025). Genetic Diversity and Conservation of Bomarea ovallei (Phil.) Ravenna: Microsatellite Markers Reveal Population Vulnerability in the Atacama Desert. Plants, 14(10), 1468. https://doi.org/10.3390/plants14101468







