LcTprxII Overexpression Enhances Physiological and Biochemical Effects in Maize Under Alkaline (Na2CO3) Stress
Abstract
1. Introduction
2. Results
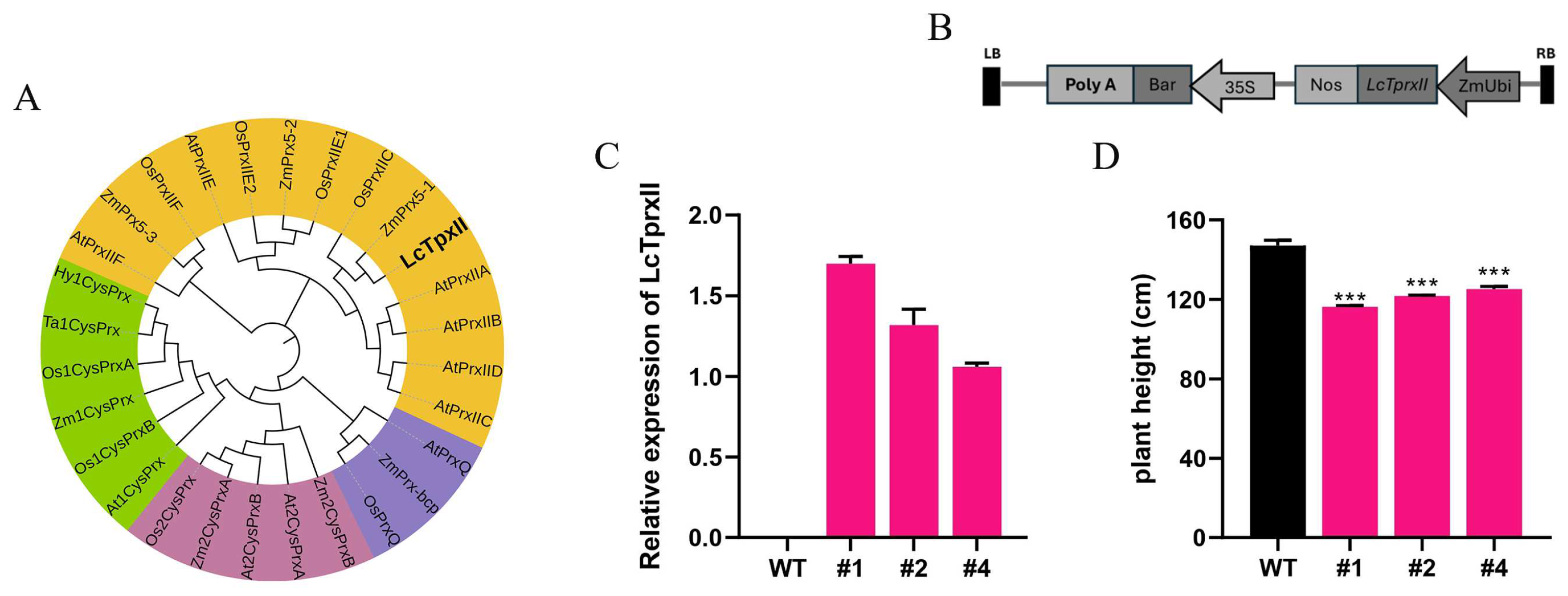
2.1. Phylogenetic Relationship and Expression Pattern of LcTprxII
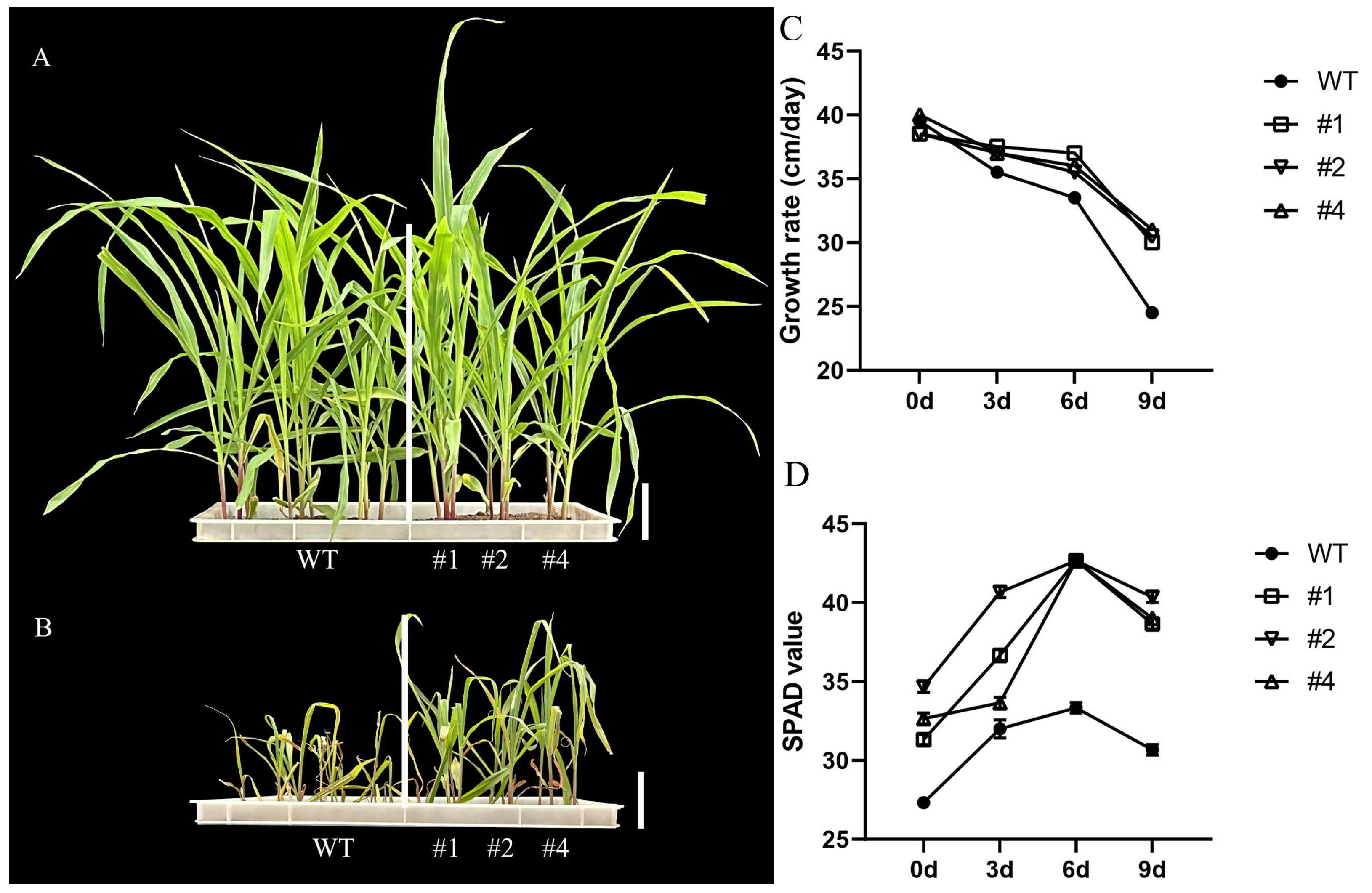
2.2. Physical and Biochemical Interactions of LcTprxII Overexpression with Alkaline Stress
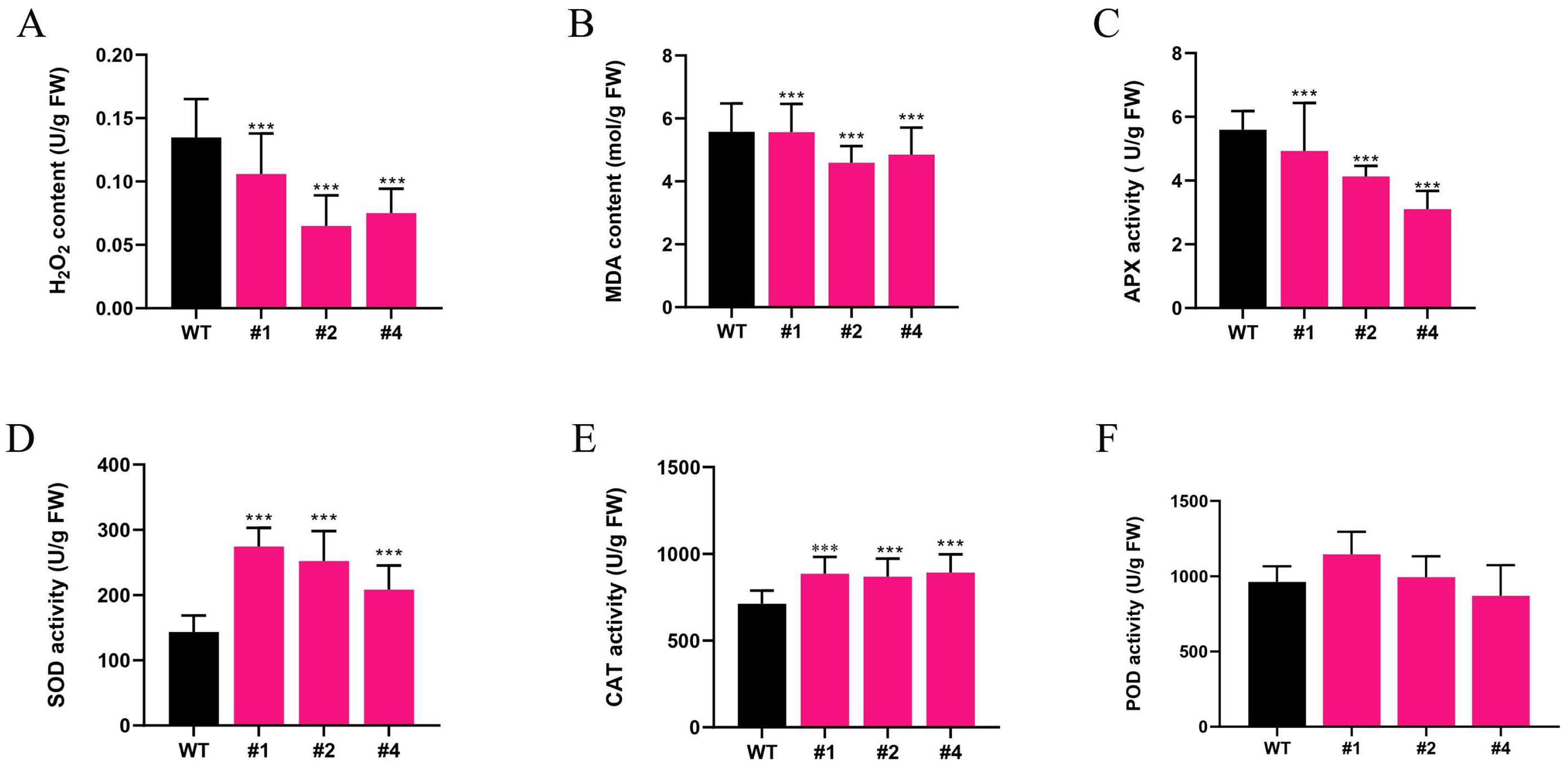
2.3. Effects of Alkaline Stress on Antioxidant Enzymes and Oxidative Markers
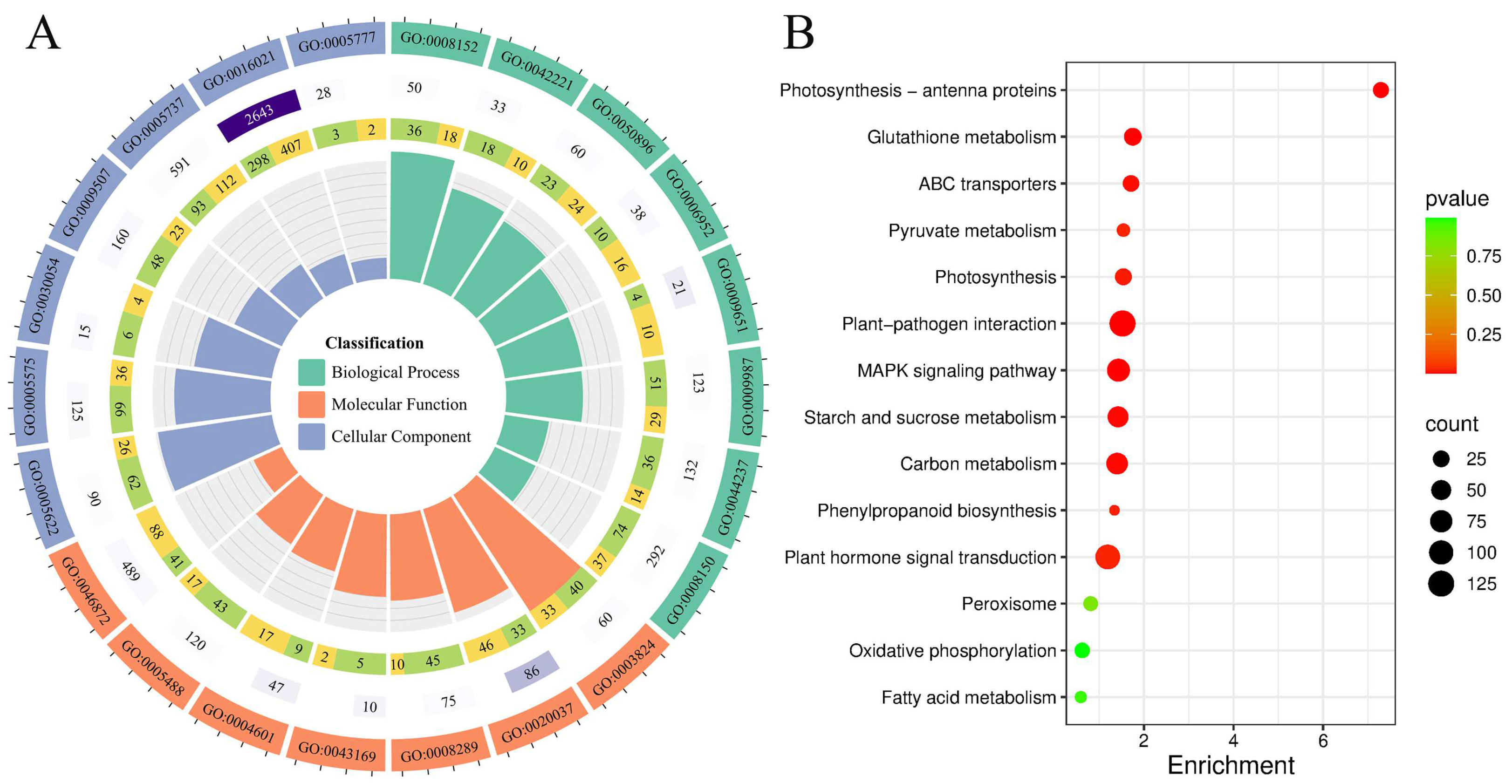
2.4. Differential Transcriptome Analysis Between Transgenic Maize and Control Plants
2.5. Differential Regulation of ABA Metabolites in LcTprxII Overexpression
2.6. Difference in Agronomic Traits Due to LcTprxII Overexpression
2.7. Differentially Expressed TFs Under Alkaline (Na2CO3) Stress
2.8. The Verification of Gene Expression Candidates
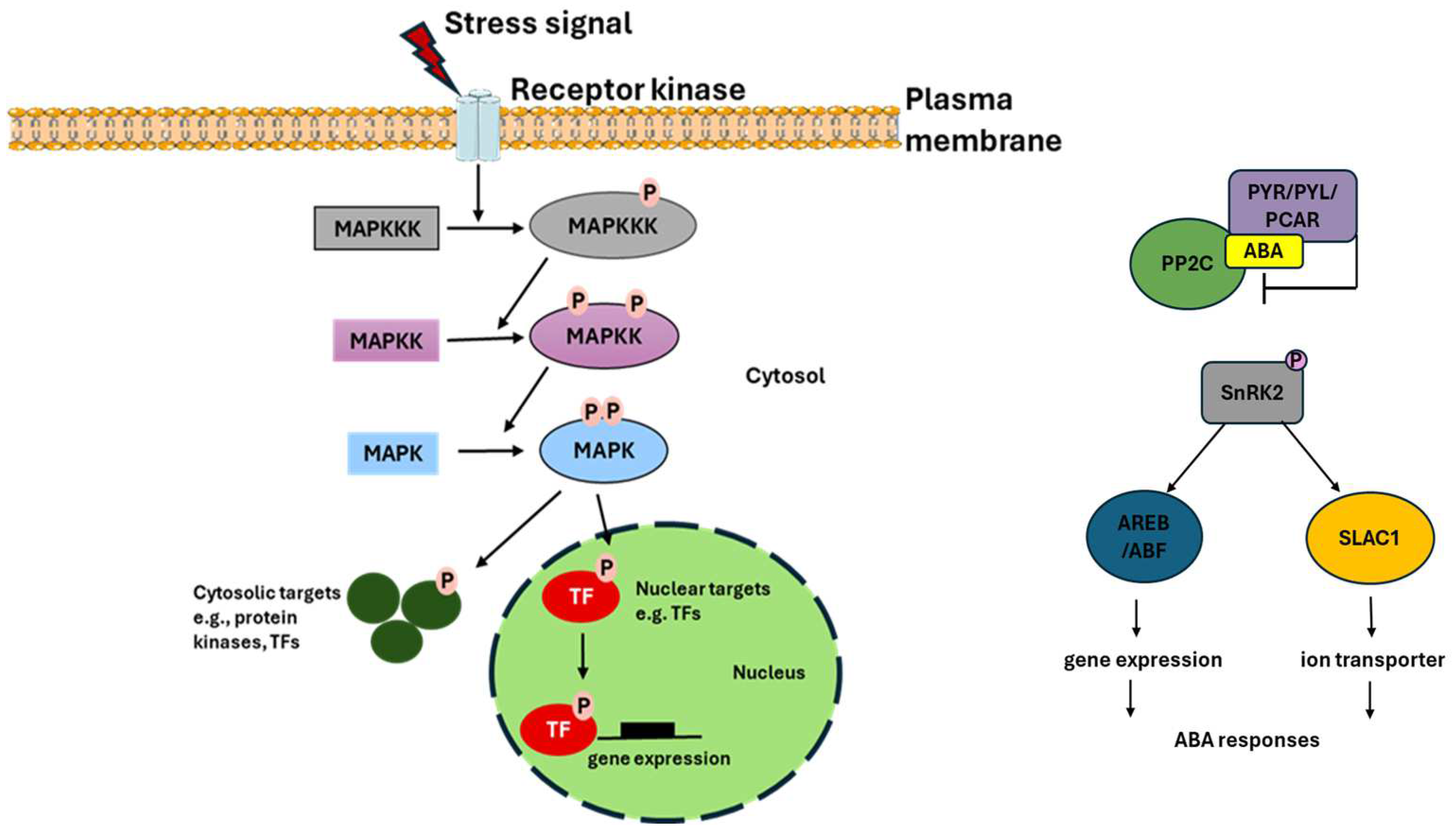
3. Discussion
4. Materials and Methods
4.1. Cloning and Transformation of LcTprxII
4.2. Phenotyping and Relative Chlorophyll Content Analysis
4.3. Physiological and Biochemical Analysis
4.4. Determination of Endogenous Plant Hormones (GA, ABA)
4.5. Field Trials and Yield Assessment of LcTprxII Overexpression
4.6. RNA-Seq Analysis and Expression Patterns of LcTprxII-OE
4.7. Real-Time PCR (qRT-PCR) Analysis
4.8. Statistical Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.; Ma, H.; Liu, G.; Xu, C.; Zhang, D.; Ban, Q. Analysis of Gene Expression Profile of Limonium bicolor under NaHCO3 Stress Using cDNA Microarray. Plant Mol. Biol. Rep. 2008, 26, 241–254. [Google Scholar] [CrossRef]
- Yu, Y.; Duan, X.; Ding, X.; Chen, C.; Zhu, D.; Yin, K.; Cao, L.; Song, X.; Zhu, P.; Li, Q.; et al. A novel AP2/ERF family transcription factor from Glycine soja, GsERF71, is a DNA binding protein that positively regulates alkaline stress tolerance in Arabidopsis. Plant Mol. Biol. 2017, 94, 509–530. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Yang, S.; Sun, M.; Wang, S.; Ding, X.; Zhu, D.; Ji, W.; Cai, H.; Zhao, C.; Wang, X.; et al. A novel Glycine soja cysteine proteinase inhibitor GsCPI14, interacting with the calcium/calmodulin-binding receptor-like kinase GsCBRLK, regulated plant tolerance to alkali stress. Plant Mol. Biol. 2014, 85, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.W.; Xu, H.H.; Wang, L.L.; Liu, J.; Shi, D.C.; Wang, D.L. Comparative effects of salt-stress and alkali-stress on the growth, photosynthesis, solute accumulation, and ion balance of barley plants. Photosynthetica 2009, 47, 79–86. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, S.; Yu, R. Saline Soil in China; Science Press Beijing: Beijing, China, 1993. (In Chinese) [Google Scholar]
- Flowers, T.J.; Munns, R.; Colmer, T.D. Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann. Bot. 2015, 115, 419–431. [Google Scholar] [CrossRef]
- Munns, R.; Schachtman, D.; Condon, A. The Significance of a Two-Phase Growth Response to Salinity in Wheat and Barley. Funct. Plant Biol. 1995, 22, 561. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.; Zulfiqar, F.; Raza, A.; Mohsin, S.; Mahmud, J.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef]
- Ben Hsouna, A.; Dhibi, S.; Dhifi, W.; Mnif, W.; Ben Nasr, H.; Hfaiedh, N. Chemical composition and hepatoprotective effect of essential oil from Myrtus communis L. flowers against CCL 4 -induced acute hepatotoxicity in rats. RSC Adv. 2019, 9, 3777–3787. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox Homeostasis and Antioxidant Signaling: A Metabolic Interface between Stress Perception and Physiological Responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef]
- Vieira Dos Santos, C.; Rey, P. Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci. 2006, 11, 329–334. [Google Scholar] [CrossRef]
- Jin, H.; Plaha, P.; Park, J.Y.; Hong, C.P.; Lee, I.S.; Yang, Z.H.; Jiang, G.B.; Kwak, S.S.; Liu, S.K.; Lee, J.S.; et al. Comparative EST profiles of leaf and root of Leymus chinensis, a xerophilous grass adapted to high pH sodic soil. Plant Sci. 2006, 170, 1081–1086. [Google Scholar] [CrossRef]
- Dietz, K.J. Plant peroxiredoxins. Annu. Rev. Plant. Biol. 2003, 54, 93. [Google Scholar] [CrossRef]
- Choi, Y.O.; Cheong, N.E.; Lee, K.O.; Jung, B.G.; Hong, C.H.; Jeong, J.H.; Chi, Y.H.; Kim, K.; Cho, M.J.; Lee, S.Y. Cloning and Expression of a New Isotype of the Peroxiredoxin Gene of Chinese Cabbage and Its Comparison to 2Cys-Peroxiredoxin Isolated from the Same Plant. Biochem. Biophys. Res. Commun. 1999, 258, 768–771. [Google Scholar] [CrossRef]
- Horling, F.; Lamkemeyer, P.; König, J.; Finkemeier, I.; Kandlbinder, A.; Baier, M.; Dietz, K.-J. Divergent Light-, Ascorbate-, and Oxidative Stress-Dependent Regulation of Expression of the Peroxiredoxin Gene Family in Arabidopsis. Plant Physiol. 2003, 131, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Rouhier, N.; Gelhaye, E.; Sautiere, P.-E.; Brun, A.; Laurent, P.; Tagu, D.; Gerard, J.; De Faÿ, E.; Meyer, Y.; Jacquot, J.-P. Isolation and Characterization of a New Peroxiredoxin from Poplar Sieve Tubes That Uses Either Glutaredoxin or Thioredoxin as a Proton Donor. Plant Physiol. 2001, 127, 1299–1309. [Google Scholar] [CrossRef]
- Neumann, C.A.; Krause, D.S.; Carman, C.V.; Das, S.; Dubey, D.P.; Abraham, J.L.; Bronson, R.T.; Fujiwara, Y.; Orkin, S.H.; Van Etten, R.A. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature 2003, 424, 561–565. [Google Scholar] [CrossRef]
- Choi, M.H.; Lee, I.K.; Kim, G.W.; Kim, B.U.; Han, Y.-H.; Yu, D.-Y.; Park, H.S.; Kim, K.Y.; Lee, J.S.; Choi, C.; et al. Regulation of PDGF signalling and vascular remodelling by peroxiredoxin II. Nature 2005, 435, 347–353. [Google Scholar] [CrossRef]
- Jang, H.H.; Lee, K.O.; Chi, Y.H.; Jung, B.G.; Park, S.K.; Park, J.H.; Lee, J.R.; Lee, S.S.; Moon, J.C.; Yun, J.W.; et al. Two Enzymes in One. Cell 2004, 117, 625–635. [Google Scholar] [CrossRef]
- Astier, J.; Rasul, S.; Koen, E.; Manzoor, H.; Besson-Bard, A.; Lamotte, O.; Jeandroz, S.; Durner, J.; Lindermayr, C.; Wendehenne, D. S-nitrosylation: An emerging post-translational protein modification in plants. Plant Sci. 2011, 181, 527–533. [Google Scholar] [CrossRef]
- Bartoli, C.G.; Casalongué, C.A.; Simontacchi, M.; Marquez-Garcia, B.; Foyer, C.H. Interactions between hormone and redox signalling pathways in the control of growth and cross tolerance to stress. Environ. Exp. Bot. 2013, 94, 73–88. [Google Scholar] [CrossRef]
- van der Est, A.; Bruce, D. (Eds.) Photosynthesis: Fundamental Aspects to Global Perspectives: Proceedings of the 13th International Congress on Photosynthesis; Alliance Communications Group: Lawrence, KS, USA, 2005; ISBN 978-1-891276-39-2. [Google Scholar]
- Chinnusamy, V.; Jagendorf, A.; Zhu, J. Understanding and Improving Salt Tolerance in Plants. Crop Sci. 2005, 45, 437–448. [Google Scholar] [CrossRef]
- Alexandrov, N.N.; Brover, V.V.; Freidin, S.; Troukhan, M.E.; Tatarinova, T.V.; Zhang, H.; Swaller, T.J.; Lu, Y.-P.; Bouck, J.; Flavell, R.B.; et al. Insights into corn genes derived from large-scale cDNA sequencing. Plant Mol. Biol. 2009, 69, 179–194. [Google Scholar] [CrossRef]
- Verdoucq, L.; Vignols, F.; Jacquot, J.-P.; Chartier, Y.; Meyer, Y. In Vivo Characterization of a Thioredoxin h Target Protein Defines a New Peroxiredoxin Family. J. Biol. Chem. 1999, 274, 19714–19722. [Google Scholar] [CrossRef]
- Nozu, Y.; Tsugita, A.; Kamijo, K. Proteomic analysis of rice leaf, stem and root tissues during growth course. Proteomics 2006, 6, 3665–3670. [Google Scholar] [CrossRef]
- Fan, X.; Jiang, H.; Meng, L.; Chen, J. Gene Mapping, Cloning and Association Analysis for Salt Tolerance in Rice. Int. J. Mol. Sci. 2021, 22, 11674. [Google Scholar] [CrossRef]
- Ding, Y.; Cao, J.; Ni, L.; Zhu, Y.; Zhang, A.; Tan, M.; Jiang, M. ZmCPK11 is involved in abscisic acid-induced antioxidant defence and functions upstream of ZmMPK5 in abscisic acid signalling in maize. J. Exp. Bot. 2013, 64, 871–884. [Google Scholar] [CrossRef]
- Liang, Y.; Jiang, Y.; Du, M.; Li, B.; Chen, L.; Chen, M.; Jin, D.; Wu, J. ZmASR3 from the Maize ASR Gene Family Positively Regulates Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2278. [Google Scholar] [CrossRef]
- Zelm, V.; Eva; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Cheng, Z.; Luan, Y.; Meng, J.; Sun, J.; Tao, J.; Zhao, D. WRKY Transcription Factor Response to High-Temperature Stress. Plants 2021, 10, 2211. [Google Scholar] [CrossRef]
- Belda-Palazón, B.; Adamo, M.; Valerio, C.; Ferreira, L.J.; Confraria, A.; Reis-Barata, D.; Rodrigues, A.; Meyer, C.; Rodriguez, P.L.; Baena-González, E. A dual function of SnRK2 kinases in the regulation of SnRK1 and plant growth. Nat. Plants 2020, 6, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Lv, B.; Yan, Z.; Tian, H.; Zhang, X.; Ding, Z. Local Auxin Biosynthesis Mediates Plant Growth and Development. Trends Plant Sci. 2019, 24, 6–9. [Google Scholar] [CrossRef]
- Manosalva Pérez, N.; Staut, J.; Ferrando, A.M.; Dissanayake, I.; Vandepoele, K. A map of integrated cis-regulatory elements enhances gene regulatory analysis in maize. bioRxiv 2024. [Google Scholar] [CrossRef]
- Li, M.; Qi, X.; Li, D.; Wu, Z.; Liu, M.; Yang, W.; Zang, Z.; Jiang, L. Comparative transcriptome analysis highlights resistance regulatory networks of maize in response to Exserohilum turcicum infection at the early stage. Physiol. Plant. 2024, 176, e14615. [Google Scholar] [CrossRef] [PubMed]
- Hayford, R.K.; Haley, O.C.; Cannon, E.K.; Portwood, J.L.; Gardiner, J.M.; Andorf, C.M.; Woodhouse, M.R. Functional annotation and meta-analysis of maize transcriptomes reveal genes involved in biotic and abiotic stress. BMC Genom. 2024, 25, 533. [Google Scholar] [CrossRef]
- Jiang, Q.; Wang, Y. Leaf angle regulation toward a maize smart canopy. Plant J. 2024, 121, e17208. [Google Scholar] [CrossRef]
- Kumar, B.; Choudhary, M.; Kumar, P.; Kumar, K.; Kumar, S.; Singh, B.K.; Lahkar, C.; Meenakshi; Kumar, P.; Dar, Z.A.; et al. Population Structure Analysis and Association Mapping for Turcicum Leaf Blight Resistance in Tropical Maize Using SSR Markers. Genes 2022, 13, 618. [Google Scholar] [CrossRef]
- Fernandez, E.C.J.; Simon-Ada, E.V.M.; Mendoza, J.-V.S.; Manohar, A.N.C.; Gardoce, R.R.; Laude, T.P.; Dela Cueva, F.M.; Lantican, D.V. Comparative Transcriptomics Reveals the Molecular Mechanisms of Maize in Response to Downy Mildew Disease Caused by Peronosclerospora philippinensis (Weston) Shaw. Plant Mol. Biol. Rep. 2024, 43, 269–285. [Google Scholar] [CrossRef]
- Ma, Z.; Jia, Y.; Min, Y.; Fang, X.; Yan, H.; Ma, Q.; Cai, R. Maize ZmWRKY71 gene positively regulates drought tolerance through reactive oxygen species homeostasis. Plant Physiol. Biochem. 2025, 219, 109399. [Google Scholar] [CrossRef]
- Torres-Rodríguez, J.V.; Li, D.; Turkus, J.; Newton, L.; Davis, J.; Lopez-Corona, L.; Ali, W.; Sun, G.; Mural, R.V.; Grzybowski, M.W.; et al. Population-level gene expression can repeatedly link genes to functions in maize. Plant J. 2024, 119, 844–860. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA 2/Ethylene Responsive Factor (AP 2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Kotak, S.; Vierling, E.; Bäumlein, H.; Koskull-Döring, P.V. A Novel Transcriptional Cascade Regulating Expression of Heat Stress Proteins during Seed Development of Arabidopsis. Plant Cell 2007, 19, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Feng, B.-R.; Yang, S.-S.; Huang, Y.-B.; Tang, Y.-X. The R2R3-MYB Transcription Factor Gene Family in Maize. PLoS ONE 2012, 7, e37463. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Suo, J.; Chen, S.; Jin, Y.; Ma, X.; Yin, Z.; Zhang, Y.; Wang, T.; Luo, J.; Jin, W.; et al. Na2CO3-responsive mechanisms in halophyte Puccinellia tenuiflora roots revealed by physiological and proteomic analyses. Sci. Rep. 2016, 6, 32717. [Google Scholar] [CrossRef]
- Ying, Y.; Liu, X.-G.; Lu, Y.; Dou, Y.; Wang, H.-Y.; Han, S.-P.; Feng, S.-D.; Hao, D.-Y. Novel Type II Peroxiredoxin Gene Homologue from Chinese Wildrye Enhancing Salt Stress Tolerance of Transgenic Yeast. Chem. Res. Chin. Univ. 2011, 27, 1000–1005. [Google Scholar]
- Ishida, Y.; Hiei, Y.; Komari, T. Agrobacterium-mediated transformation of maize. Nat. Protoc. 2007, 2, 1614–1621. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Meng, H.; Wen, H.; Fan, Y.; Zhao, J. Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol. Biol. 2011, 75, 365–378. [Google Scholar] [CrossRef]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants. Plant Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide Dismutases: I. Occurrence in Higher Plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef]
- Hernández, J.A.; Jiménez, A.; Mullineaux, P.; Sevilia, F. Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell Environ. 2000, 23, 853–862. [Google Scholar] [CrossRef]
- Aebi, H. [13] Catalase in vitro. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1984; Volume 105, pp. 121–126. ISBN 978-0-12-182005-3. [Google Scholar] [CrossRef]
- Nakano, Y.; Asada, K. Hydrogen Peroxide is Scavenged by Ascorbate-specific Peroxidase in Spinach Chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar] [CrossRef]
- Altaf, M.A.; Hao, Y.; He, C.; Mumtaz, M.A.; Shu, H.; Fu, H.; Wang, Z. Physiological and Biochemical Responses of Pepper (Capsicum annuum L.) Seedlings to Nickel Toxicity. Front. Plant Sci. 2022, 13, 950392. [Google Scholar] [CrossRef]
- Chen, M.-L.; Fu, X.-M.; Liu, J.-Q.; Ye, T.-T.; Hou, S.-Y.; Huang, Y.-Q.; Yuan, B.-F.; Wu, Y.; Feng, Y.-Q. Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC–ESI-Q-TOF-MS analysis. J. Chromatogr. B 2012, 905, 67–74. [Google Scholar] [CrossRef]
- Cheng, H.; Song, S.; Xiao, L.; Soo, H.M.; Cheng, Z.; Xie, D.; Peng, J. Gibberellin Acts through Jasmonate to Control the Expression of MYB21, MYB24, and MYB57 to Promote Stamen Filament Growth in Arabidopsis. PLoS Genet. 2009, 5, e1000440. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- SAS Institute. SAS/IML User’s Guide: Release 6.03 Edition; SAS Institute: Cary, NC, USA, 1988; ISBN 978-1-55544-094-7. [Google Scholar]

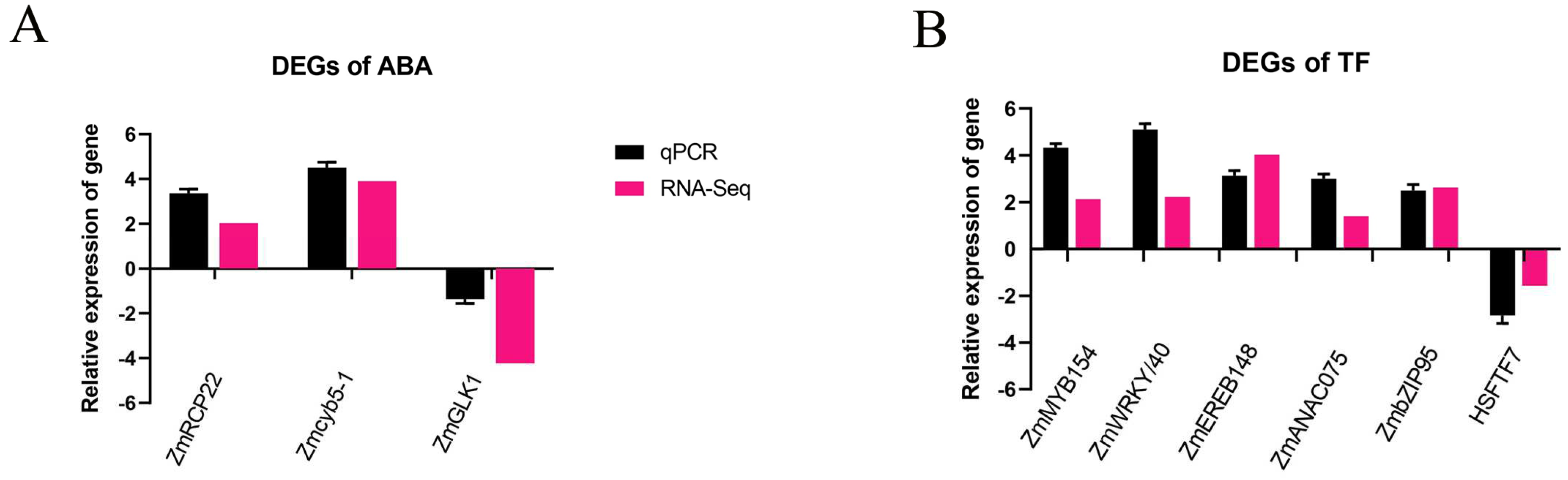
| Group | Gene ID | Log2FC | Swiss_Prot Annotation | NR_Annotation | Reference |
|---|---|---|---|---|---|
| MYB | Zm00001eb396390 | 1.09 | Myb-related protein P OS = Zea mays OX = 4577 GN = P PE = 2 SV = 1 | myb-related protein Myb4 [Zea mays] | [35] |
| WRKY/40 | Zm00001eb419370 | 2.68 | WRKY transcription factor SUSIBA2 OS = Hordeum vulgare OX = 4513 GN = WRKY46 PE = 1 SV = 1 | WRKY transcription factor SUSIBA2-like [Zea mays] | [36] |
| Zm00001eb149570 | 1.24 | WRKY DNA-binding transcription factor 70 OS = Solanum lycopersicum OX = 4081 GN = WRKY70 PE = 2 SV = 1 | WRKY DNA binding domain-containing protein [Zea mays] | [37] | |
| NAC | Zm00001eb121380 | 1.01 | NAC domain-containing protein 75 OS = Arabidopsis thaliana OX = 3702 GN = NAC075 PE = 2 SV = 1 | ANAC075 isoform X2 [Zea mays] | [38] |
| bZIP | Zm00001eb212940 | 1.03 | bZIP transcription factor RISBZ2 OS = Oryza sativa subsp. japonica OX = 39947 GN = RISBZ2 PE = 1 SV = 1 | unknown [Zea mays] | [39] |
| AP2/ERF | Zm00001eb124740 | −1.42 | AP2/ERF and B3 domain-containing protein Os01g0141000 OS = Oryza sativa subsp. japonica OX = 39947 GN = Os01g0141000 PE = 2 SV = 1 | AP2/ERF and B3 domain-containing protein Os01g0141000 [Zea mays] | [36,40] |
| HSF | Zm00001eb198620 | −1.38 | Heat stress transcription factor B-2b OS = Oryza sativa subsp. japonica OX = 39947 GN = HSFB2B PE = 2 SV = 1 | uncharacterized protein LOC100283530 isoform X1 [Zea mays] | [37] |
| Zm00001eb239380 | 1.47 | Heat stress transcription factor C-2a OS = Oryza sativa subsp. japonica OX = 39947 GN = HSFC2A PE = 2 SV = 1 | Unknown [Zea mays] | [41,42] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michael, D.P.J.; Liu, Q.; Yin, Y.; Wei, X.; Lu, J.; Rehman, F.U.; Temitope, A.; Qian, B.; Xia, H.; Han, J.; et al. LcTprxII Overexpression Enhances Physiological and Biochemical Effects in Maize Under Alkaline (Na2CO3) Stress. Plants 2025, 14, 1467. https://doi.org/10.3390/plants14101467
Michael DPJ, Liu Q, Yin Y, Wei X, Lu J, Rehman FU, Temitope A, Qian B, Xia H, Han J, et al. LcTprxII Overexpression Enhances Physiological and Biochemical Effects in Maize Under Alkaline (Na2CO3) Stress. Plants. 2025; 14(10):1467. https://doi.org/10.3390/plants14101467
Chicago/Turabian StyleMichael, David Pitia Julius, Qing Liu, Yuejia Yin, Xuancheng Wei, Jainyu Lu, Faiz Ur Rehman, Aroge Temitope, Buxuan Qian, Hanchao Xia, Jiarui Han, and et al. 2025. "LcTprxII Overexpression Enhances Physiological and Biochemical Effects in Maize Under Alkaline (Na2CO3) Stress" Plants 14, no. 10: 1467. https://doi.org/10.3390/plants14101467
APA StyleMichael, D. P. J., Liu, Q., Yin, Y., Wei, X., Lu, J., Rehman, F. U., Temitope, A., Qian, B., Xia, H., Han, J., Liu, X., Jiang, L., Qi, X., Sun, R., Chen, Z., & Zhang, J. (2025). LcTprxII Overexpression Enhances Physiological and Biochemical Effects in Maize Under Alkaline (Na2CO3) Stress. Plants, 14(10), 1467. https://doi.org/10.3390/plants14101467







