Generation and Functional Characteristics of CRISPR/Cas9-Edited PtrPHOTs Triple-Gene Mutants in Poplar
Abstract
1. Introduction
2. Results
2.1. Characterization of PHOT Genes in Populus
2.2. CRISPR/Cas9-Mediated Generation of PtrPHOT1/2.1/2.2 Gene Mutants in P. trichocarpa
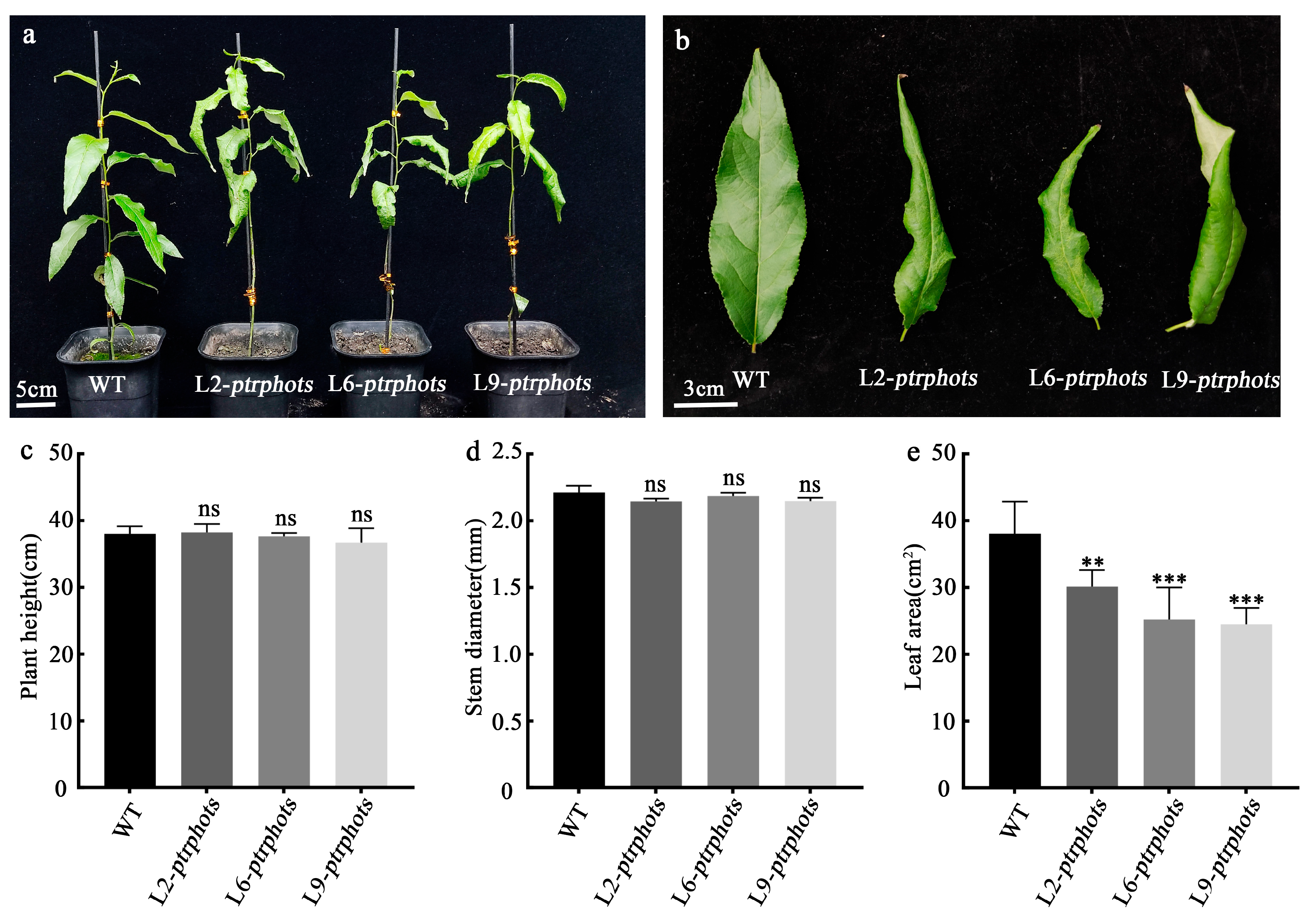
2.3. Phenotypes of Ptrphots Triple-Gene Mutants
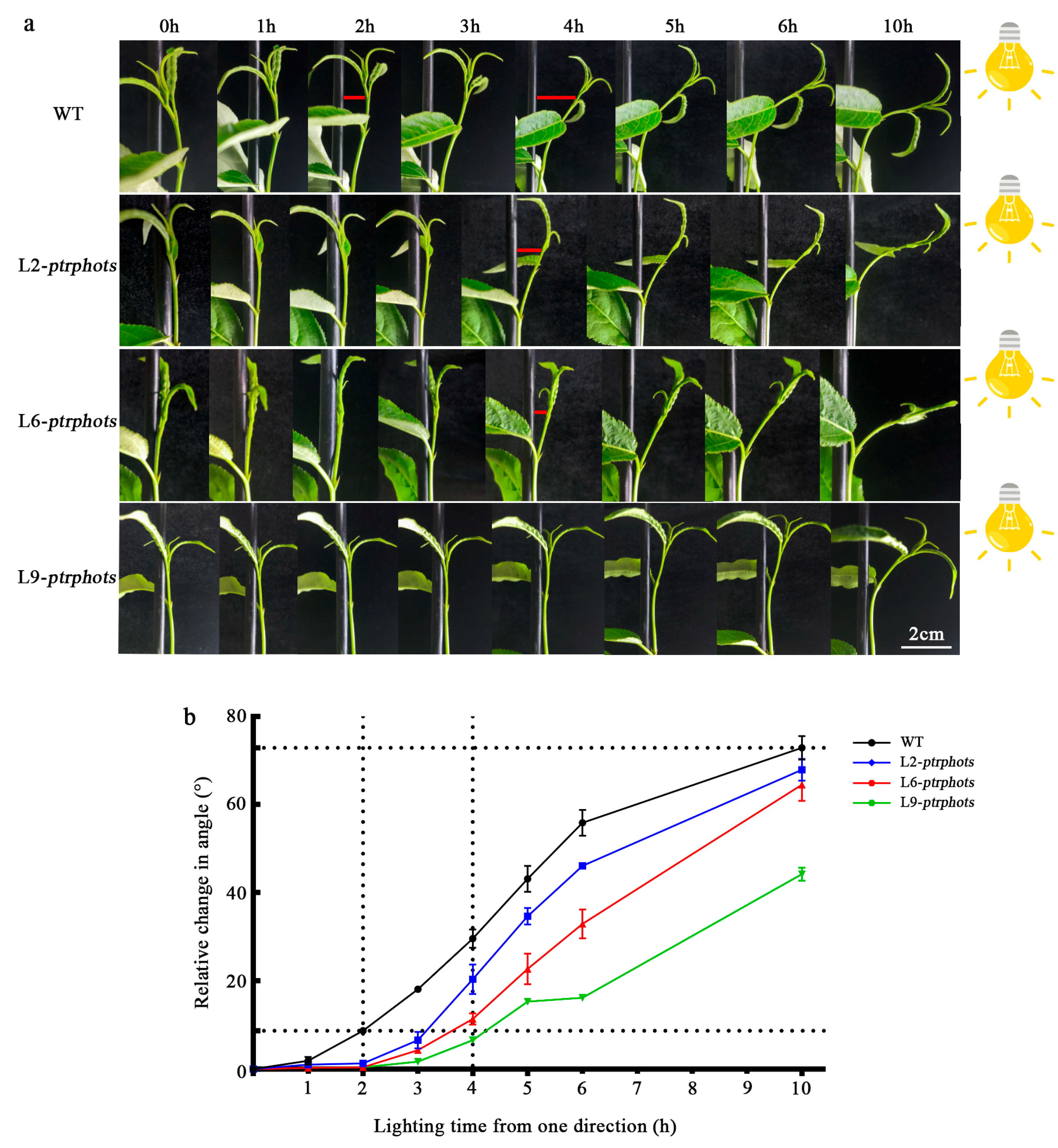
2.4. Knockout of PtrPHOTs Impairs Phototropic Responses in Populus
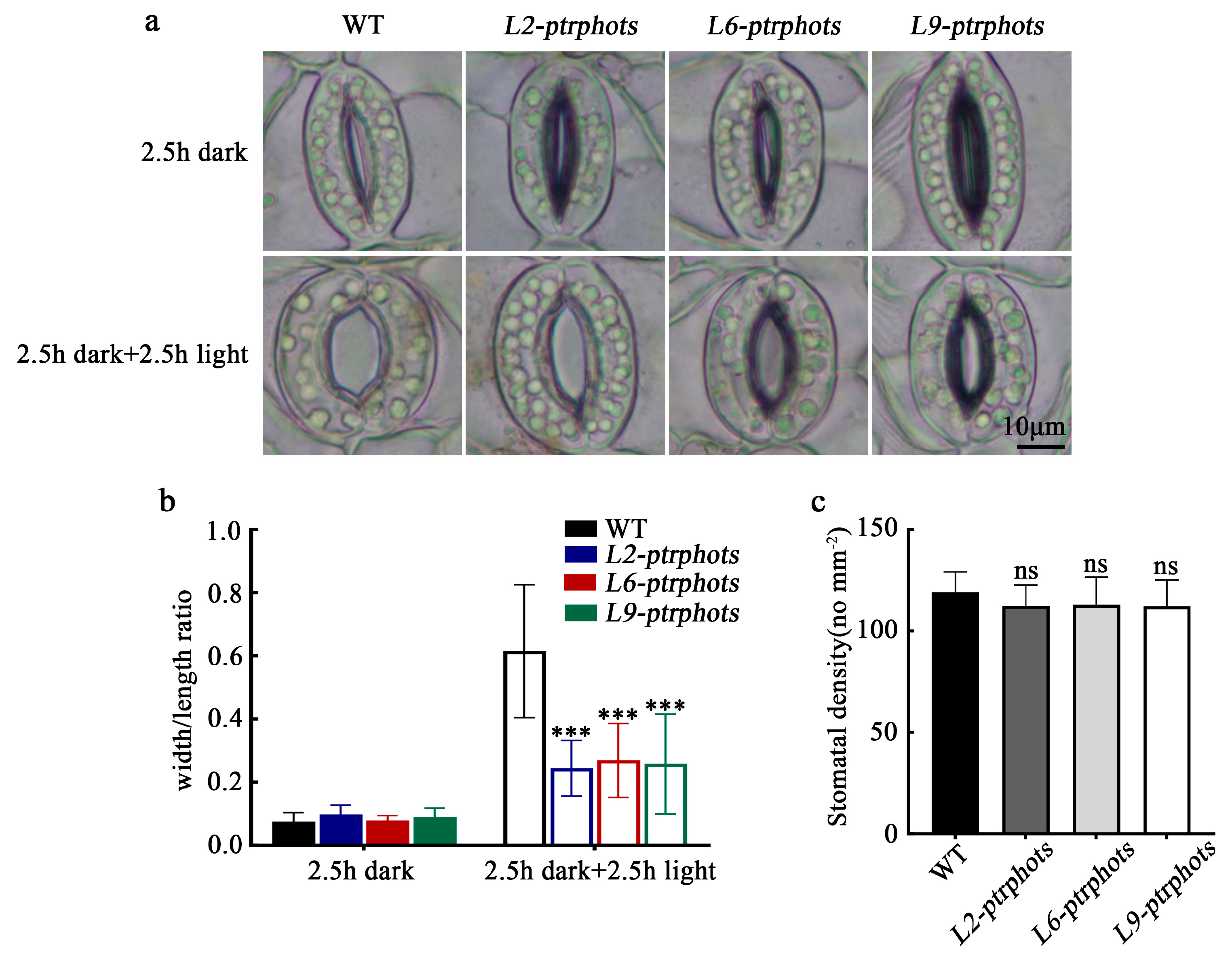
2.5. Effect of PtrPHOTs Knockout on Light-Induced Stomatal Opening in Populus
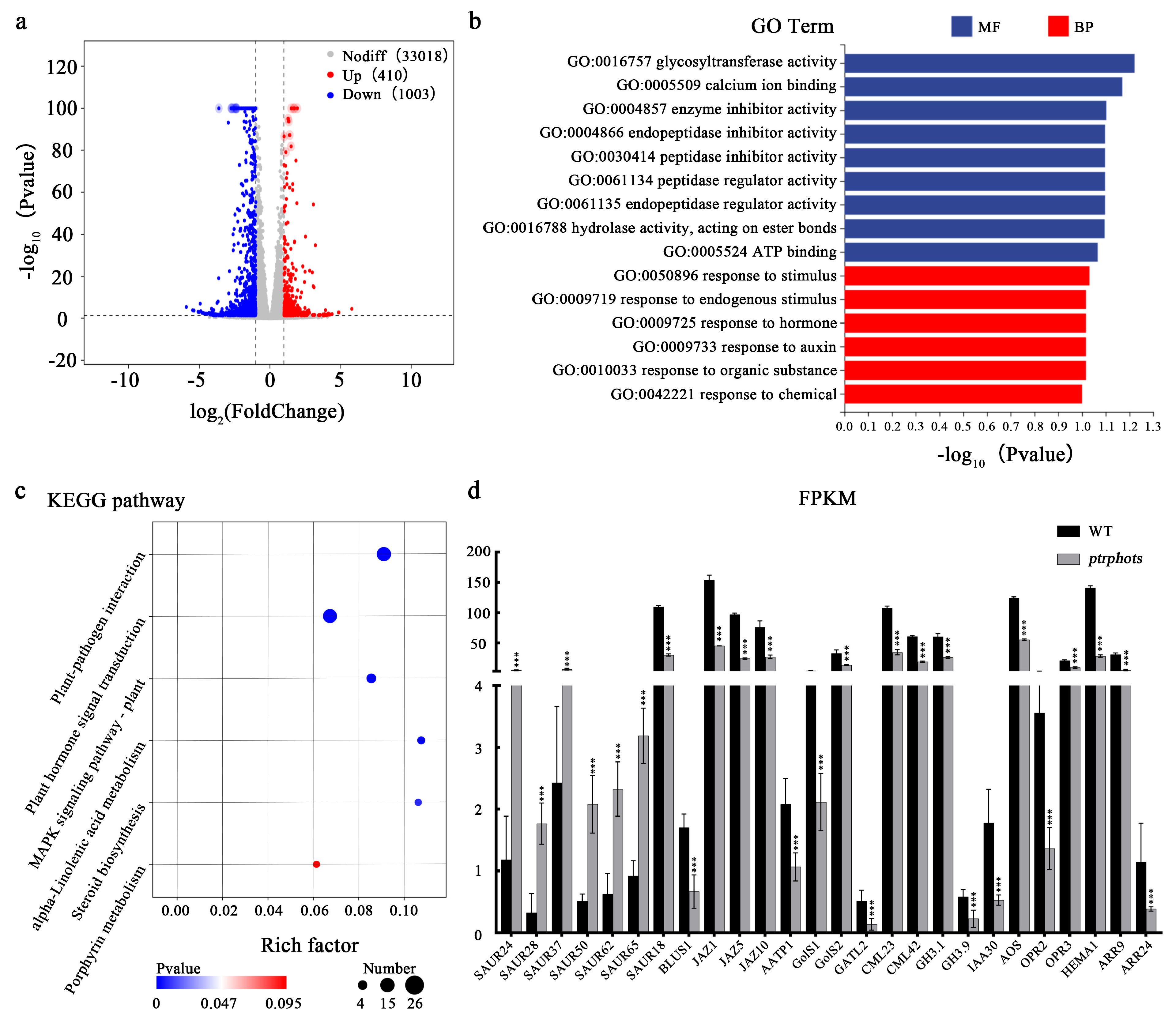
2.6. Transcriptome Analysis Reveals the Impacts of PtrPHOTs Gene Knockout on Leaf Development and Hormonal Homeostasis in Poplar
3. Discussion
3.1. PtrPHOTs Genes Knockout Causes Abnormal Leaf Development and Delayed Phototropic Response
3.2. Stomatal Aperture Defects in the Ptrphots Mutants and the Conservation of the PHOT-BLUS1-H⁺-ATPase Signaling Axis
3.3. Transcriptome Analysis Reveals Potential Mechanisms Underlying Leaf Developmental Changes Induced by PtrPHOTs Knockout
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification of the PHOT Genes in P. trichocarpa
4.3. gRNA Design and Vector Construction
4.4. Generation of Transgenic P. trichocarpa
4.5. Identification of Mutations
4.6. Analyses of Growth and Leaf Area
4.7. Extraction of RNA and RT-PCR and qRT-PCR
4.8. Stomatal Aperture Measurements
4.9. Transcriptome Sequencing
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kami, C.; Lorrain, S.; Hornitschek, P.; Fankhauser, C. Light-Regulated Plant Growth and Development. Curr. Top. Dev. Biol. 2010, 91, 29–66. [Google Scholar] [PubMed]
- de Wit, M.; Galvão, V.C.; Fankhauser, C. Light-Mediated Hormonal Regulation of Plant Growth and Development. Annu. Rev. Plant Biol. 2016, 67, 513–537. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.N.; Kathare, P.K.; Huq, E. Phytochromes and Phytochrome Interacting Factors. Plant Physiol. 2018, 176, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Ponnu, J.; Hoecker, U. Signaling Mechanisms by Arabidopsis Cryptochromes. Front. Plant Sci. 2022, 13, 844714. [Google Scholar] [CrossRef]
- Briggs, W.R.; Christie, J.M. Phototropins 1 and 2: Versatile Plant Blue-Light Receptors. Trends Plant Sci. 2002, 7, 204–210. [Google Scholar] [CrossRef]
- Suetsugu, N.; Wada, M. Evolution of Three LOV Blue Light Receptor Families in Green Plants and Photosynthetic Stramenopiles: Phototropin, ZTL/FKF1/LKP2 and Aureochrome. Plant Cell Physiol. 2013, 54, 8–23. [Google Scholar] [CrossRef]
- Liang, T.; Yang, Y.; Liu, H. Signal Transduction Mediated by the Plant UV-B Photoreceptor UVR8. New Phytol. 2019, 221, 1247–1252. [Google Scholar] [CrossRef]
- Goyal, A.; Szarzynska, B.; Fankhauser, C. Phototropism: At the Crossroads of Light-Signaling Pathways. Trends Plant Sci. 2013, 18, 393–401. [Google Scholar] [CrossRef]
- Fankhauser, C.; Christie, J.M. Plant Phototropic Growth. Curr. Biol. 2015, 25, R384–R389. [Google Scholar] [CrossRef]
- Yang, J.; Li, C.; Kong, D.; Guo, F.; Wei, H. Light-Mediated Signaling and Metabolic Changes Coordinate Stomatal Opening and Closure. Front. Plant Sci. 2020, 11, 601478. [Google Scholar] [CrossRef]
- Kinoshita, T.; Doi, M.; Suetsugu, N.; Kagawa, T.; Wada, M.; Shimazaki, K.I. phot1 and phot2 Mediate Blue Light Regulation of Stomatal Opening. Nature 2001, 414, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Wada, M. Chloroplast Movement. Plant Sci. 2013, 210, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Łabuz, J.; Sztatelman, O.; Hermanowicz, P. Molecular Insights into the Phototropin Control of Chloroplast Movements. J. Exp. Bot. 2022, 73, 6034–6051. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, T.; Sakai, T.; Suetsugu, N.; Oikawa, K.; Ishiguro, S.; Kato, T.; Tabata, S.; Okada, K.; Wada, M. Arabidopsis NPL1: A Phototropin Homolog Controlling the Chloroplast High-Light Avoidance Response. Science 2001, 291, 2138–2141. [Google Scholar] [CrossRef]
- Flores-Ibarra, A.; Maia, R.N.A.; Olasz, B.; Church, J.R.; Gotthard, G.; Schapiro, I.; Heberle, J.; Nogly, P. Light-Oxygen-Voltage (LOV)-Sensing Domains: Activation Mechanism and Optogenetic Stimulation. J. Mol. Biol. 2024, 436, 168356. [Google Scholar] [CrossRef]
- Sakai, T.; Kagawa, T.; Kasahara, M.; Swartz, T.E.; Christie, J.M.; Briggs, W.R.; Wada, M.; Okada, K. Arabidopsis nph1 and npl1: Blue Light Receptors that Mediate both Phototropism and Chloroplast Relocation. Proc. Natl. Acad. Sci. USA 2001, 98, 6969–6974. [Google Scholar] [CrossRef]
- Hart, J.E.; Gardner, K.H. Lighting the Way: Recent Insights into the Structure and Regulation of Phototropin Blue Light Receptors. J. Biol. Chem. 2021, 296, 100594. [Google Scholar] [CrossRef]
- Hohm, T.; Preuten, T.; Fankhauser, C. Phototropism: Translating Light into Directional Growth. Am. J. Bot. 2013, 100, 47–59. [Google Scholar] [CrossRef]
- Goh, C.H.; Jang, S.; Jung, S.; Kim, H.S.; Kang, H.G.; Park, Y.I.; Bae, H.J.; Lee, C.H.; An, G. Rice phot1a Mutation Reduces Plant Growth by Affecting Photosynthetic Responses to Light during Early Seedling Growth. Plant Mol. Biol. 2009, 69, 605–619. [Google Scholar] [CrossRef]
- Kanegae, H.; Tahir, M.; Savazzini, F.; Yamamoto, K.; Yano, M.; Sasaki, T.; Kanegae, T.; Wada, M.; Takano, M. Rice NPH1 Homologues, OsNPH1a and OsNPH1b, are Differently Photoregulated. Plant Cell Physiol. 2000, 41, 415–423. [Google Scholar] [CrossRef]
- Takemiya, A.; Sugiyama, N.; Fujimoto, H.; Tsutsumi, T.; Yamauchi, S.; Hiyama, A.; Tada, Y.; Christie, J.M.; Shimazaki, K.I. Phosphorylation of BLUS1 Kinase by Phototropins is a Primary Step in Stomatal Opening. Nat. Commun. 2013, 4, 2094. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Gray, W.M. SAUR Proteins as Effectors of Hormonal and Environmental Signals in Plant Growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jin, D.; Deng, Z.; Zheng, L.; Guo, P.; Ji, Y.; Song, Z.; Zeng, H.Y.; Kinoshita, T.; Liao, Z.; et al. The Apoplastic pH is a Key Determinant in the Hypocotyl Growth Response to Auxin Dosage and Light. Nat. Plants 2025, 11, 279–294. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar]
- Xu, W.; Cheng, H.; Zhu, S.; Cheng, J.; Ji, H.; Zhang, B.; Cao, S.; Wang, C.; Tong, G.; Zhen, C.; et al. Functional Understanding of Secondary Cell Wall Cellulose Synthases in Populus trichocarpa via the Cas9/gRNA-Induced Gene Knockouts. New Phytol. 2021, 231, 1478–1495. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential Gene and Transcript Expression Analysis of RNA-seq Experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Demarsy, E.; Fankhauser, C. Higher Plants use LOV to Perceive Blue Light. Curr. Opin. Plant Biol. 2009, 12, 69–74. [Google Scholar] [CrossRef]
- de Carbonnel, M.; Davis, P.; Roelfsema, M.R.; Inoue, S.I.; Schepens, I.; Lariguet, P.; Geisler, M.; Shimazaki, K.I.; Hangarter, R.; Fankhauser, C. The Arabidopsis PHYTOCHROME KINASE SUBSTRATE2 Protein is a Phototropin Signaling Element that Regulates Leaf Flattening and Leaf Positioning. Plant Physiol. 2010, 152, 1391–1405. [Google Scholar] [CrossRef]
- Harada, A.; Takemiya, A.; Inoue, S.; Sakai, T.; Shimazaki, K.I. Role of RPT2 in Leaf Positioning and Flattening and a Possible Inhibition of phot2 Signaling by phot1. Plant Cell Physiol. 2013, 54, 36–47. [Google Scholar] [CrossRef]
- Kong, Y.; Zheng, Y. Phototropin is Partly Involved in Blue-Light-Mediated Stem Elongation, Flower Initiation, and Leaf Expansion: A Comparison of Phenotypic Responses between Wild Arabidopsis and its Phototropin Mutants. Environ. Exp. Bot. 2020, 171, 103967. [Google Scholar] [CrossRef]
- Tsuchida-Mayama, T.; Sakai, T.; Hanada, A.; Uehara, Y.; Asami, T.; Yamaguchi, S. Role of the Phytochrome and Cryptochrome Signaling Pathways in Hypocotyl Phototropism. Plant J. 2010, 62, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Ohgishi, M.; Saji, K.; Okada, K.; Sakai, T. Functional Analysis of Each Blue Light Receptor, cry1, cry2, phot1, and phot2, by using Combinatorial Multiple Mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 2223–2228. [Google Scholar] [CrossRef] [PubMed]
- Mott, K.A.; Sibbernsen, E.D.; Shope, J.C. The Role of the Mesophyll in Stomatal Responses to Light and CO2. Plant Cell Environ. 2008, 31, 1299–1306. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, N.; Takami, T.; Ebisu, Y.; Watanabe, H.; Iiboshi, C.; Doi, M.; Shimazaki, K.I. Guard Cell Chloroplasts are Essential for Blue Light-Dependent Stomatal Opening in Arabidopsis. PLoS ONE 2014, 9, e108374. [Google Scholar] [CrossRef]
- Inoue, S.I.; Kinoshita, T. Blue Light Regulation of Stomatal Opening and the Plasma Membrane H (+)-ATPase. Plant Physiol. 2017, 174, 531–538. [Google Scholar] [CrossRef]
- Mao, J.; Zhang, Y.C.; Sang, Y.; Li, Q.H.; Yang, H.Q. A Role for Arabidopsis Cryptochromes and COP1 in the Regulation of Stomatal Opening. Proc. Natl. Acad. Sci. USA 2005, 102, 12270–12275. [Google Scholar] [CrossRef]
- Wang, F.F.; Lian, H.L.; Kang, C.Y.; Yang, H.Q. Phytochrome B is Involved in Mediating Red Light-Induced Stomatal Opening in Arabidopsis thaliana. Mol. Plant 2010, 3, 246–259. [Google Scholar] [CrossRef]
- Liu, C.; Wang, Y. Plant Auxin Responsive Gene SAUR: A Review. Chin. Agric. Sci. Bull. 2024, 40, 83–89. [Google Scholar]
- Du, M.; Spalding, E.P.; Gray, W.M. Rapid Auxin-Mediated Cell Expansion. Annu. Rev. Plant Biol. 2020, 71, 379–402. [Google Scholar] [CrossRef]
- Pauwels, L.; Goossens, A. The JAZ Proteins: A Crucial Interface in the Jasmonate Signaling Cascade. Plant cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic Acid Signaling Pathway in Plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xiao, L.; Qin, S.; Kuang, Z.; Wan, M.; Li, Z.; Li, L. Heterogeneity in Mechanical Properties of Plant Cell Walls. Plants 2024, 13, 3561. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, H.E.; Thom, E.; Möhlmann, T.; Steup, M.; Kampfenkel, K. Characterization of a Novel Eukaryotic ATP/ADP Translocator Located in the Plastid Envelope of Arabidopsis thaliana L. Plant J. 1997, 11, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhen, C.; Xu, W.; Wang, C.; Cheng, Y. Simple, Rapid and Efficient Transformation of Genotype Nisqually-1: A Basic Tool for the First Sequenced Model Tree. Sci. Rep. 2017, 7, 2638. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent Updates, New Developments and Status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 Toolkit for Multiplex Genome Editing in Plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef]
- Xu, C.; Fu, X.; Liu, R.; Guo, L.; Ran, L.; Li, C.; Tian, Q.; Jiao, B.; Wang, B.; Luo, K. PtoMYB170 Positively Regulates Lignin Deposition during Wood Formation in Poplar and Confers Drought Tolerance in Transgenic Arabidopsis. Tree Physiol. 2017, 37, 1713–1726. [Google Scholar] [CrossRef]


| Name | Gene ID | Protein Length (aa) | LOV Number | STK Number |
|---|---|---|---|---|
| AtPHOT1 | AT3G45780 | 996 | 2 | 1 |
| AtPHOT2 | AT5G58140 | 915 | 2 | 1 |
| PtrPHOT1 | Potri.001G342000 | 991 | 2 | 1 |
| PtrPHOT2.1 | Potri.009G170600 | 985 | 2 | 1 |
| PtrPHOT2.2 | Potri.004G209700 | 1000 | 2 | 1 |
| ZmPHOT1 | Zm00001d044599 | 911 | 2 | 1 |
| ZmPHOT2 | Zm00001d032353 | 932 | 2 | 1 |
| OsPHOT1a | LOC_Os11g01140 | 921 | 2 | 1 |
| OsPHOT1b | LOC_Os12g01140 | 921 | 2 | 1 |
| OsPHOT2 | LOC_Os04g23890 | 907 | 2 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, H.; Cheng, J.; Jing, Y.; Zhu, S.; Wang, C.; Cheng, Y. Generation and Functional Characteristics of CRISPR/Cas9-Edited PtrPHOTs Triple-Gene Mutants in Poplar. Plants 2025, 14, 1455. https://doi.org/10.3390/plants14101455
Yao H, Cheng J, Jing Y, Zhu S, Wang C, Cheng Y. Generation and Functional Characteristics of CRISPR/Cas9-Edited PtrPHOTs Triple-Gene Mutants in Poplar. Plants. 2025; 14(10):1455. https://doi.org/10.3390/plants14101455
Chicago/Turabian StyleYao, Hongtao, Jiyao Cheng, Yuning Jing, Siran Zhu, Chong Wang, and Yuxiang Cheng. 2025. "Generation and Functional Characteristics of CRISPR/Cas9-Edited PtrPHOTs Triple-Gene Mutants in Poplar" Plants 14, no. 10: 1455. https://doi.org/10.3390/plants14101455
APA StyleYao, H., Cheng, J., Jing, Y., Zhu, S., Wang, C., & Cheng, Y. (2025). Generation and Functional Characteristics of CRISPR/Cas9-Edited PtrPHOTs Triple-Gene Mutants in Poplar. Plants, 14(10), 1455. https://doi.org/10.3390/plants14101455






