Research Progress of CLE and Its Prospects in Woody Plants
Abstract
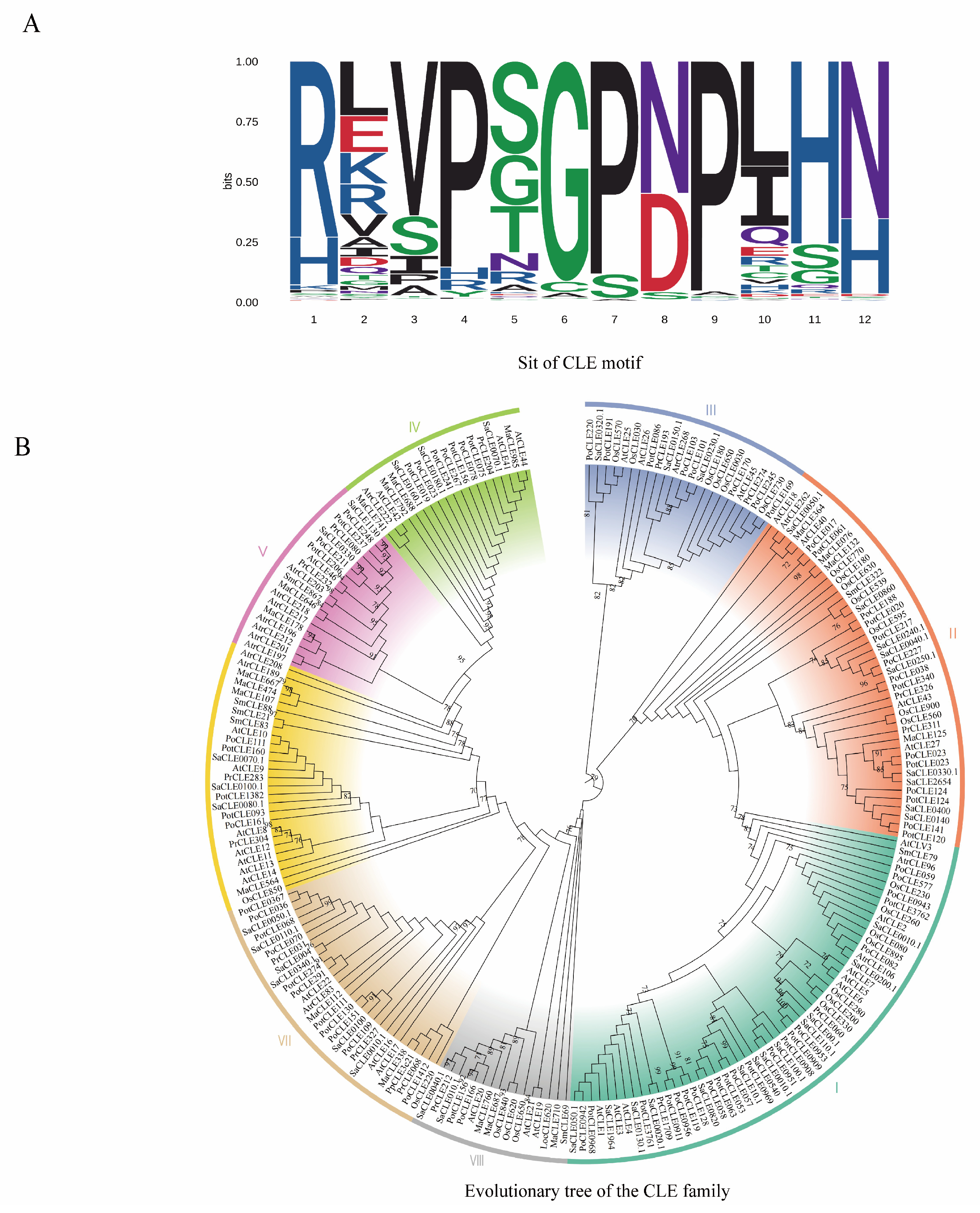
1. Introduction
2. Development and Maintenance of CLEs in Root Apical Meristem
3. Development and Maintenance of CLEs in Shoot Apical Meristem
3.1. CLV3-CLV1
3.2. CLV3-CLV2-CRN/SOL2
3.3. CLV3-RPK2
3.4. CLV3-BAMs
4. Development and Maintenance of CLEs in Stem and Root Cambium
5. Development and Maintenance of CLEs in Leaf
6. Development and Maintenance of CLEs in Floral Meristem
7. Compensation Mechanism of CLEs in Plant
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CLAVATA 1 | CLV1 | CLE7 | CLV3/ESR1-LIKE7 | CLE14 | CLV3/ESR1-LIKE14 | CLE40 | CLV3/ESR1-LIKE 40 |
| CLAVATA 2 | CLV2(CRN) | CLE9 | CLV3/ESR1-LIKE9 | CLE19 | CLV3/ESR1-LIKE19 | PtCLE41 | Populus trichocarpa CLV3/ESR1-LIKE41 |
| CLV3 | CLAVATA3 | CLE10 | CLV3/ESR1-LIKE10 | PtCLE20 | Populus trichocarpa CLV3/ESR1-LIKE20 | PtCLE42 | Populus trichocarpa CLV3/ESR1-LIKE42 |
| CLE4 | CLV3/ESR1-LIKE4 | CLE11 | CLV3/ESR1-LIKE11 | CLE25 | CLV3/ESR1-LIKE25 | PtCLE44 | Populus trichocarpa CLV3/ESR1-LIKE44 |
| CLE5 | CLV3/ESR1-LIKE5 | CLE12 | CLV3/ESR1-LIKE12 | CLE26 | CLV3/ESR1-LIKE26 | CLE45 | CLV3/ESR1-LIKE45 |
| CLE6 | CLV3/ESR1-LIKE6 | CLE13 | CLV3/ESR1-LIKE13 | CLE33 | CLV3/ESR1-LIKE33 | PtCLE47 | Populus trichocarpa CLV3/ESR1-LIKE47 |
| FAS1/FAS2 | FASCIATA1 and FASCIATA1 | STM | SHOOT MERISTEMLESS | WUS | WUSCHEL | TDIF | tracheary element differentiation inhibitory factor |
| RAM | Root Apical Meristem | SCN | stem cell niche | QC | quiescent center | CSC | columella stem cells |
| CC | columella cells | CIKs | CLAVATA3 INSENSITIVE RECEPTOR KINASES | ACR4 | ARABIDOPSIS CRINKLY4 | WOX5 | WUS-RELATED HOMEOBOX5 |
| SAN | Shoot Apical Meristem | OC | organizing center | PZ | peripheral zone | RM | rib meristem |
| HAM | HAIRY MERISRTEM | LRR-RLKs | LEUCINE-RICH REPEAT RECEPTOR-LIKE KINASES | SOL2 | SUPPRESSOR OF LLP1 2 | PM | plasma membrane |
| ER | endoplasmic reticulum | ER | the transmembrane | BAM | BARELY ANY MERISTEM | POL | POLTERGEIST |
| RPK2 | RECEPTOR-LIKE PROTEIN KINASE2 | PC | procambium | PPh | protophloem | Pxy | protoxylem |
| MC | metacambium | TDR/PXY | TDIF RECEPTOR/PHLOEM INTERCALATED WITH XYLEM | GSK3s | GLYCOGEN SYNTHASE KINASE 3 PROTEINS | BES1 | BRI1-EMS SUPPRESSOR 1 |
| VCM2/PIN5 | VASCULAR CAMBIUM-RELATED MADS2/PIN-FORMED5 | CLERK-CLV2 | TOR KINASE -CLV2 | MMCs | meristemoid mother cells | PIN1 | PIN-FORMED1 |
| TEs | tracheary elements | BRX | BREVIS RADIX | RPK2 | RECEPTOR-LIKE PROTEIN KINASE2 | SEs | sieve element |
| ARFs | AUXIN RESPONSE FACTORS | BRM | BRAHMA | ESR1/DRN | ENHANCER OF SHOOT REGENERATION1/DORNROSCHEN | MAPK | MEMBRANE-ASSOCIATED KINASE REGULATOR 5 |
| HSL1 | HAESA-LIKE1 | AM | axillary meristem | ACS | THE ENZYME 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE | EBF1/2 | ETHYLENE-INSENSITIVE3 (EIN3)-binding F-BOX1/2 |
| EIN3 | ETH-YLENE-INSENSITIVE3 | FM | Floral Meristem | IM | inflorescence meristem | PLL1 | POL-LIKE1 |
| TD1 | THICK TASSEL DWARF | ERF2 | FASCIATED EAR2 | YUC | YUCCA | CLE42 | CLV3/ESR1-LIKE42 |
References
- Khavinson, V.; Linkova, N.; Diatlova, A.; Dudkov, A. Peptide regulation of plant cells differentiation and growth. BIO Web Conf. 2024, 82, 02003. [Google Scholar] [CrossRef]
- Baskin, C.C.; Baskin, J.M. The rudimentary embryo: An early angiosperm invention that contributed to their dominance over gymnosperms. Seed Sci. Res. 2023, 33, 63–74. [Google Scholar] [CrossRef]
- Kathryn Barton, M. Cell type specification and self renewal in the vegetative shoot apical meristem. Curr. Opin. Plant Biol. 1998, 1, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Khan, N.; Xie, L. Molecular and hormonal regulation of leaf morphogenesis in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 5132. [Google Scholar] [CrossRef]
- Dinneny, J.R.; Benfey, P.N. Plant stem cell niches: Standing the test of time. Cell 2008, 132, 553–557. [Google Scholar] [CrossRef]
- Scofield, S.; Murray, J.A.H. KNOX gene function in plant stem cell niches. Plant Mol. Biol. 2006, 60, 929–946. [Google Scholar] [CrossRef]
- Scofield, S.; Dewitte, W.; Murray, J.A. STM sustains stem cell function in the Arabidopsis shoot apical meristem and controls KNOX gene expression independently of the transcriptional repressor AS1. Plant Signal. Behav. 2014, 9, e28934. [Google Scholar] [CrossRef]
- Kwon, C.S.; Chen, C.; Wagner, D. WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 2005, 19, 992–1003. [Google Scholar] [CrossRef]
- Singh, S.; Singh, A.; Singh, A.; Yadav, S.; Bajaj, I.; Kumar, S.; Jain, A.; Sarkar, A.K. Role of chromatin modification and remodeling in stem cell regulation and meristem maintenance in Arabidopsis. J. Exp. Bot. 2020, 71, 778–792. [Google Scholar] [CrossRef]
- Kaya, H.; Shibahara, K.I.; Taoka, K.I.; Iwabuchi, M.; Stillman, B.; Araki, T. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 2001, 104, 131–142. [Google Scholar] [CrossRef]
- Cock, J.M.; McCormick, S. A large family of genes that share homology with CLAVATA3. Plant Physiol. 2001, 126, 939–942. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, Y.; Yu, X.; Wu, Q.; Man, X.; Diao, Z.; You, H.; Shen, J.; Cai, Y. Identification and application of CLE peptides for drought resistance in Solanaceae Crops. J. Agric. Food Chem. 2024, 72, 13869–13884. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Gao, X.; Wen, L.; Deng, Z.; Liu, T.; Guo, Y. Characterization of the CLE Family in three nicotiana species and potential roles of CLE peptides in osmotic and salt stress responses. Agronomy 2023, 13, 1480. [Google Scholar] [CrossRef]
- Gao, X.; Guo, Y. CLE Peptides in Plants: Proteolytic processing, structure-activity relationship, and ligand-receptor interaction. J. Integr. Plant Biol. 2012, 54, 738–745. [Google Scholar] [CrossRef]
- Murphy, E.; Smith, S.; De Smet, I. Small signaling peptides in Arabidopsis development: How cells communicate over a short distance. Plant Cell 2012, 24, 3198–3217. [Google Scholar] [CrossRef]
- Olsen, A. Ligand mimicry? Plant-parasitic nematode polypeptide with similarity to CLAVATA3. Trends Plant Sci. 2003, 8, 55–57. [Google Scholar] [CrossRef]
- Clark, S.E.; Running, M.P.; Meyerowitz, E.M. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 1993, 119, 397–418. [Google Scholar] [CrossRef]
- Clark, S.E.; Running, M.P.; Meyerowitz, E.M. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 1995, 121, 2057–2067. [Google Scholar] [CrossRef]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Rojo, E.; Sharma, V.K.; Kovaleva, V.; Raikhel, N.V.; Fletcher, J.C. CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 2002, 14, 969–977. [Google Scholar] [CrossRef]
- Ito, Y.; Nakanomyo, I.; Motose, H.; Iwamoto, K.; Sawa, S.; Dohmae, N.; Fukuda, H. Dodeca-CLE Peptides as suppressors of plant stem cell differentiation. Science 2006, 313, 842–845. [Google Scholar] [CrossRef] [PubMed]
- Somssich, M.; Je, B.I.; Simon, R.; Jackson, D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development 2016, 143, 3238–3248. [Google Scholar] [CrossRef]
- Lopes, F.L.; Galvan-Ampudia, C.; Landrein, B. WUSCHEL in the shoot apical meristem: Old player, new tricks. J. Exp. Bot. 2021, 72, 1527–1535. [Google Scholar] [CrossRef]
- Stahl, Y.; Wink, R.H.; Ingram, G.C.; Simon, R. A Signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 2009, 19, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Whitford, R.; Fernandez, A.; De Groodt, R.; Ortega, E.; Hilson, P. Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc. Natl. Acad. Sci. USA 2008, 105, 18625–18630. [Google Scholar] [CrossRef]
- Fukuda, H.; Hirakawa, Y.; Sawa, S. Peptide signaling in vascular development. Curr. Opin. Plant Biol. 2007, 10, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Gancheva, M.S.; Losev, M.R.; Dodueva, I.E.; Lutova, L.A. Phloem-expressed CLAVATA3/ESR-like genes in Potato. Horticulturae 2023, 9, 1265. [Google Scholar] [CrossRef]
- Skripnikov, A. Bioassays for Identifying and Characterizing Plant Regulatory Peptides. Biomolecules 2023, 13, 1795. [Google Scholar] [CrossRef]
- Xu, S.; Zhai, X. Research progress on genetic transformation of woody plants. Henan For. Sci. Technol. 2021, 41, 10–13. [Google Scholar] [CrossRef]
- Li, W.F.; Ding, Q.; Chen, J.J.; Cui, K.M.; He, X.Q. Induction of PtoCDKB and PtoCYCB transcription by temperature during cambium reactivation in Populus tomentosa Carr. J. Exp. Bot. 2009, 60, 2621–2630. [Google Scholar] [CrossRef]
- Baba, K.; Karlberg, A.; Schmidt, J.; Schrader, J.; Hvidsten, T.R.; Bako, L.; Bhalerao, R.P. Activity-dormancy transition in the cambial meristem involves stage-specific modulation of auxin response in hybrid aspen. Proc. Natl. Acad. Sci. USA 2011, 108, 3418–3423. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Ye, Z.H. Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol. 2010, 152, 1044–1055. [Google Scholar] [CrossRef]
- Liu, X.; Li, J.; Huang, M.; Chen, J. Mechanisms for the influence of citrus rootstocks on fruit size. J. Agric. Food Chem. 2015, 63, 2618–2627. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Wang, K.; Wang, Y.; Hu, Z.; Yan, C.; Huang, H.; Ma, X.; Cao, Y.; Long, W.; Liu, W.; et al. The genome of oil-Camellia and population genomics analysis provide insights into seed oil domestication. Genome Biol. 2022, 23, 14. [Google Scholar] [CrossRef] [PubMed]
- Trujillo-Moya, C.; Ganthaler, A.; Stöggl, W.; Kranner, I.; Schüler, S.; Ertl, R.; Schlosser, S.; George, J.P.; Mayr, S. RNA-Seq and secondary metabolite analyses reveal a putative defence-transcriptome in Norway spruce (Picea abies) against needle bladder rust (Chrysomyxa rhododendri) infection. BMC Genom. 2020, 21, 336. [Google Scholar] [CrossRef]
- Wang, L.Q.; Wen, S.S.; Wang, R.; Wang, C.; Gao, B.; Lu, M.Z. PagWOX11/12a activates PagCYP736A12 gene that facilitates salt tolerance in poplar. Plant Biotechnol. J. 2021, 19, 2249–2260. [Google Scholar] [CrossRef]
- Han, H.; Zhang, G.; Wu, M.; Wang, G. Identification and characterization of the Populus trichocarpa CLE family. BMC Genom. 2016, 17, 174. [Google Scholar] [CrossRef]
- Zhao, S. Identification of the CLE gene family in Camellia oleifera. South For. Sci. 2023, 51, 7–11. [Google Scholar] [CrossRef]
- Cheng, M.; Li, X.; Wang, P.; Zhang, H.; Zhang, S.; Wu, J. Identification of the CLE peptide family in Rosaceae fruit trees and functional analysis of PbrCLE31 in regulating pollen tube growth in pear. J. Nanjing Agric. Univ. 2021, 44, 850–861. [Google Scholar]
- Brumfield, R.T. Cell-lineage studies in root meristems by means of chromosome rearrangements induced by x-rays. Am. J. Bot. 1943, 30, 101–110. [Google Scholar]
- Zhang, H.; Mu, Y.; Zhang, H.; Yu, C. Maintenance of stem cell activity in plant development and stress responses. Front. Plant Sci. 2023, 14, 1302046. [Google Scholar] [CrossRef]
- Lee, Y.; Lee, W.S.; Kim, S.-H. Hormonal regulation of stem cell maintenance in roots. J. Exp. Bot. 2013, 64, 1153–1165. [Google Scholar] [CrossRef] [PubMed]
- Kumpf, R.P.; Nowack, M.K. The root cap: A short story of life and death. J. Exp. Bot. 2015, 66, 5651–5662. [Google Scholar] [CrossRef]
- Dolan, L.; Janmaat, K.; Willemsen, V.; Linstead, P.; Poethig, S.; Roberts, K.; Scheres, B. Cellular organisation of the Arabidopsis thaliana root. Development 1993, 119, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.P.; Sozzani, R. Uncovering the networks involved in stem cell maintenance and asymmetric cell division in the Arabidopsis root. Curr. Opin. Plant Biol. 2015, 29, 38–43. [Google Scholar] [CrossRef]
- Han, H.; Liu, X.; Zhou, Y. Transcriptional circuits in control of shoot stem cell homeostasis. Curr. Opin. Plant Biol. 2019, 53, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Song, X.-F.; Hou, X.-L.; Liu, C.-M. CLE peptides: Critical regulators for stem cell maintenance in plants. Planta 2021, 255, 5. [Google Scholar] [CrossRef]
- Wen, Y.; Yang, Y.; Liu, J.; Han, H. CLV3-CLV1 signaling governs flower primordia outgrowth across environmental temperatures. Trends Plant Sci. 2024, 29, 400–402. [Google Scholar] [CrossRef]
- Olt, P.; Ding, W.; Schulze, W.X.; Ludewig, U. The LaCLE35 peptide modifies rootlet density and length in cluster roots of white lupin. Plant Cell Environ. 2024, 47, 1416–1431. [Google Scholar] [CrossRef]
- Hobe, M.; Müller, R.; Grünewald, M.; Brand, U.; Simon, R. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev. Genes Evol. 2003, 213, 371–381. [Google Scholar] [CrossRef]
- Zhu, Y.; Hu, C.; Cui, Y.; Zeng, L.; Li, S.; Zhu, M.; Meng, F.; Huang, S.; Long, L.; Yi, J.; et al. Conserved and differentiated functions of CIK receptor kinases in modulating stem cell signaling in Arabidopsis. Mol. Plant 2021, 14, 1119–1134. [Google Scholar] [CrossRef] [PubMed]
- Berckmans, B.; Kirschner, G.; Gerlitz, N.; Stadler, R.; Simon, R. CLE40 signaling regulates root stem cell fate. Plant Physiol. 2020, 182, 1776–1792. [Google Scholar] [CrossRef] [PubMed]
- Gancheva, M.S.; Lutova, L.A. Nitrogen-activated CLV3/ESR-Related 4 (CLE4) regulates shoot, root, and stolon growth in Potato. Plants 2023, 12, 3468. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, S.; Aoyama, T.; Sato, Y.; Kajiwara, T.; Ishida, T.; Sawa, S. CLE3 and its homologs share overlapping functions in the modulation of lateral root formation through CLV1 and BAM1 in Arabidopsis thaliana. Plant J. 2023, 113, 1176–1191. [Google Scholar] [CrossRef]
- Miwa, H.; Betsuyaku, S.; Iwamoto, K.; Kinoshita, A.; Fukuda, H.; Sawa, S. The receptor-like kinase SOL2 mediates CLE Signaling in Arabidopsis. Plant Cell Physiol. 2008, 49, 1752–1757. [Google Scholar] [CrossRef]
- Fiers, M.; Golemiec, E.; Xu, J.; van der Geest, L.; Heidstra, R.; Stiekema, W.; Liu, C.-M. The 14–amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through aCLAVATA2-Dependent Pathway. Plant Cell 2005, 17, 2542–2553. [Google Scholar] [CrossRef]
- Casamitjana-Martínez, E.; Hofhuis, H.F.; Xu, J.; Liu, C.-M.; Heidstra, R.; Scheres, B. Root-specific CLE19 overexpression and the SOL1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 2003, 13, 1435–1441. [Google Scholar] [CrossRef]
- Fiers, M.; Hause, G.; Boutilier, K.; Casamitjana-Martinez, E.; Weijers, D.; Offringa, R.; van der Geest, L.; van Lookeren Campagne, M.; Liu, C.-M. Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 2004, 327, 37–49. [Google Scholar] [CrossRef]
- Barton, M.K. Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 2010, 341, 95–113. [Google Scholar] [CrossRef]
- Hirakawa, Y. Evolution of meristem zonation by CLE gene duplication in land plants. Nat. Plants 2022, 8, 735–740. [Google Scholar] [CrossRef]
- Steffensen, D.M. A reconstruction of cell development in the shoot apex of maize. Am. J. Bot. 1968, 55, 354–369. [Google Scholar] [CrossRef]
- Itoh, J.I.; Kitano, H.; Matsuoka, M.; Nagato, Y. Shoot organization genes regulate shoot apical meristem organization and the pattern of leaf primordium initiation in rice. Plant Cell 2000, 12, 2161–2174. [Google Scholar] [CrossRef]
- Meyerowitz, E.M. Genetic control of cell division patterns in developing plants. Cell 1997, 88, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Gallois, J.-L.; Woodward, C.; Reddy, G.V.; Sablowski, R. Combined shoot meristemless and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 2002, 129, 3207–3217. [Google Scholar] [CrossRef]
- Vernoux, T.; Autran, D.; Traas, J. Developmental control of cell division patterns in the shoot apex. Plant Mol. Biol. 2000, 43, 569–581. [Google Scholar] [CrossRef]
- Fletcher, J.C.; Brand, U.; Running, M.P.; Simon, R.; Meyerowitz, E.M. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 1999, 283, 1911–1914. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jürgens, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.E.; Jacobsen, S.E.; Levin, J.Z.; Meyerowitz, E.M. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 1996, 122, 1567–1575. [Google Scholar] [CrossRef]
- Yadav, R.K.; Perales, M.; Gruel, J.; Girke, T.; Jönsson, H.; Reddy, G.V. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011, 25, 2025–2030. [Google Scholar] [CrossRef]
- Su, Y.H.; Zhou, C.; Li, Y.J.; Yu, Y.; Tang, L.P.; Zhang, W.J.; Yao, W.J.; Huang, R.; Laux, T.; Zhang, X.S. Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 2020, 117, 22561–22571. [Google Scholar] [CrossRef]
- Li, R.; Wei, Z.; Li, Y.; Shang, X.; Cao, Y.; Duan, L.; Ma, L. Ski-interacting protein interacts with shoot meristemless to regulate shoot apical meristem formation. Plant Physiol. 2022, 189, 2193–2209. [Google Scholar] [CrossRef]
- Jun, J.; Fiume, E.; Roeder, A.H.K.; Meng, L.; Sharma, V.K.; Osmont, K.S.; Baker, C.; Ha, C.M.; Meyerowitz, E.M.; Feldman, L.J.; et al. Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 2010, 154, 1721–1736. [Google Scholar] [CrossRef]
- Xu, T.-T.; Song, X.-F.; Ren, S.-C.; Liu, C.-M. The sequence flanking the N-terminus of the CLV3 peptide is critical for its cleavage and activity in stem cell regulation in Arabidopsis. BMC Plant Biol. 2013, 13, 225. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y. CLAVATA3, a plant peptide controlling stem cell fate in the meristem. Peptides 2021, 142, 170579. [Google Scholar] [CrossRef] [PubMed]
- Stuurman, J.; Jäggi, F.; Kuhlemeier, C. Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 2002, 16, 2213–2218. [Google Scholar] [CrossRef]
- Zhou, Y.; Yan, A.; Han, H.; Li, T.; Geng, Y.; Liu, X.; Meyerowitz, E.M. HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers. Science 2018, 361, 502–506. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, X.; Engstrom, E.M.; Nimchuk, Z.L.; Pruneda-Paz, J.L.; Tarr, P.T.; Yan, A.; Kay, S.A.; Meyerowitz, E.M. Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 2014, 517, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Perales, M.; Rodriguez, K.; Snipes, S.; Yadav, R.K.; Diaz-Mendoza, M.; Reddy, G.V. Threshold-dependent transcriptional discrimination underlies stem cell homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, E6298–E6306. [Google Scholar] [CrossRef]
- Engstrom, E.M.; Andersen, C.M.; Gumulak-Smith, J.; Hu, J.; Orlova, E.; Sozzani, R.; Bowman, J.L. Arabidopsis homologs of the petunia hairy meristem gene are required for maintenance of shoot and root indeterminacy. Plant Physiol. 2010, 155, 735–750. [Google Scholar] [CrossRef]
- Brand, U.; Grünewald, M.; Hobe, M.; Simon, R. Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 2002, 129, 565–575. [Google Scholar] [CrossRef]
- Brand, U.; Fletcher, J.C.; Hobe, M.; Meyerowitz, E.M.; Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 2000, 289, 617–619. [Google Scholar] [CrossRef]
- Hu, C.; Zhu, Y.; Cui, Y.; Cheng, K.; Liang, W.; Wei, Z.; Zhu, M.; Yin, H.; Zeng, L.; Xiao, Y. A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants 2018, 4, 205–211. [Google Scholar] [CrossRef]
- Wang, Y.; Jiao, Y. Cell signaling in the shoot apical meristem. Plant Physiol. 2023, 193, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.E.; Williams, R.W.; Meyerowitz, E.M. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 1997, 89, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Nimchuk, Z.L.; Tarr, P.T.; Ohno, C.; Qu, X.; Meyerowitz, E.M. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr. Biol. 2011, 21, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Shinohara, H.; Sakagami, Y.; Matsubayashi, Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 2008, 319, 294. [Google Scholar] [CrossRef]
- Ni, J.U.N.; Clark, S.E. Chapter 3–CLAVATA3: A putative peptide ligand controlling Arabidopsis stem cell specification. In Handbook of Biologically Active Peptides; Kastin, A.J., Ed.; Academic Press: Burlington, NJ, USA, 2006; pp. 9–15. [Google Scholar]
- Stone, J.M.; Trotochaud, A.E.; Walker, J.C.; Clark, S.E. Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol. 1998, 117, 1217–1225. [Google Scholar] [CrossRef]
- Nimchuk, Z.L.; Tarr, P.T.; Meyerowitz, E.M. An evolutionarily conserved pseudokinase mediates stem cell production in plants. Plant Cell 2011, 23, 851–854. [Google Scholar] [CrossRef]
- Bleckmann, A.; Weidtkamp-Peters, S.; Seidel, C.A.M.; Simon, R. Stem Cell Signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 2010, 152, 166–176. [Google Scholar] [CrossRef]
- Jeong, S.; Trotochaud, A.E.; Clark, S.E. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Cell 1999, 11, 1925–1933. [Google Scholar]
- Müller, R.; Bleckmann, A.; Simon, R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell–limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 2008, 20, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Diévart, A.; Dalal, M.; Tax, F.E.; Lacey, A.D.; Huttly, A.; Li, J.; Clark, S.E. CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 2003, 15, 1198–1211. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, S.; Osakabe, Y.; Maruyama, K.; Ito, T.; Osakabe, K.; Sato, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 2007, 50, 751–766. [Google Scholar] [CrossRef] [PubMed]
- Betsuyaku, S.; Takahashi, F.; Kinoshita, A.; Miwa, H.; Shinozaki, K.; Fukuda, H.; Sawa, S. Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 2011, 52, 14–29. [Google Scholar] [CrossRef]
- Shinohara, H.; Matsubayashi, Y. Reevaluation of the CLV3-receptor interaction in the shoot apical meristem: Dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. Plant J. 2015, 82, 328–336. [Google Scholar] [CrossRef]
- DeYoung, B.J.; Bickle, K.L.; Schrage, K.J.; Muskett, P.; Patel, K.; Clark, S.E. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 2006, 45, 1–16. [Google Scholar] [CrossRef]
- Guo, Y.; Han, L.; Hymes, M.; Denver, R.; Clark, S.E. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 2010, 63, 889–900. [Google Scholar] [CrossRef]
- Lee, H.; Jun, Y.S.; Cha, O.-K.; Sheen, J. Mitogen-activated protein kinases MPK3 and MPK6 are required for stem cell maintenance in the Arabidopsis shoot apical meristem. Plant Cell Rep. 2018, 38, 311–319. [Google Scholar] [CrossRef]
- Yu, L.P.; Miller, A.K.; Clark, S.E. POLTERGEIST encodes a PROTEIN PHOSPHATASE 2C that regulates CLAVATA pathways controlling stem cell identity at Arabidopsis shoot and flower meristems. Curr. Biol. 2003, 13, 179–188. [Google Scholar] [CrossRef]
- Chaffey, N. Esau’s Plant anatomy, meristems, cells, and tissues of the plant body: Their structure, function, and development. 3rd edn. Ann. Bot. 2006, 99, 785–786. [Google Scholar] [CrossRef]
- Du, J.; Wang, Y.; Chen, W.; Xu, M.; Zhou, R.; Shou, H.; Chen, J. High-resolution anatomical and spatial transcriptome analyses reveal two types of meristematic cell pools within the secondary vascular tissue of poplar stem. Mol. Plant 2023, 16, 809–828. [Google Scholar] [CrossRef] [PubMed]
- Larson, P.R. Procambium vs. Cambium and Protoxylem vs. Metaxylem in Populus deltoides seedlings. Am. J. Bot. 1976, 63, 1332–1348. [Google Scholar] [CrossRef]
- Lucas, W.J.; Groover, A.; Lichtenberger, R.; Furuta, K.; Yadav, S.-R.; Helariutta, Y.; He, X.-Q.; Fukuda, H.; Kang, J.; Brady, S.M.; et al. The plant vascular system: Evolution, development and functions. J. Integr. Plant Biol. 2013, 55, 294–388. [Google Scholar] [CrossRef] [PubMed]
- Etchells, J.P.; Turner, S.R. The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 2010, 137, 767–774. [Google Scholar] [CrossRef]
- Hirakawa, Y.; Shinohara, H.; Kondo, Y.; Inoue, A.; Nakanomyo, I.; Ogawa, M.; Sawa, S.; Ohashi-Ito, K.; Matsubayashi, Y.; Fukuda, H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 2008, 105, 15208–15213. [Google Scholar] [CrossRef]
- Kondo, Y.; Ito, T.; Nakagami, H.; Hirakawa, Y.; Saito, M.; Tamaki, T.; Shirasu, K.; Fukuda, H. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signalling. Nat. Commun. 2014, 5, 3504. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, S.; Xu, L.; Chu, S.; Yan, X.; Lin, L.; Wen, J.; Zheng, B.; Chen, S.; Li, Q. Transcription factor PagMYB31 positively regulates cambium activity and negatively regulates xylem development in poplar. Plant Cell 2024, 36, 1806–1828. [Google Scholar] [CrossRef]
- Han, S.; Cho, H.; Noh, J.; Qi, J.; Jung, H.-J.; Nam, H.; Lee, S.; Hwang, D.; Greb, T.; Hwang, I. BIL1-mediated MP phosphorylation integrates PXY and cytokinin signalling in secondary growth. Nat. Plants 2018, 4, 605–614. [Google Scholar] [CrossRef]
- Zhu, Y.; Song, D.; Zhang, R.; Luo, L.; Cao, S.; Huang, C.; Sun, J.; Gui, J.; Li, L. A xylem-produced peptide PtrCLE20 inhibits vascular cambium activity in Populus. Plant Biotechnol. J. 2019, 18, 195–206. [Google Scholar] [CrossRef]
- Kucukoglu, M.; Chaabouni, S.; Zheng, B.; Mähönen, A.P.; Helariutta, Y.; Nilsson, O. Peptide encoding Populus CLV3/ESR-RELATED 47 (PtCLE47) promotes cambial development and secondary xylem formation in hybrid aspen. New Phytol. 2019, 226, 75–85. [Google Scholar] [CrossRef]
- Bauby, H.; Divol, F.; Truernit, E.; Grandjean, O.; Palauqui, J.-C. Protophloem differentiation in early Arabidopsis thaliana development. Plant Cell Physiol. 2007, 48, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Qian, P.; Song, W.; Yokoo, T.; Minobe, A.; Wang, G.; Ishida, T.; Sawa, S.; Chai, J.; Kakimoto, T. The CLE9/10 secretory peptide regulates stomatal and vascular development through distinct receptors. Nat. Plants 2018, 4, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Carbonnel, S.; Cornelis, S.; Hazak, O. The CLE33 peptide represses phloem differentiation via autocrine and paracrine signaling in Arabidopsis. Commun. Biol. 2023, 6, 588. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.-C.; Song, X.-F.; Chen, W.-Q.; Lu, R.; Lucas, W.J.; Liu, C.-M. CLE25 peptide regulates phloem initiation in Arabidopsis through a CLERK-CLV2 receptor complex. J. Integr. Plant Biol. 2019, 61, 1043–1061. [Google Scholar] [CrossRef]
- Anne, P.; Amiguet-Vercher, A.; Brandt, B.; Kalmbach, L.; Geldner, N.; Hothorn, M.; Hardtke, C.S. CLERK is a novel receptor kinase required for sensing of root-active CLE peptides in Arabidopsis. Development 2018, 145, dev162354. [Google Scholar] [CrossRef]
- Depuydt, S.; Rodriguez-Villalon, A.; Santuari, L.; Wyser-Rmili, C.; Ragni, L.; Hardtke, C.S. Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. Proc. Natl. Acad. Sci. USA 2013, 110, 7074–7079. [Google Scholar] [CrossRef]
- Kang, Y.H.; Hardtke, C.S. Arabidopsis MAKR5 is a positive effector of BAM3-dependent CLE45 signaling. EMBO Rep. 2016, 17, 1145–1154. [Google Scholar] [CrossRef]
- Hang, Z.; Qian, W.; Noel, B.-T.; Christian, S.H. Antagonistic CLE peptide pathways shape root meristem tissue patterning. Nat. Plants 2024, 10, 1900–1908. [Google Scholar] [CrossRef]
- Qian, P.; Song, W.; Zaizen-Iida, M.; Kume, S.; Wang, G.; Zhang, Y.; Kinoshita-Tsujimura, K.; Chai, J.; Kakimoto, T. A Dof-CLE circuit controls phloem organization. Nat. Plants 2022, 8, 817–827. [Google Scholar] [CrossRef]
- Araya, T.; Miyamoto, M.; Wibowo, J.; Suzuki, A.; Kojima, S.; Tsuchiya, Y.N.; Sawa, S.; Fukuda, H.; von Wirén, N.; Takahashi, H. CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. USA 2014, 111, 2029–2034. [Google Scholar] [CrossRef]
- Pazourek, J.T.A. Steeves & I.M. Sussex patterns in plant development. Folia Geobot. Phytotaxon. 1992, 27, 136. [Google Scholar] [CrossRef]
- Berná, G.; Robles, P.; Micol, J.L. A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 1999, 152, 729–742. [Google Scholar] [CrossRef]
- Pillitteri, L.J.; Torii, K.U. Mechanisms of stomatal development. Annu. Rev. Plant Biol. 2012, 63, 591–614. [Google Scholar] [CrossRef] [PubMed]
- Geisler, M.; Nadeau, J.; Sack, F.D. Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 2000, 12, 2075–2086. [Google Scholar] [CrossRef]
- Cho, K.H.; Jun, S.E.; Jeong, S.J.; Lee, Y.K.; Kim, G.T. Developmental processes of leaf morphogenesis in Arabidopsis. J. Plant Biol. 2007, 50, 282–290. [Google Scholar] [CrossRef]
- Kalve, S.; De Vos, D.; Beemster, G.T.S. Leaf development: A cellular perspective. Front. Plant Sci. 2014, 5, 362. [Google Scholar] [CrossRef]
- Bennett, T.; Hines, G.; van Rongen, M.; Waldie, T.; Sawchuk, M.G.; Scarpella, E.; Ljung, K.; Leyser, O. Connective auxin transport in the shoot facilitates communication between shoot apices. PLOS Biol. 2016, 14, e1002446. [Google Scholar] [CrossRef]
- De Reuille, P.B.; Bohn-Courseau, I.; Ljung, K.; Morin, H.; Carraro, N.; Godin, C.; Traas, J. Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 1627–1632. [Google Scholar] [CrossRef]
- Vidaurre, D.P.; Ploense, S.; Krogan, N.T.; Berleth, T. AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development 2007, 134, 2561–2567. [Google Scholar] [CrossRef]
- Zhao, Z.; Andersen, S.U.; Ljung, K.; Dolezal, K.; Miotk, A.; Schultheiss, S.J.; Lohmann, J.U. Hormonal control of the shoot stem-cell niche. Nature 2010, 465, 1089–1092. [Google Scholar] [CrossRef]
- Rademacher, E.H.; Möller, B.; Lokerse, A.S.; Llavata-Peris, C.I.; van den Berg, W.; Weijers, D. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 2011, 68, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Zhao, W.; Kong, S.; Li, L.; Lin, S. Overview of molecular mechanisms of plant leaf development: A systematic review. Front. Plant Sci. 2023, 14, 1293424. [Google Scholar] [CrossRef]
- Luo, L.; Zeng, J.; Wu, H.; Tian, Z.; Zhao, Z. A molecular framework for auxin-controlled homeostasis of shoot stem cells in Arabidopsis. Mol. Plant 2018, 11, 899–913. [Google Scholar] [CrossRef]
- Matsuo, N.; Makino, M.; Banno, H. Arabidopsis ENHANCER OF SHOOT REGENERATION (ESR) 1 and ESR2 regulate in vitro shoot regeneration and their expressions are differentially regulated. Plant Sci. 2011, 181, 39–46. [Google Scholar] [CrossRef]
- Luo, L.; Liu, L.; She, L.; Zhang, H.; Zhang, N.; Wang, Y.; Zhao, Z. DRN facilitates WUS transcriptional regulatory activity by chromatin remodeling to regulate shoot stem cell homeostasis in Arabidopsis. PLoS Biol. 2024, 22, e3002878. [Google Scholar] [CrossRef] [PubMed]
- Kusnandar, A.S.; Itoh, J.I.; Sato, Y.; Honda, E.; Hibara, K.I.; Kyozuka, J.; Naramoto, S. NARROW and DWARF LEAF 1, the ortholog of Arabidopsis enhancer of shoot regeneration1/dornröschen, mediates leaf development and maintenance of the shoot apical meristem in Oryza sativa L. Plant Cell Physiol. 2022, 63, 265–278. [Google Scholar] [CrossRef] [PubMed]
- DiGennaro, P.; Grienenberger, E.; Dao, T.Q.; Jun, J.H.; Fletcher, J.C. Peptide signaling molecules CLE5 and CLE6 affect Arabidopsis leaf shape downstream of leaf patterning transcription factors and auxin. Plant Direct 2018, 2, e00103. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, S.; Gao, Y.; Kan, C.; Wang, H.-L.; Yang, Q.; Xia, X.; Ishida, T.; Sawa, S.; Guo, H.; et al. CLE42 delays leaf senescence by antagonizing ethylene pathway in Arabidopsis. New Phytol. 2022, 235, 550–562. [Google Scholar] [CrossRef]
- Han, H.; Zhuang, K.; Qiu, Z. CLE peptides join the plant longevity club. Trends Plant Sci. 2022, 27, 961–963. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, C.; Li, K.; Li, X.; Xu, M.; Guo, Y. CLE14 functions as a “brake signal” to suppress age-dependent and stress-induced leaf senescence by promoting JUB1-mediated ROS scavenging in Arabidopsis. Mol. Plant 2021, 15, 179–188. [Google Scholar] [CrossRef]
- Dennis, L.; Peacock, J. Genes directing flower development in Arabidopsis. Plant Cell 2019, 31, 1192–1193. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhang, K.; Guo, L.; Liu, X.; Zhang, Z. AUXIN RESPONSE FACTOR3 plays distinct role during early flower development. Plant Signal. Behav. 2018, 13, e1467690. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Benfey, P.N. Signaling in and out: Control of cell division and differentiation in the shoot and root. Plant Cell 2002, 14, S265–S276. [Google Scholar] [CrossRef]
- Takeda, S.; Iwasaki, A.; Matsumoto, N.; Uemura, T.; Tatematsu, K.; Okada, K. Physical interaction of floral organs controls petal morphogenesis in Arabidopsis. Plant Physiol. 2013, 161, 1242–1250. [Google Scholar] [CrossRef]
- Liu, H.; Yang, L.; Tu, Z.; Zhu, S.; Zhang, C.; Li, H. Genome-wide identification of MIKC-type genes related to stamen and gynoecium development in Liriodendron. Sci. Rep. 2021, 11, 6585. [Google Scholar] [CrossRef]
- Jones, D.S.; John, A.; VanDerMolen, K.R.; Nimchuk, Z.L. CLAVATA signaling ensures reproductive development in plants across thermal environments. Curr. Biol. 2021, 31, 220–227.e5. [Google Scholar] [CrossRef] [PubMed]
- Kayes, J.M.; Clark, S.E. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 1998, 125, 3843–3851. [Google Scholar] [CrossRef]
- Nidhi, S.; Preciado, J.; Tie, L. Knox homologs shoot meristemless (STM) and KNAT6 are epistatic to CLAVATA3 (CLV3) during shoot meristem development in Arabidopsis thaliana. Mol. Biol. Rep. 2021, 48, 6291–6302. [Google Scholar] [CrossRef]
- Bommert, P.; Lunde, C.; Nardmann, J.; Vollbrecht, E.; Running, M.; Jackson, D.; Hake, S.; Werr, W. thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 2005, 132, 1235–1245. [Google Scholar] [CrossRef]
- Taguchi-Shiobara, F.; Yuan, Z.; Hake, S.; Jackson, D. The fasciated EAR2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 2001, 15, 2755–2766. [Google Scholar] [CrossRef]
- Chu, H.; Qian, Q.; Liang, W.; Yin, C.; Tan, H.; Yao, X.; Yuan, Z.; Yang, J.; Huang, H.; Luo, D.; et al. The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 2006, 142, 1039–1052. [Google Scholar] [CrossRef]
- Box, M.S.; Huang, B.E.; Domijan, M.; Jaeger, K.E.; Khattak, A.K.; Yoo, S.J.; Sedivy, E.L.; Jones, D.M.; Hearn, T.J.; Webb, A.A.R.; et al. ELF3 Controls thermoresponsive growth in Arabidopsis. Curr. Biol. 2014, 25, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Barbosa, A.D.; Hutin, S.; Kumita, J.R.; Gao, M.; Derwort, D.; Silva, C.S.; Lai, X.; Pierre, E.; Geng, F.; et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 2020, 585, 256–260. [Google Scholar] [CrossRef]
- Lindsay, R.J.; Stelzl, L.S.; Pietrek, L.; Hummer, G.; Wigge, P.A.; Hanson, S.M. Helical region near poly-Q tract in prion-like domain of Arabidopsis ELF3 plays role in temperature-sensing mechanism. Biophys. J. 2022, 121, 355a–356a. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef]
- John, A.; Smith, E.S.; Jones, D.S.; Soyars, C.L.; Nimchuk, Z.L. A network of CLAVATA receptors buffers auxin-dependent meristem maintenance. Nat. Plants 2023, 9, 1306–1317. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Leal, D.; Xu, C.; Kwon, C.-T.; Soyars, C.; Demesa-Arevalo, E.; Man, J.; Liu, L.; Lemmon, Z.H.; Jones, D.S.; Van Eck, J.; et al. Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat. Genet. 2019, 51, 786–792. [Google Scholar] [CrossRef] [PubMed]
- Bashyal, S.; Gautam, C.K.; Müller, L.M. CLAVATA signaling in plant–environment interactions. Plant Physiol. 2023, 194, 1336–1357. [Google Scholar] [CrossRef]
- Diss, G.; Ascencio, D.; DeLuna, A.; Landry, C.R. Molecular mechanisms of paralogous compensation and the robustness of cellular networks. J. Exp. Zool. Part B Mol. Dev. Evol. 2013, 322, 488–499. [Google Scholar] [CrossRef]
- Hanada, K.; Sawada, Y.; Kuromori, T.; Klausnitzer, R.; Saito, K.; Toyoda, T.; Shinozaki, K.; Li, W.H.; Hirai, M.Y. Functional compensation of primary and secondary metabolites by duplicate genes in Arab. thaliana. Mol. Biol. Evol. 2010, 28, 377–382. [Google Scholar] [CrossRef]
- Moens, C.; El-Brolosy, M.A.; Stainier, D.Y.R. Genetic compensation: A phenomenon in search of mechanisms. PLOS Genet. 2017, 13, e1006780. [Google Scholar] [CrossRef]
- Goad, D.M.; Zhu, C.; Kellogg, E.A. Comprehensive identification and clustering of CLV3/ESR-related (CLE) genes in plants finds groups with potentially shared function. New Phytol. 2016, 216, 605–616. [Google Scholar] [CrossRef]
- Dao, T.Q.; Weksler, N.; Liu, H.M.H.; Leiboff, S.; Fletcher, J.C. Interactive CLV3, CLE16, and CLE17 signaling mediates stem cell homeostasis in the Arabidopsis shoot apical meristem. Development 2022, 149, dev200787. [Google Scholar] [CrossRef]
- Nimchuk, Z.L.; Zhou, Y.; Tarr, P.T.; Peterson, B.A.; Meyerowitz, E.M. Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 2015, 142, 1043–1049. [Google Scholar] [CrossRef]
- Shimizu, N.; Ishida, T.; Yamada, M.; Shigenobu, S.; Tabata, R.; Kinoshita, A.; Yamaguchi, K.; Hasebe, M.; Mitsumasu, K.; Sawa, S. BAM 1 and RECEPTOR-LIKE PROTEIN KINASE 2 constitute a signaling pathway and modulate CLE peptide-triggered growth inhibition in Arabidopsis root. New Phytol. 2015, 208, 1104–1113. [Google Scholar] [CrossRef]
- Ni, J.; Clark, S.E. Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 2006, 140, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Gallagher, J.; Arevalo, E.D.; Chen, R.; Skopelitis, T.; Wu, Q.; Bartlett, M.; Jackson, D. Enhancing grain-yield-related traits by CRISPR–Cas9 promoter editing of maize CLE genes. Nat. Plants 2021, 7, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Selby, R.; Jones, D.S. Complex peptide hormone signaling in plant stem cells. Curr. Opin. Plant Biol. 2023, 75, 102442. [Google Scholar] [CrossRef]
- Strabala, T.J.; Phillips, L.; West, M.; Stanbra, L. Bioinformatic and phylogenetic analysis of the CLAVATA3/EMBRYO-SURROUNDING REGION (CLE) and the CLE-LIKE signal peptide genes in the Pinophyta. BMC Plant Biol. 2014, 14, 47. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, L.; Kucukoglu, M.; Tian, D.; Larkin, R.M.; Shi, X.; Zheng, B. Predicting and clustering plant CLE genes with a new method developed specifically for short amino acid sequences. BMC Genom. 2020, 21, 709. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Z.; Zhou, W.; Jiang, H.; Duan, Y. Research Progress of CLE and Its Prospects in Woody Plants. Plants 2025, 14, 1424. https://doi.org/10.3390/plants14101424
Song Z, Zhou W, Jiang H, Duan Y. Research Progress of CLE and Its Prospects in Woody Plants. Plants. 2025; 14(10):1424. https://doi.org/10.3390/plants14101424
Chicago/Turabian StyleSong, Zewen, Wenjun Zhou, Hanyu Jiang, and Yifan Duan. 2025. "Research Progress of CLE and Its Prospects in Woody Plants" Plants 14, no. 10: 1424. https://doi.org/10.3390/plants14101424
APA StyleSong, Z., Zhou, W., Jiang, H., & Duan, Y. (2025). Research Progress of CLE and Its Prospects in Woody Plants. Plants, 14(10), 1424. https://doi.org/10.3390/plants14101424




