Transcriptome and Physiological Analysis Reveals the Mechanism of Abscisic Acid in Regulating Cadmium Uptake and Accumulation in the Hyperaccumulator Phytolacca acinosa Roxb.
Abstract
1. Introduction
2. Results
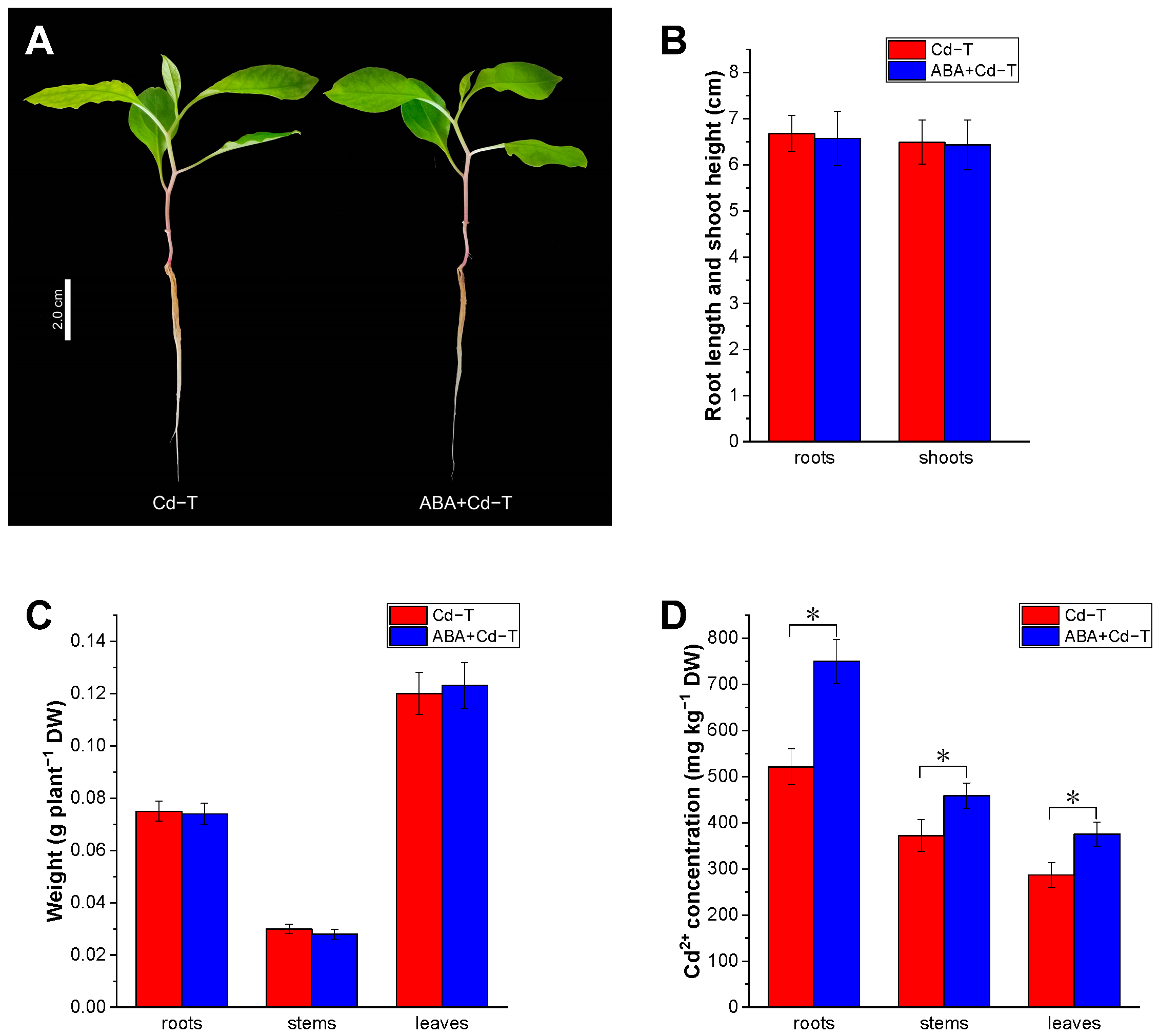
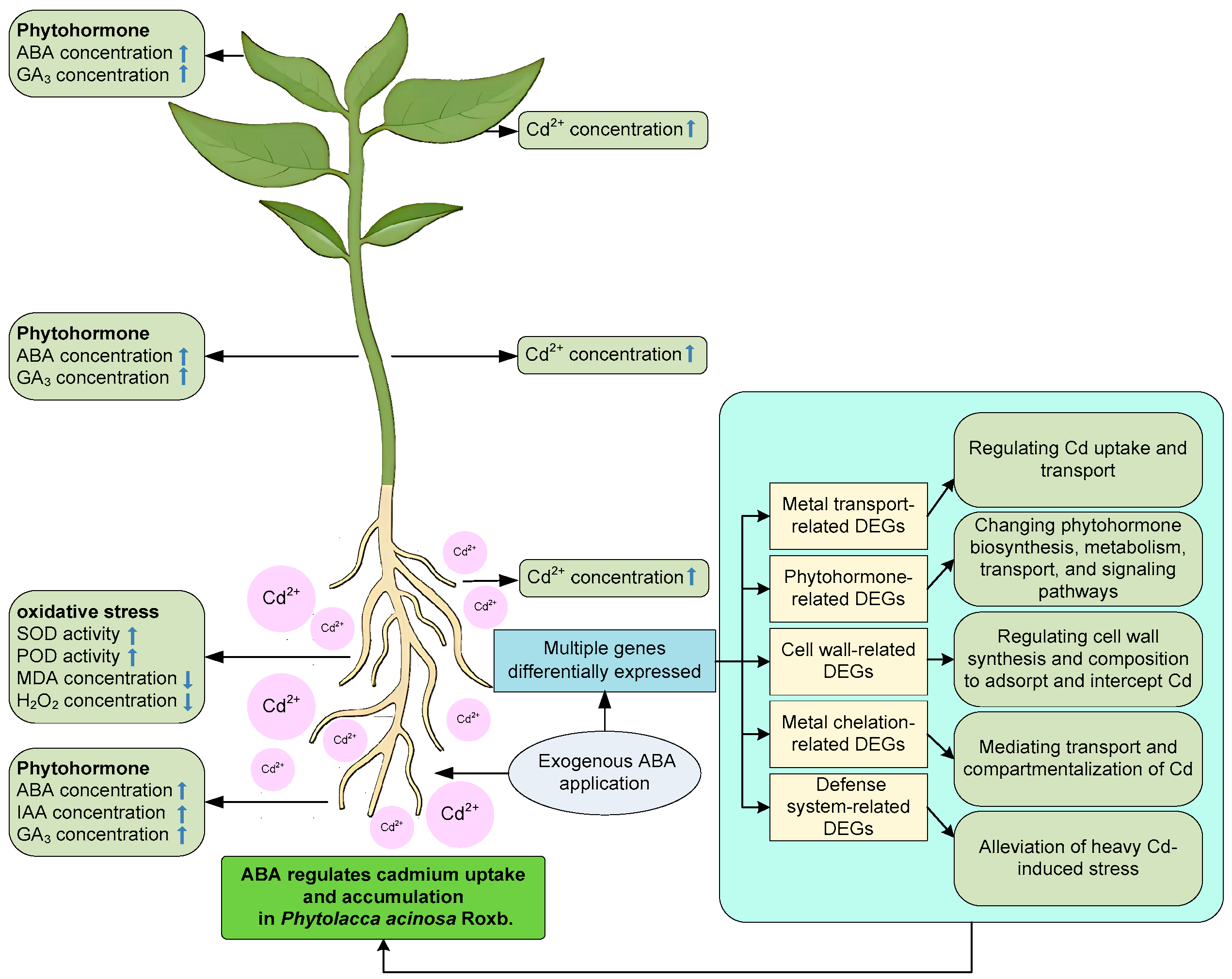
2.1. Effects of ABA Treatment on Cd Uptake and Accumulation in P. acinosa Under Cd Stress
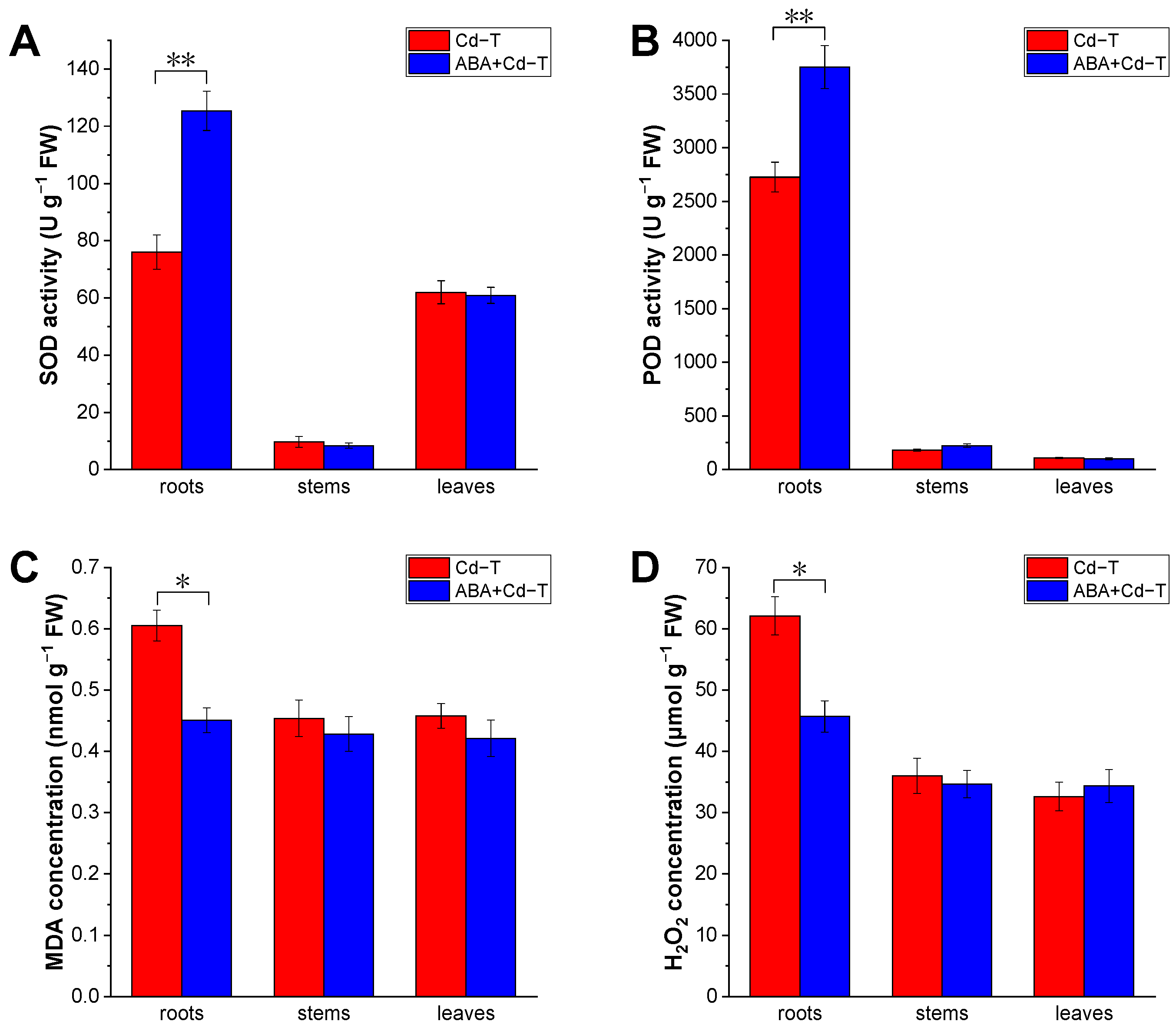
2.2. Effects of ABA Treatment on Oxidative Stress of P. acinosa Under Cd Stress
2.3. Effects of ABA Treatment on Phytohormone Concentration of P. acinosa Under Cd Stress
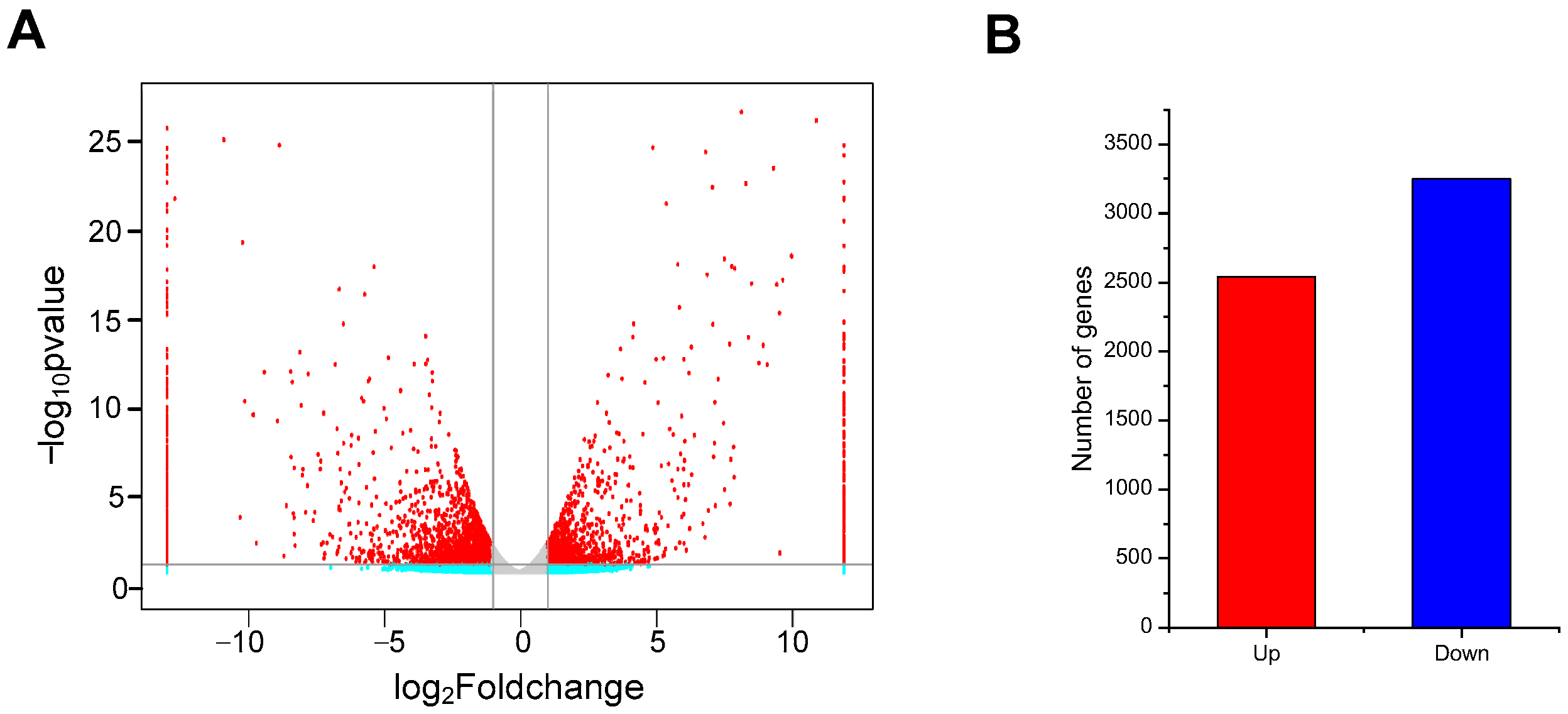
2.4. De Novo Transcriptome Sequencing and Analysis of DEGs Between ABA+Cd−T and Cd−T Samples in P. acinosa Roots
2.5. Metal Transport-Related Gene Expression Differences Between ABA+Cd−T and Cd−T Samples in P. acinosa Roots
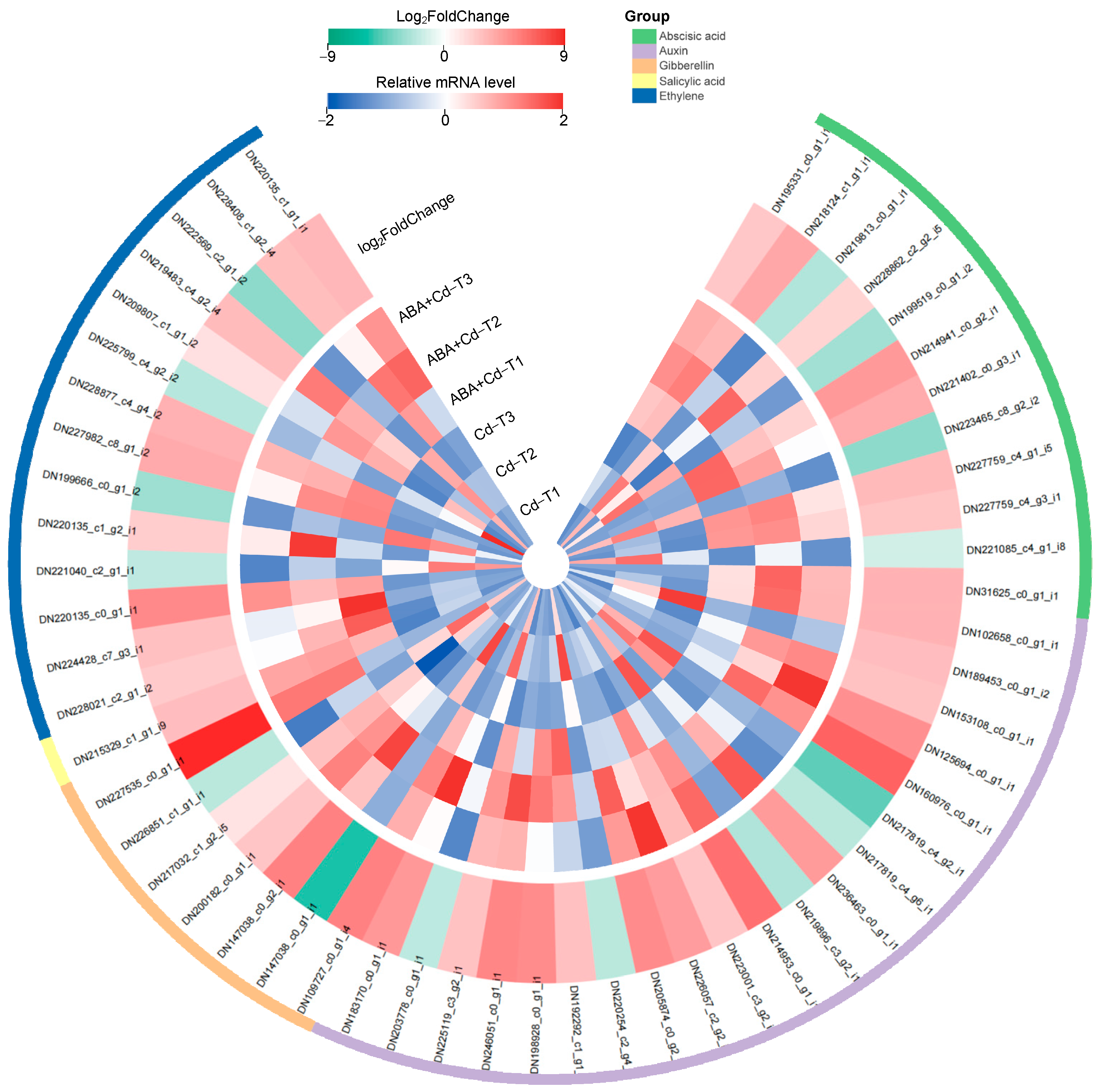
2.6. Phytohormone-Related Gene Expression Differences Between ABA+Cd−T and Cd−T Samples in P. acinosa Roots
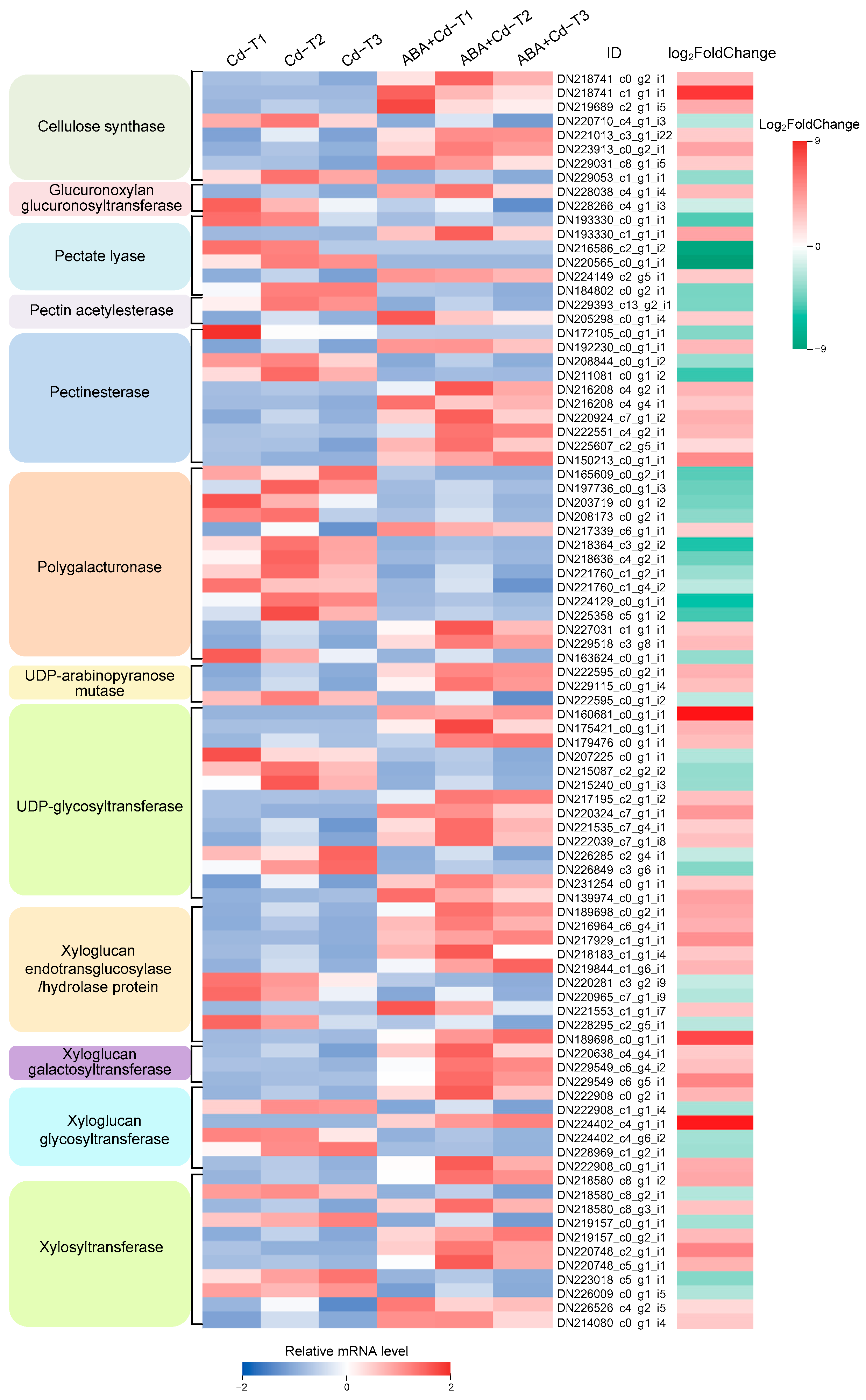
2.7. Cell Wall-Related Gene Expression Differences Between ABA+Cd−T and Cd−T Samples in P. acinosa Roots
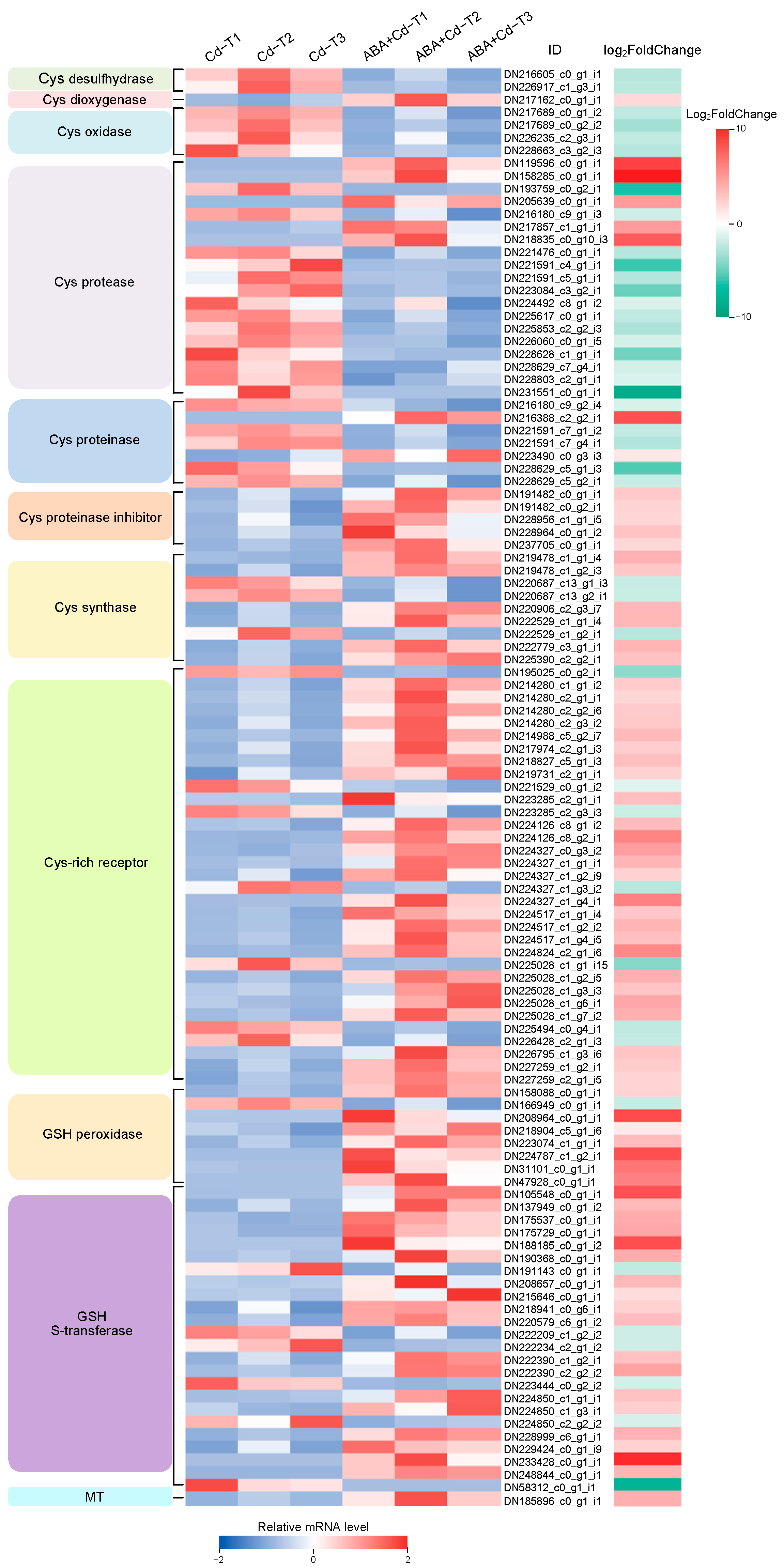
2.8. Metal Chelation-Related Gene Expression Differences Between ABA+Cd−T and Cd−T Samples in P. acinosa Roots
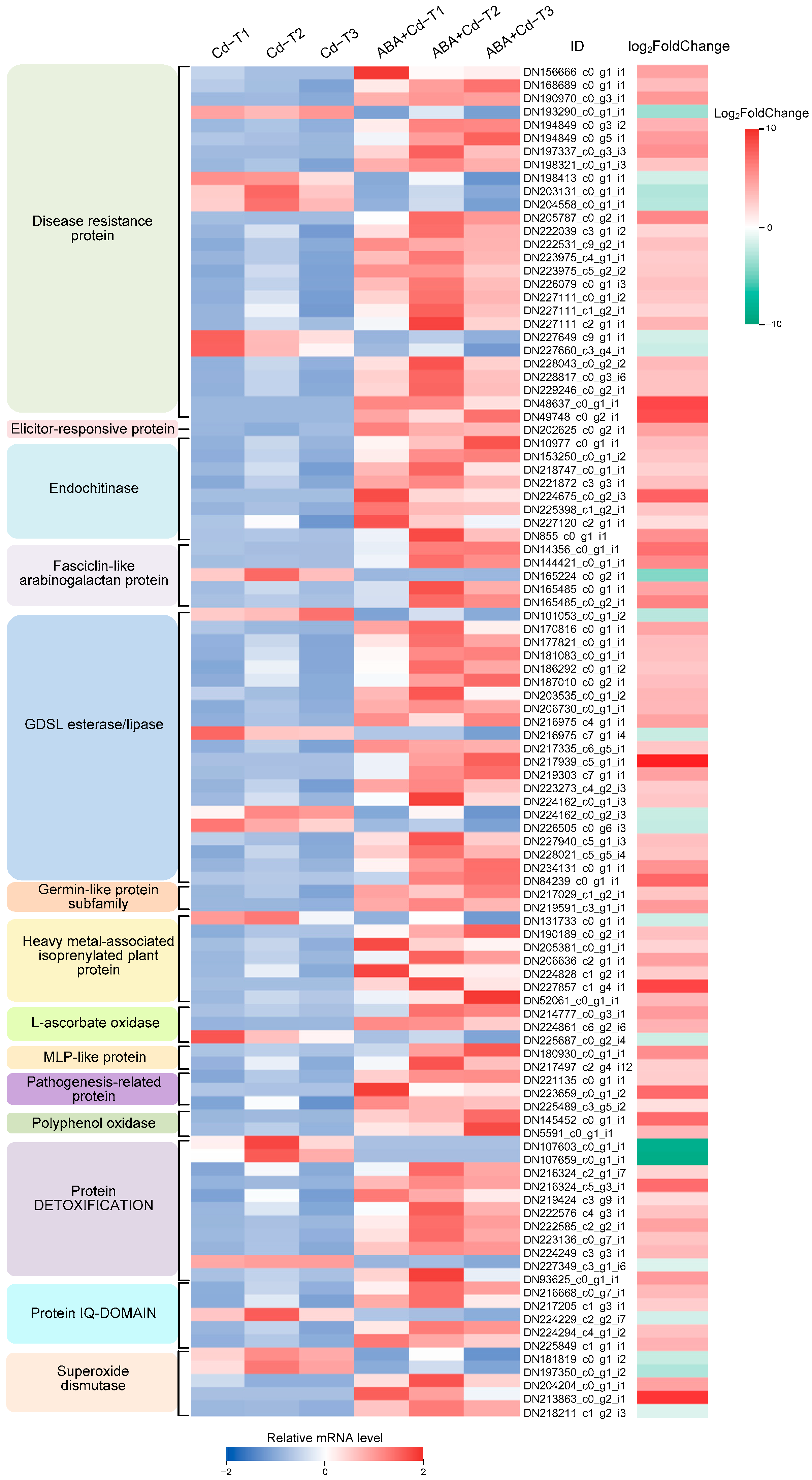
2.9. Defense System-Related Gene Expression Differences Between ABA+Cd−T and Cd−T Samples in P. acinosa Roots
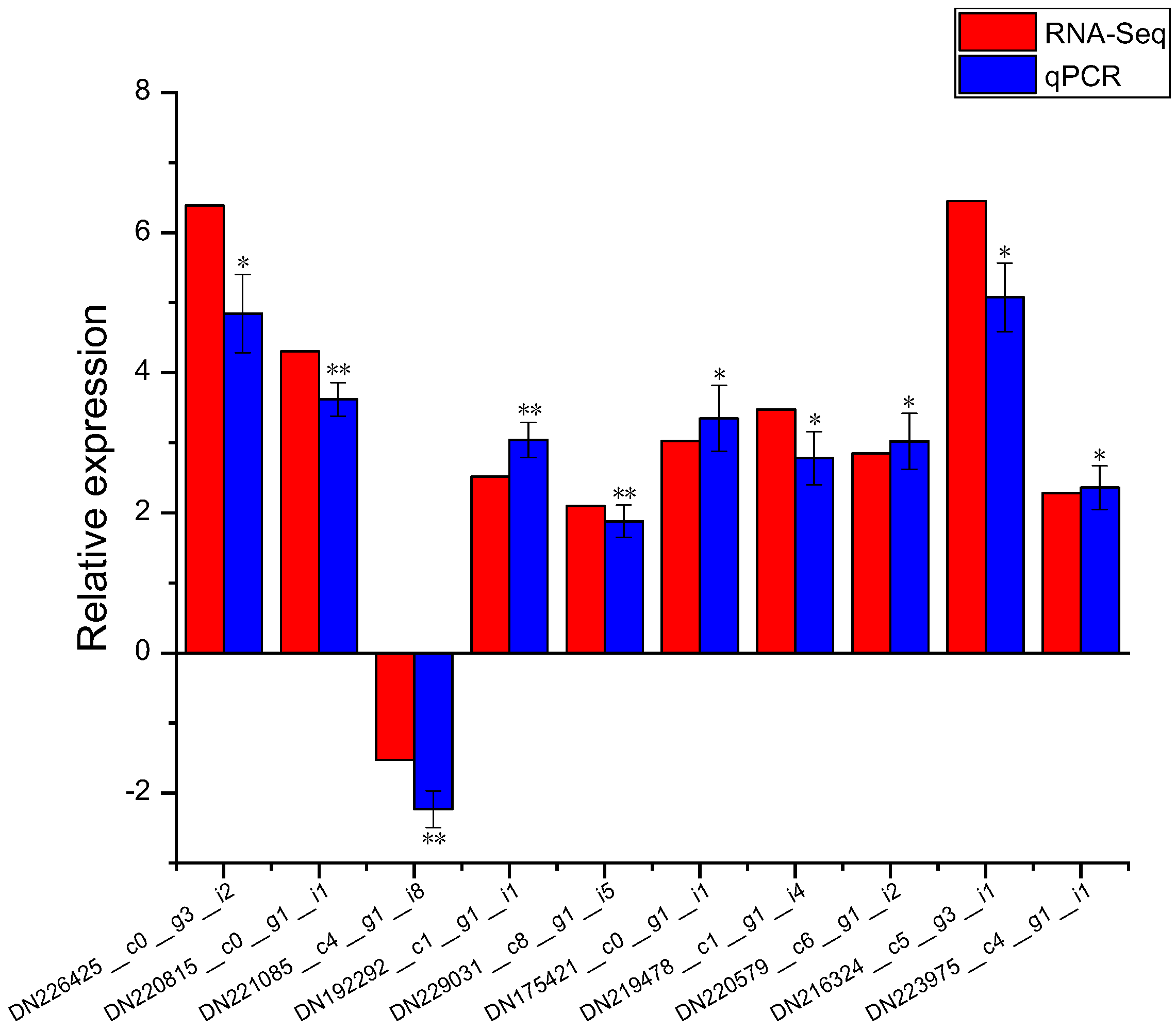
2.10. Real Time-PCR Validation of DEGs
3. Discussion
3.1. ABA Treatment Regulated Physiological and Biochemical Processes in P. acinosa Under Cd Stress
3.2. Phytohormone Signal Transduction Pathway Positively Responds to Cd Stress
3.3. Metal Transporter Proteins Play an Important Role in Cd Uptake and Accumulation in P. acinosa
3.4. ABA Treatment May Enhance the Cell Wall Structure of the Root System
3.5. Phytohormone Signaling May Regulate Chelating Agents Involved in Cd Stress Response
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Cd Treatments and ABA+Cd Treatments
4.3. Measurement of Antioxidative Enzyme Activity
4.4. Measurement of MDA Concentration
4.5. Measurement of H2O2 Concentration
4.6. Measurement of Phytohormone Concentration
4.7. Measurement of Cd2+ Concentration
4.8. Total RNA Extraction, Library Preparation, and De Novo Sequencing
4.9. Functional Annotation and Classification
4.10. Analysis and Functional Enrichment of DEGs
4.11. Real-Time PCR Analysis of Gene Expression
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kim, J.J.; Kim, Y.S.; Kumar, V. Heavy metal toxicity: An update of chelating therapeutic strategies. J. Trace Elem. Med. Biol. 2019, 54, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Chen, H.; Chen, Y.; Zhang, C.; He, Y. Rapid detection of cadmium and its distribution in Miscanthus sacchariflorus based on visible and near-infrared hyperspectral imaging. Sci. Total Environ. 2019, 659, 1021–1031. [Google Scholar] [CrossRef] [PubMed]
- Beans, C. Core Concept: Phytoremediation advances in the lab but lags in the field. Proc. Natl. Acad. Sci. USA 2017, 114, 7475–7477. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.R.; Wu, Z.H.; Luo, J. Transfer Mechanism, Uptake Kinetic Process, and Bioavailability of P, Cu, Cd, Pb, and Zn in Macrophyte Rhizosphere Using Diffusive Gradients in Thin Films. Environ. Sci. Technol. 2018, 52, 1096–1108. [Google Scholar] [CrossRef]
- Zeng, P.; Guo, Z.; Xiao, X.; Peng, C.; Feng, W.; Xin, L.; Xu, Z. Phytoextraction potential of Pteris vittata L. co-planted with woody species for As, Cd, Pb and Zn in contaminated soil. Sci. Total Environ. 2019, 650, 594–603. [Google Scholar] [CrossRef]
- Manzoor, M.; Gul, I.; Ahmed, I.; Zeeshan, M.; Hashmi, I.; Amin, B.A.Z.; Kallerhoff, J.; Arshad, M. Metal tolerant bacteria enhanced phytoextraction of lead by two accumulator ornamental species. Chemosphere 2019, 227, 561–569. [Google Scholar] [CrossRef]
- Fasani, E.; Manara, A.; Martini, F.; Furini, A.; DalCorso, G. The Potential of Genetic Engineering of Plants for the Remediation of Soils Contaminated with Heavy Metals. Plant Cell Environ. 2018, 41, 1201–1232. [Google Scholar] [CrossRef]
- Barbafieri, M.; Morelli, E.; Tassi, E.; Pedron, F.; Remorini, D.; Petruzzelli, G. Overcoming limitation of “recalcitrant areas” to phytoextraction process: The synergistic effects of exogenous cytokinins and nitrogen treatments. Sci. Total Environ. 2018, 639, 1520–1529. [Google Scholar] [CrossRef]
- Cassina, L.; Tassi, E.; Pedron, F.; Petruzzelli, G.; Ambrosini, P.; Barbafieri, M. Using a plant hormone and a thioligand to improve phytoremediation of Hg-contaminated soil from a petrochemical plant. J. Hazard. Mater. 2012, 231, 36–42. [Google Scholar] [CrossRef]
- Hauser, F.; Li, Z.; Waadt, R.; Schroeder, J.I. SnapShot: Abscisic Acid Signaling. Cell 2017, 171, 1708–1708.e0. [Google Scholar] [CrossRef]
- Dixit, A.; Tomar, P.; Vaine, E.; Abdullah, H.; Hazen, S.; Dhankher, O.P. A stress-associated protein, AtSAP13, from Arabidopsis thaliana provides tolerance to multiple abiotic stresses. Plant Cell Environ. 2018, 41, 1171–1185. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.S.; Wang, S.J.; Zhao, N.; Deng, S.R.; Zhao, C.J.; Li, N.F.; Sun, J.; Zhao, R.; Yi, H.L.; Shen, X. Exogenous Abscisic Acid Alleviates Cadmium Toxicity by Restricting Cd2+ Influx in Populus euphratica Cells. J. Plant Growth Regul. 2016, 35, 827–837. [Google Scholar] [CrossRef]
- Gilliham, M.; Tester, M. The Regulation of Anion Loading to the Maize Root Xylem. Plant Physiol. 2005, 137, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.; Niu, J.; Deng, Z. Abscisic acid treatment alleviates cadmium toxicity in purple flowering stalk (Brassica campestris L. ssp. chinensis var. purpurea Hort.) seedlings. Plant Physiol. Biochem. 2017, 118, 471–478. [Google Scholar] [CrossRef]
- Wang, J.; Lin, L.; Luo, L.; Liao, M.a.; Lv, X.; Wang, Z.; Liang, D.; Xia, H.; Wang, X.; Lai, Y.; et al. The effects of abscisic acid (ABA) addition on cadmium accumulation of two ecotypes of Solanum photeinocarpum. Environ. Monit. Assess. 2016, 188, 182. [Google Scholar] [CrossRef]
- Shi, W.G.; Liu, W.; Yu, W.; Zhang, Y.; Ding, S.; Li, H.; Mrak, T.; Kraigher, H.; Luo, Z.B. Abscisic acid enhances lead translocation from the roots to the leaves and alleviates its toxicity in Populus × canescens. J. Hazard. Mater. 2019, 362, 275–285. [Google Scholar] [CrossRef]
- Shi, W.G.; Li, H.; Liu, T.X.; Polle, A.; Peng, C.H.; Luo, Z.B. Exogenous abscisic acid alleviates zinc uptake and accumulation in Populus × canescens exposed to excess zinc. Plant Cell Environ. 2015, 38, 207–223. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, L.; Xing, W.; Ran, S.; Wei, Z.; Amee, M.; Wassie, M.; Niu, H.; Tang, D.; Sun, J.; et al. Phytohormones-induced senescence efficiently promotes the transport of cadmium from roots into shoots of plants: A novel strategy for strengthening of phytoremediation. J. Hazard. Mater. 2020, 388, 122080. [Google Scholar] [CrossRef]
- Nie, F.H. Cd hyper-accumulator Phytolacca acinosa Roxb. and Cd-accumulative characteristics. Ecol. Environ. 2006, 15, 303–306. [Google Scholar] [CrossRef]
- Xie, Q.; Deng, W.T.; Su, Y.; Ma, L.Y.; Yang, H.J.; Yao, F.H.; Lin, W.H. Transcriptome Analysis Reveals Novel Insights into the Hyperaccumulator Phytolacca acinosa Roxb. Responses to Cadmium Stress. Plants 2024, 13, 297. [Google Scholar] [CrossRef]
- Lu, Q.Y.; Chen, S.M.; Li, Y.Y.; Zheng, F.H.; He, B.; Gu, M.H. Exogenous abscisic acid (ABA) promotes cadmium (Cd) accumulation in Sedum alfredii Hance by regulating the expression of Cd stress response genes. Environ. Sci. Pollut. Res. 2020, 27, 8719–8731. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Takahashi, R.; Bashir, K.; Shimo, H.; Nishizawa, N.K. Characterizing the role of rice NRAMP5 in Manganese, Iron and Cadmium Transport. Sci. Rep. 2012, 2, 286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.F.; Lu, T.T.; Miao, W.W.; Sun, L.R.; Tian, M.; Wang, J.; Hao, F.S. Genome-wide identification of ABA receptor PYL family and expression analysis of PYLs in response to ABA and osmotic stress in Gossypium. PeerJ 2017, 5, e4126. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Li, Y.; Ma, X.; Guo, L.; He, Y.; Ren, Z.; Kuang, Z.; Zhang, X.; Zhang, Z. Analysis of potential strategies for cadmium stress tolerance revealed by transcriptome analysis of upland cotton. Sci. Rep. 2019, 9, 86. [Google Scholar] [CrossRef]
- Xu, Y.F. Transcriptome Analysis and Screening of Key Genes in Symphytum officinale L. Under Different Heavy Metal Stresses. Master’s Thesis, Northwest Normal University, Lanzhou, China, 2024. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Hu, B.; Deng, F.; Chen, G.; Chen, X.; Chen, Z.H. Evolution of Abscisic Acid Signaling for Stress Responses to Toxic Metals and Metalloids. Front. Plant. Sci. 2020, 11, 909. [Google Scholar] [CrossRef]
- Mittler, R.; Blumwald, E. The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 2015, 27, 64–70. [Google Scholar] [CrossRef]
- Chen, G.; Wang, Y.Y.; Wang, X.L.; Yang, Q.; Quan, X.Y.; Zeng, J.B.; Dai, F.; Zeng, F.R.; Wu, F.B.; Zhang, G.P. Leaf epidermis transcriptome reveals drought-Induced hormonal signaling for stomatal regulation in wild barley. Plant Growth Regul. 2019, 87, 39–54. [Google Scholar] [CrossRef]
- Chmielowska-Bak, J.; Gzyl, J.; Ruciska-Sobkowiak, R.; Arasimowicz-Jelonek, M.; Deckert, J. The new insights into cadmium sensing. Front. Plant. Sci. 2014, 5, 245. [Google Scholar] [CrossRef]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef]
- Kyndt, T.; Denil, S.; Haegeman, A.; Trooskens, G.; Meyer, T.D.; Criekinge, W.V.; Gheysen, G. Transcriptome analysis of rice mature root tissue and root tips in early development by massive parallel sequencing. J. Exp. Bot. 2012, 63, 2141. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Pan, Y.J.; Zhao, X.Q.; Zhu, L.H.; Li, Z.K. Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genom. 2011, 12, 149. [Google Scholar] [CrossRef] [PubMed]
- Saini, S.; Kaur, N.; Pati, P.K. Phytohormones: Key players in the modulation of heavy metal stress tolerance in plants. Ecotoxicol. Environ. Saf. 2021, 223, 112578. [Google Scholar] [CrossRef] [PubMed]
- Zhen, C. Effects of salicyic acid on seeds germination and seedlings growth of cauliflower under cadmium stress. Seed 2009, 28, 39–43. [Google Scholar] [CrossRef]
- Emamverdian, A.; Ding, Y.L.; Mokhberdoran, F. The role of salicylic acid and gibberellin signaling in plant responses to abiotic stress with an emphasis on heavy metals. Plant Signal. Behav. 2020, 15, 1777372. [Google Scholar] [CrossRef]
- Zheng, Y.X.; Zhang, R.; Zhu, Y.; Ao, Q.M.; Liu, H.; Li, A.H.; Lin, L.J.; Wang, L. Salicylic acid improves Nasturtium officinale phytoremediation capability for cadmium-contaminated paddy soils. Front. Plant. Sci. 2022, 13, 1059175. [Google Scholar] [CrossRef]
- Chen, L.; Long, X.H.; Jin, L.; Jin, S.Z.; Zheng, X.T.; Liu, Z.P. Mitigation effects of exogenous salicylic acid on the seedling growth of two Helianthus tuberosus varieties under Cd stress. Chin. J. Ecol. 2011, 30, 2155–2164. [Google Scholar] [CrossRef]
- Falkowska, M.; Pietryczuk, A.; Piotrowska, A.; Bajguz, A.; Grygoruk, A.; Czerpak, R. The Effect of Gibberellic Acid (GA(3)) on Growth, Metal Biosorption and Metabolism of the Green Algae Chlorella vulgaris (Chlorophyceae) Beijerinck Exposed to Cadmium and Lead Stress. Pol. J. Environ. Stud. 2011, 20, 53–59. [Google Scholar] [CrossRef]
- Chen, H.; Yang, R.X.; Zhang, X.; Chen, Y.H.; Xia, Y.; Xu, X.M. Foliar application of gibberellin inhibits the cadmium uptake and xylem transport in lettuce (Lactuca sativa L.). Sci. Hortic. 2021, 288, 110410. [Google Scholar] [CrossRef]
- Hsu, Y.T.; Kao, C.H. Role of abscisic acid in cadmium tolerance of rice (Oryza sativa L.) seedlings. Plant Cell Environ. 2010, 26, 867–874. [Google Scholar] [CrossRef]
- Stroiński, A.; Giewska, K.; Zielezińska, M. Abscisic acid is required in transduction of cadmium signal to potato roots. Biol. Plant. 2012, 57, 121–127. [Google Scholar] [CrossRef]
- Liu, Y.K.; Tao, Q.; Li, J.X.; Guo, X.Y.; Li, T.Q. Ethylene-mediated apoplastic barriers development involved in cadmium accumulation in root of hyperaccumulator Sedum alfredii. J. Hazard. Mater. 2020, 403, 123729. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.H.; Wu, L.W.; Wang, Q.; Wang, Y.; Luo, H.; Song, J.Y.; Yang, M.H.; Yao, H.; Chen, S.L. Physiological and molecular mechanisms of medicinal plants in response to cadmium stress: Current status and future perspective. J. Hazard. Mater. 2023, 450, 131008. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.L.; Tian, S.K.; Yang, X.E.; Li, T.Q.; He, Z.L. Cadmium uptake and xylem loading are active processes in the hyperaccumulator Sedum alfredii. J. Plant Physiol. 2009, 166, 579–587. [Google Scholar] [CrossRef]
- Sterckeman, T.; Thomine, S. Mechanisms of Cadmium Accumulation in Plants. Crit. Rev. Plant Sci. 2020, 39, 322–359. [Google Scholar] [CrossRef]
- Plaza, S.; Tearall, K.L.; Jie, Z.F.; Buchner, P.; McGrath, S.P.; Hawkesford, M.J. Expression and functional analysis of metal transporter genes in two contrasting ecotypes of the hyperaccumulator Thlaspi caerulescens. J. Exp. Bot. 2007, 58, 1717–1728. [Google Scholar] [CrossRef]
- Yang, Q.Y.; Ma, X.X.; Luo, S.; Gao, J.; Yang, X.E.; Yang, Y. SaZIP4, an uptake transporter of Zn/Cd hyperaccumulator Sedum alfredii Hance. Environ. Exp. Bot. 2018, 155, 107–117. [Google Scholar] [CrossRef]
- Takahashi, R.; Ishimaru, Y.; Senoura, T.; Shimo, H.; Ishikawa, S.; Arao, T.; Nakanishi, H.; Nishizawa, N.K. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J. Exp. Bot. 2011, 62, 4843–4850. [Google Scholar] [CrossRef]
- Hassinen, V.H.; Tervahauta, A.I.; Halimaa, P.; Plessl, M.; Peräniemi, S.; Schat, H.; Aarts, M.G.M.; Servomaa, K.; Kärenlampi, S.O. Isolation of Zn-responsive genes from two accessions of the hyperaccumulator plant Thlaspi caerulescens. Planta 2007, 225, 977–989. [Google Scholar] [CrossRef]
- Van de Mortel, J.E.; Schat, H.; Moerland, P.D.; Ver Loren van Themaat, E.; Van der Ent, S.; Blankestijn, H.; Ghandilyan, A.; Tsiatsiani, S.; Aarts, M.G. Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 2008, 31, 301–324. [Google Scholar] [CrossRef]
- Fu, X.Z.; Tong, Y.H.; Zhou, X.; Ling, L.L.; Chun, C.P.; Cao, L.; Zeng, M.; Peng, L.Z. Genome-wide identification of sweet orange (Citrus sinensis) metal tolerance proteins and analysis of their expression patterns under zinc, manganese, copper, and cadmium toxicity. Gene 2017, 629, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, D.F.; Ruscitti, T.; Mccue, K.F.; Ow, D.W. Transport of Metal-binding Peptides by HMT1, A Fission Yeast ABC-type Vacuolar Membrane Protein. J. Biol. Chem. 1995, 270, 4721–4728. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Sun, L.; Yang, X.E.; Liu, J.X. Transcriptomic Analysis of Cadmium Stress Response in the Heavy Metal Hyperaccumulator Sedum alfredii Hance. PLoS ONE 2013, 8, e64643. [Google Scholar] [CrossRef]
- Zhang, Z.L.; Zhong, L.L.; Xiao, W.T.; Du, Y.P.; Han, G.Q.; Yan, Z.Y.; He, D.M.; Zheng, C. Transcriptomics combined with physiological analysis reveals the mechanism of cadmium uptake and tolerance in Ligusticum chuanxiong Hort. under cadmium treatment. Front. Plant. Sci. 2023, 14, 1263981. [Google Scholar] [CrossRef]
- Park, J.; Song, W.Y.; Ko, D.; Eom, Y.; Hansen, T.H.; Schiller, M.; Lee, T.G.; Martinoia, E.; Lee, Y. The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 2012, 69, 278–288. [Google Scholar] [CrossRef]
- Zhu, X.F.; Lei, G.J.; Jiang, T.; Liu, Y.; Li, G.X.; Zheng, S.J. Cell wall polysaccharides are involved in P-deficiency-induced Cd exclusion in Arabidopsis thaliana. Planta 2012, 236, 989–997. [Google Scholar] [CrossRef]
- Hall, J.L. Cellular mechanisms for heavy metal detoxification and tolerance. J. Exp. Bot. 2002, 53, 1–11. [Google Scholar] [CrossRef]
- Lampugnani, E.R.; Khan, G.A.; Somssich, M.; Persson, S. Building a plant cell wall at a glance. J. Cell Sci. 2018, 131, 207373. [Google Scholar] [CrossRef]
- Lu, Q.Y. Study on the Promoting Effect of ABA on Cd Accumulation of Sedum alfredii Based on Omics Technology. Ph.D. Thesis, Guangxi University, Nanning, China, 2020. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Lehman, T.A.; Smertenko, A.; Sanguinet, K.A. Auxin, microtubules, and vesicle trafficking: Conspirators behind the cell wall. J. Exp. Bot. 2017, 68, 3321–3329. [Google Scholar] [CrossRef]
- Xiong, J.; An, L.; Lu, H.; Zhu, C. Exogenous nitric oxide enhances cadmium tolerance of rice by increasing pectin and hemicellulose contents in root cell wall. Planta 2009, 230, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Bücker-Neto, L.; Paiva, A.L.S.; Machado, R.D.; Arenhart, R.A.; Margis-Pinheiro, M. Interactions between plant hormones and heavy metals responses. Genet. Mol. Biol. 2017, 40, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.H.; Yin, X.M.; Gao, K.H.; Ge, Y.; Cheng, W.D. Non-protein thiols and glutathione S-transferase alleviate Cd stress and reduce root-to-shoot translocation of Cd in rice. J. Plant Nutr. Soil Sci. 2013, 176, 626–633. [Google Scholar] [CrossRef]
- Nguyen, T.Q.; Sesin, V.; Kisiala, A.; Emery, R.J.N. Phytohormonal Roles in Plant Responses to Heavy Metal Stress: Implications for Using Macrophytes in Phytoremediation of Aquatic Ecosystems. Environ. Toxicol. Chem. 2021, 40, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Bajguz, A. Brassinosteroids and lead as stimulators of phytochelatins synthesis in Chlorella vulgaris. J. Plant Physiol. 2002, 159, 321–324. [Google Scholar] [CrossRef]
- Stroiński, A.; Chadzinikolau, T.; Giżewska, K.; Zielezińska, M. ABA or cadmium induced phytochelatin synthesis in potato tubers. Biol. Plant. 2010, 54, 117–120. [Google Scholar] [CrossRef]
- Afzal, J.; Hu, C.; Imtiaz, M.; Elyamine, A.M.; Rana, M.S.; Imran, M.; Farag, M.A. Cadmium tolerance in rice cultivars associated with antioxidant enzymes activities and Fe/Zn concentrations. Int. J. Environ. Sci. Technol. 2018, 16, 4241–4252. [Google Scholar] [CrossRef]
- Chen, Y.K.; Zhi, J.K.; Zhang, H.; Li, J.; Zhao, Q.H.; Xu, J.C. Transcriptome analysis of Phytolacca americana L. in response to cadmium stress. PLoS ONE 2017, 12, e0184681. [Google Scholar] [CrossRef]
- Zhang, F.X.; Zhang, X.Z.; Wang, Y.; Han, Z.H. Changes of H2O2 content and activities of related enzymes in leaf among ontogenetic phases in apple (Malus ssp.). J. Fruit Sci. 2013, 30, 759–764. [Google Scholar] [CrossRef]
- You, C.C.; Zhu, H.L.; Xu, B.B.; Huang, W.X.; Wang, S.H.; Ding, Y.F.; Liu, Z.H.; Li, G.H.; Chen, L.; Ding, C.Q.; et al. Effect of Removing Superior Spikelets on Grain Filling of Inferior Spikelets in Rice. Front. Plant. Sci. 2016, 7, 1161. [Google Scholar] [CrossRef]
- Xie, Q.; Ma, L.Y.; Tan, P.; Deng, W.T.; Huang, C.; Liu, D.M.; Lin, W.H.; Su, Y. Multiple High-Affinity K+ Transporters and ABC Transporters Involved in K+ Uptake/Transport in the Potassium-Hyperaccumulator Plant Phytolacca acinosa Roxb. Plants 2020, 9, 470. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Xu, J.; Huang, K.; Su, Y.; Tong, J.; Huang, Z.; Huang, C.; Wei, M.; Lin, W.; Xiao, L. Dynamic formation and transcriptional regulation mediated by phytohormones during chalkiness formation in rice. BMC Plant Biol. 2021, 21, 308. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, 182–185. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression of RNA-Seq data at the gene level–the DESeq package. Genome Res. 2012, 11, R106. [Google Scholar] [CrossRef]


| Sample | Raw Read Number | Raw Base Number | Clean Read Number | Clean Base Number | Valid Bases (%) | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|---|---|
| Cd−T1 | 37,516,010 | 5,627,401,500 | 37,184,932 | 5,549,209,598 | 98.61% | 97.68% | 93.16% | 46.00% |
| Cd−T2 | 43,508,604 | 6,526,290,600 | 42,969,078 | 6,411,399,243 | 98.23% | 97.91% | 93.73% | 46.00% |
| Cd−T3 | 42,372,036 | 6,355,805,400 | 41,966,768 | 6,268,020,644 | 98.61% | 97.84% | 93.58% | 46.00% |
| ABA+Cd−T1 | 37,543,856 | 5,631,578,400 | 37,209,394 | 5,536,724,230 | 98.31% | 97.88% | 93.68% | 45.00% |
| ABA+Cd−T2 | 39,293,478 | 5,894,021,700 | 38,817,700 | 5,787,819,153 | 98.19% | 97.89% | 93.70% | 47.50% |
| ABA+Cd−T3 | 46,979,258 | 7,046,888,700 | 46,448,560 | 6,935,119,640 | 98.41% | 97.89% | 93.70% | 48.00% |
| Subfamily | ID | Gene | Log2FoldChange | Regulation |
|---|---|---|---|---|
| ABCA | DN226612_c1_g3_i5 | ABCA1 | 2.64 | Up |
| DN218393_c2_g1_i5 | ABCA2 | 1.97 | Up | |
| DN227994_c8_g1_i3 | ABCA7 | 2.34 | Up | |
| ABCB | DN194339_c0_g1_i1 | ABCB1 | −3.19 | Down |
| DN228060_c3_g2_i2 | ABCB4 | −2.80 | Down | |
| DN220815_c0_g1_i1 | ABCB8 | 4.31 | Up | |
| DN220558_c0_g3_i2 | ABCB15 | 2.39 | Up | |
| DN189123_c0_g1_i1 | ABCB17 | 4.94 | Up | |
| DN173660_c0_g1_i1 | ABCB19 | 4.42 | Up | |
| DN225659_c2_g1_i1 | ABCB20 | −2.13 | Down | |
| DN228060_c3_g1_i9 | ABCB21 | 1.49 | Up | |
| DN132622_c0_g1_i1 | ABCB25 | 3.76 | Up | |
| DN225703_c4_g1_i1 | ABCB26 | 2.93 | Up | |
| DN226871_c9_g1_i19 | ABCB28 | 1.87 | Up | |
| ABCC | DN225934_c6_g2_i1 | ABCC1 | 3.45 | Up |
| DN228942_c1_g1_i6 | ABCC2 | 3.12 | Up | |
| DN46026_c0_g1_i1 | ABCC3 | −3.79 | Down | |
| DN222948_c9_g1_i1 | ABCC4 | 2.00 | Up | |
| DN228734_c0_g1_i7 | ABCC5 | 3.19 | Up | |
| DN218095_c0_g1_i2 | ABCC9 | 3.60 | Up | |
| DN226038_c0_g5_i1 | ABCC10 | 3.42 | Up | |
| DN224101_c1_g1_i7 | ABCC12 | 2.60 | Up | |
| DN200068_c0_g1_i4 | ABCC13 | −3.86 | Down | |
| DN219104_c0_g3_i2 | ABCC14 | 3.40 | Up | |
| ABCD | DN224069_c2_g1_i11 | ABCD1 | 2.39 | Up |
| ABCE | DN228480_c2_g1_i12 | ABCE2 | 2.61 | Up |
| ABCF | DN226310_c0_g5_i1 | ABCF1 | 2.45 | Up |
| DN221793_c0_g1_i1 | ABCF3 | 3.47 | Up | |
| DN216277_c0_g1_i1 | ABCF4 | 2.57 | Up | |
| DN216043_c1_g3_i1 | ABCF5 | 1.77 | Up | |
| ABCG | DN197065_c0_g1_i1 | ABCG2 | 2.31 | Up |
| DN203839_c0_g1_i1 | ABCG3 | 2.98 | Up | |
| DN227456_c0_g8_i1 | ABCG4 | 3.69 | Up | |
| DN200185_c0_g4_i1 | ABCG6 | 2.66 | Up | |
| DN228592_c0_g1_i1 | ABCG7 | 1.44 | Up | |
| DN218943_c3_g2_i10 | ABCG11 | −2.34 | Down | |
| DN203802_c0_g1_i1 | ABCG14 | 3.95 | Up | |
| DN196886_c0_g1_i2 | ABCG15 | 1.91 | Up | |
| DN221450_c0_g1_i2 | ABCG25 | 2.26 | Up | |
| DN221568_c5_g1_i7 | ABCG28 | −2.19 | Down | |
| DN226694_c0_g1_i5 | ABCG31 | 2.47 | Up | |
| DN228476_c1_g5_i1 | ABCG42 | 4.41 | Up | |
| DN224679_c1_g2_i1 | ABCG53 | −1.82 | Down | |
| ABCI | DN227970_c1_g1_i6 | ABCI1 | 2.14 | Up |
| DN126655_c0_g2_i1 | ABCI6 | 3.28 | Up | |
| DN219020_c7_g2_i2 | ABCI17 | 2.35 | Up | |
| DN195569_c0_g1_i1 | ABCI19 | −2.28 | Down | |
| DN221951_c1_g1_i2 | ABCI20 | 2.78 | Up |
| ID | Primer Sequences 5′–3′ |
|---|---|
| DN226425_c0_g3_i2 | F: CGGGTGGTTGGTTCACAGTA |
| R: CCAAGCTAAGTGCCCCATCA | |
| DN220815_c0_g1_i1 | F: GTCGTCTTCTCAGGGTGGTG |
| R: ATTGGCGTAGGATCGGGTTC | |
| DN221085_c4_g1_i8 | F: ATTGCCTCTTGCTCTTCCGT |
| R: GCTAGCCTCCTTCATGGGTG | |
| DN192292_c1_g1_i1 | F: CACGTGACGTCTTTGGCTAC |
| R: AGTATTCCTTCTCGATCCGCC | |
| DN229031_c8_g1_i5 | F: TCAACACCGCGCTCATTACT |
| R: CCCACCCCTGATAACCAACC | |
| DN175421_c0_g1_i1 | F: CTGCAGGTGTCCCGATGATT |
| R:CCCATTGCATAGCCACCTCA | |
| DN219478_c1_g1_i4 | F: TATCCCCCTGGCGTTTGTTC |
| R: GCATTGGGTTGGCGTTCATT | |
| DN220579_c6_g1_i2 | F: CCTGCTGATGTTTCCAAGCG |
| R: ACTTCTACCCGAGTCCGAGA | |
| DN216324_c5_g3_i1 | F: TCGAGAAAGAGGCGAACCTG |
| R: GGCGATCAACGGTGGAGATT | |
| DN223975_c4_g1_i1 | F: TACTCATGGGGGAGGGGAAG |
| R: CCACACTCGATCTGGCATGT | |
| Actin | F: TTGAGCAGGAATCGGAG |
| R: TGCTGCTTCCATACCTATC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Q.; Xu, W.; Wang, Q.; Yao, F.; Jiang, Y.; Cao, H.; Lin, W. Transcriptome and Physiological Analysis Reveals the Mechanism of Abscisic Acid in Regulating Cadmium Uptake and Accumulation in the Hyperaccumulator Phytolacca acinosa Roxb. Plants 2025, 14, 1405. https://doi.org/10.3390/plants14101405
Xie Q, Xu W, Wang Q, Yao F, Jiang Y, Cao H, Lin W. Transcriptome and Physiological Analysis Reveals the Mechanism of Abscisic Acid in Regulating Cadmium Uptake and Accumulation in the Hyperaccumulator Phytolacca acinosa Roxb. Plants. 2025; 14(10):1405. https://doi.org/10.3390/plants14101405
Chicago/Turabian StyleXie, Qin, Wenting Xu, Qing Wang, Feihong Yao, Yachao Jiang, Haijia Cao, and Wanhuang Lin. 2025. "Transcriptome and Physiological Analysis Reveals the Mechanism of Abscisic Acid in Regulating Cadmium Uptake and Accumulation in the Hyperaccumulator Phytolacca acinosa Roxb." Plants 14, no. 10: 1405. https://doi.org/10.3390/plants14101405
APA StyleXie, Q., Xu, W., Wang, Q., Yao, F., Jiang, Y., Cao, H., & Lin, W. (2025). Transcriptome and Physiological Analysis Reveals the Mechanism of Abscisic Acid in Regulating Cadmium Uptake and Accumulation in the Hyperaccumulator Phytolacca acinosa Roxb. Plants, 14(10), 1405. https://doi.org/10.3390/plants14101405






