Receptor-like Kinase GOM1 Regulates Glume-Opening in Rice
Abstract
1. Introduction
2. Results
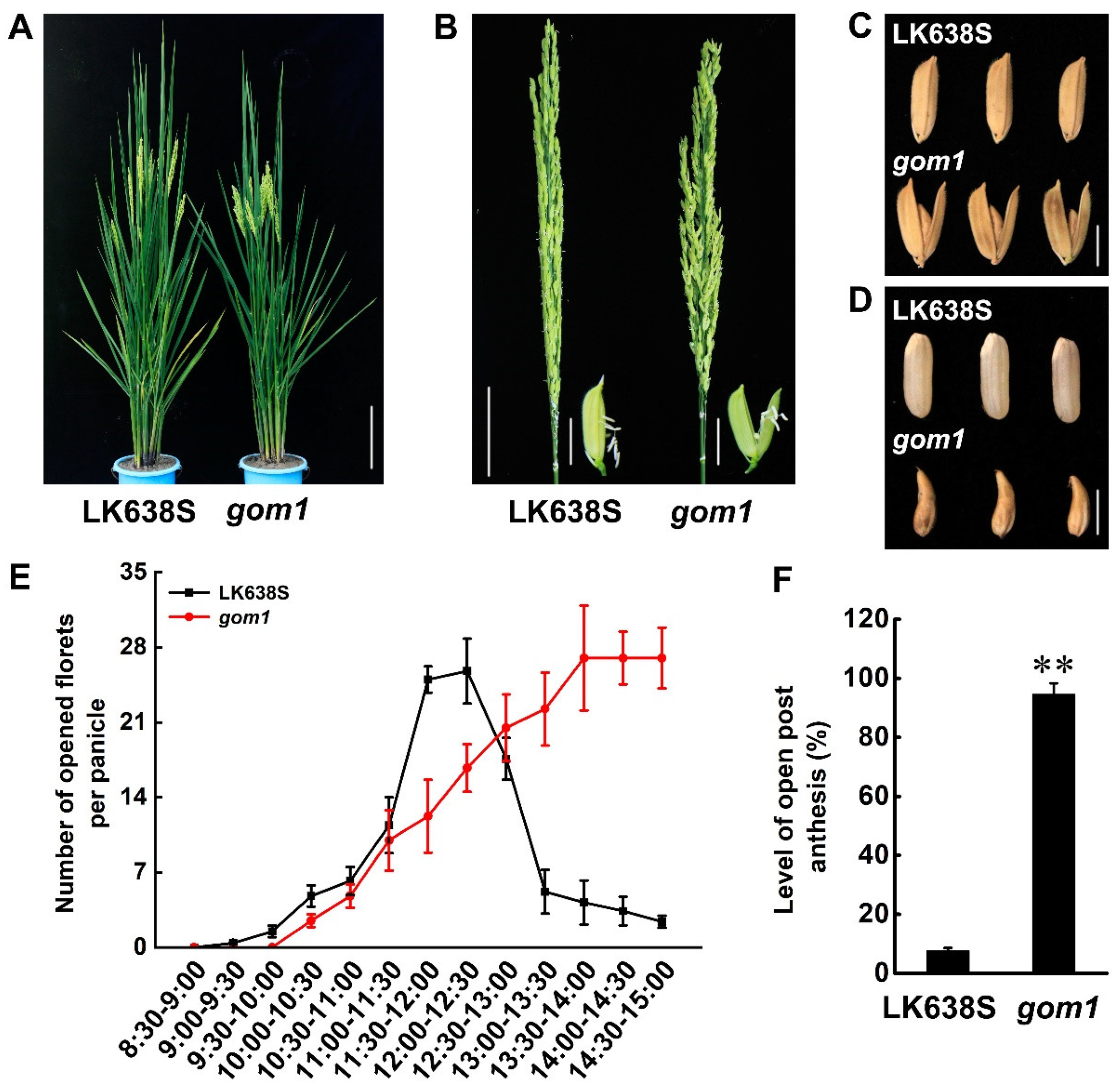
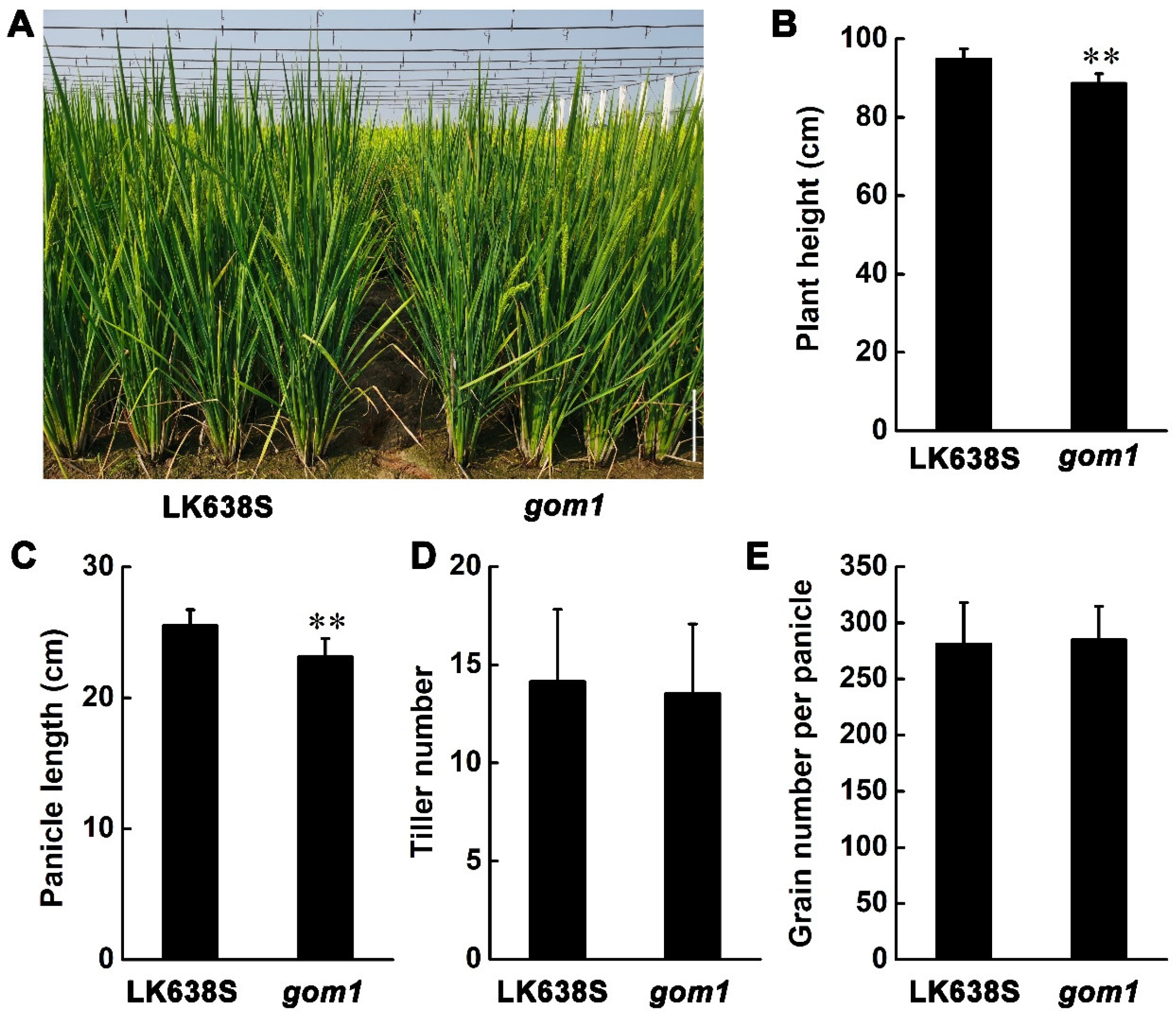
2.1. Characterization of the gom1 Mutant
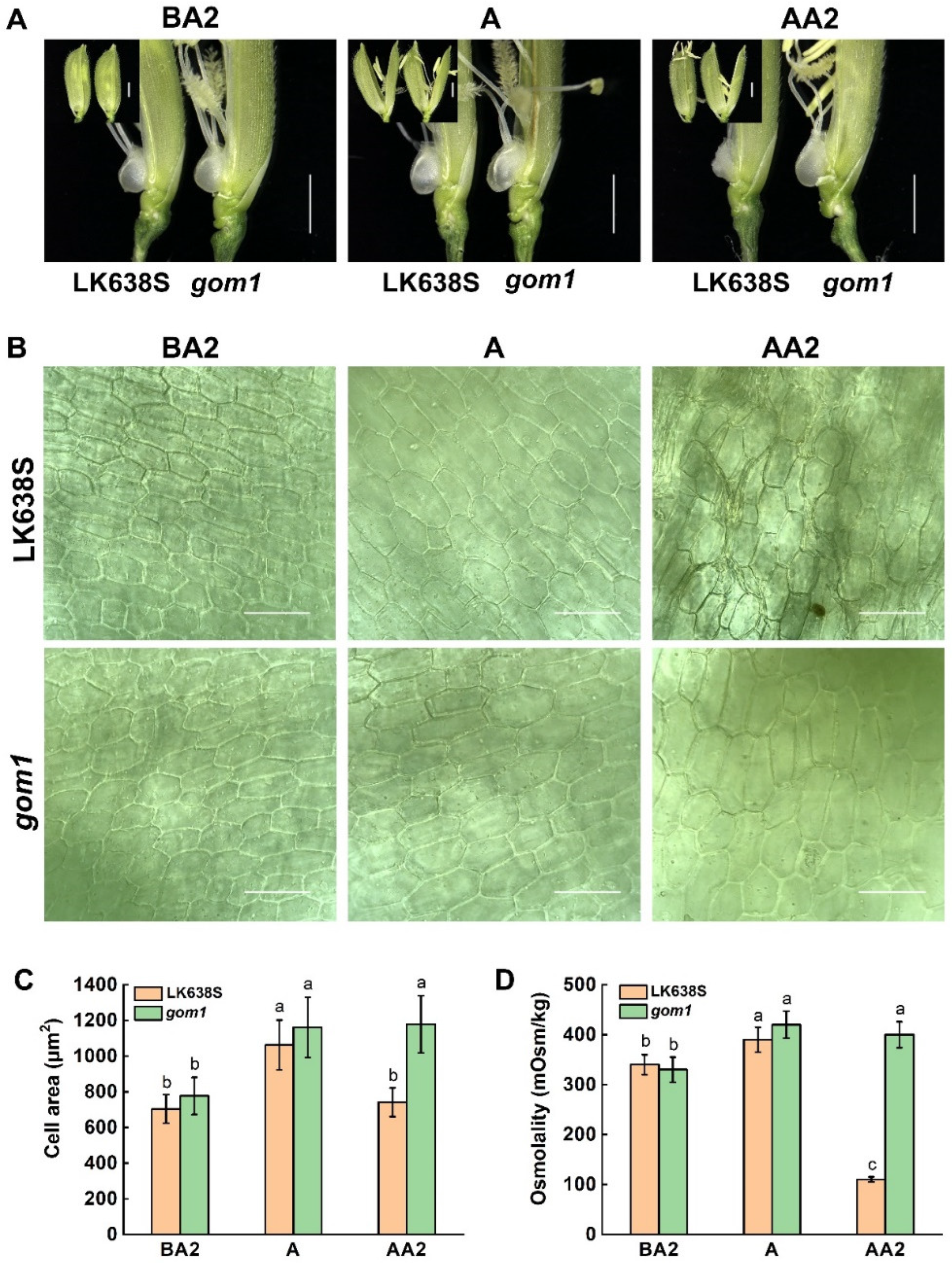
2.2. The Lodicules of gom1 Mutant Delayed Withering After Anthesis
2.3. Map-Based Cloning of the GOM1 Locus
2.4. The Expression Profiles of GOM1
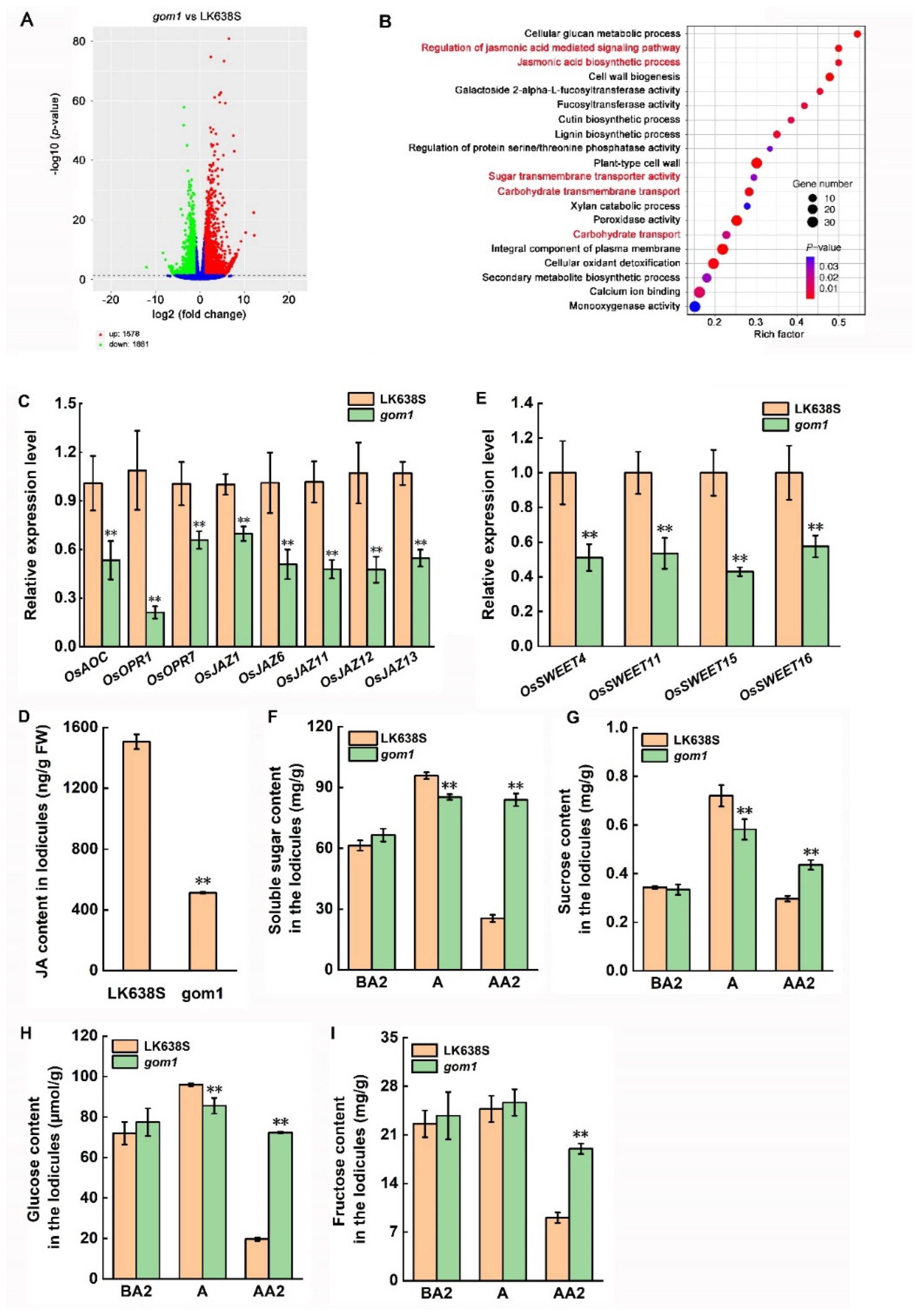
2.5. Mutation of GOM1 Compromises the Expression of Genes Related to JA Signal Pathway
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Map-Based Cloning of GOM1
4.3. Vector Construction and Transgenic Analysis
4.4. Promoter-GUS Analysis
4.5. JA Measurement
4.6. Determination of Soluble Sugar Contents and Osmolality
4.7. RNA Sequencing (RNA-Seq) Analysis
4.8. RNA Isolation and RT-qPCR
4.9. Dynamic Observation and Analysis of Lodicule Cell Size
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, L.; Liu, Y.G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 2014, 65, 579–606. [Google Scholar] [CrossRef]
- Ma, G.; Yuan, L. Hybrid rice achievements, development and prospect in China. J. Integr. Agric. 2015, 14, 197–205. [Google Scholar] [CrossRef]
- Yuan, L. Purification and production of foundation seed of rice PGMS and TGMS lines. Hybrid Rice 1994, 2, 15–16. [Google Scholar]
- Pak, H.; Wang, H.; Kim, Y.; Song, U.; Tu, M.; Wu, D.; Jiang, L. 2021 Creation of male-sterile lines that can be restored to fertility by exogenous methyl jasmonate for the establishment of a two-line system for the hybrid production of rice (Oryza sativa L.). Plant Biotechnol. J. 2021, 19, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, S.; Jiang, N.; Zhao, X.; Bai, Z.; Liu, J.; Yao, W.; Tang, Q.; Xiao, G.; Lv, C.; et al. Engineering of rice varieties with enhanced resistances to both blast and bacterial blight diseases via CRISPR/Cas9. Plant Biotechnol. J. 2022, 20, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Pu, C.X.; Ma, Y.; Wang, J.; Zhang, Y.C.; Jiao, X.W.; Hu, Y.H.; Wang, L.L.; Zhu, Z.Z.; Sun, D.; Sun, Y. Crinkly4 receptor-like kinase is required to maintain the interlocking of the palea and lemma, and fertility in rice, by promoting epidermal cell differentiation. Plant J. 2012, 70, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, E.A. Evolutionary history of the grasses. Plant Physiol. 2001, 125, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Sunohara, H.; Nagato, Y. Developmental course of inflorescence and spikelet in rice. Breed. Sci. 2004, 54, 147–156. [Google Scholar] [CrossRef]
- Itoh, J.; Nonomura, K.; Ikeda, K.; Yamaki, S.; Inukai, Y.; Yamagishi, H.; Kitano, H.; Nagato, Y. Rice plant development: From zygote to spikelet. Plant Cell Physiol. 2005, 46, 23–47. [Google Scholar] [CrossRef]
- Wang, J.; Shen, Z.; Shi, S.Y. The flowering habit of indica and japonica rices and its inheritance. Hybrid Rice 1991, 5, 39–42. [Google Scholar]
- Liu, L.; Zou, Z.; Qian, K.; Xia, C.; He, Y.; Zeng, H.; Zhou, X.; Riemann, M.; Yin, C. Jasmonic acid deficiency leads to scattered floret opening time in 893 cytoplasmic male sterile rice Zhenshan 97A. J. Exp. Bot. 2017, 68, 4613–4625. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Cai, Q.; Yuan, Z.; Chen, M.; Yin, C.; Luo, Z.; Zhao, X.; Liang, W.; Hu, J.; Zhang, D. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 2014, 5, 3476. [Google Scholar] [CrossRef]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Tian, J.; Liu, Y.; Chen, X.; Li, S.; Persson, S.; Lu, D.; Chen, M.; Luo, Z.; Zhang, D.; et al. Ectopic expression of OsJAZ6, which interacts with OsJAZ1, alters JA signaling and spikelet development in rice. Plant J. 2021, 8, 1083–1096. [Google Scholar] [CrossRef] [PubMed]
- Howe, G.A.; Major, I.T.; Koo, A.J. Modularity in jasmonate signaling for multistress resilience. Annu. Rev. Plant Biol. 2018, 69, 387–415. [Google Scholar] [CrossRef]
- Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Katsir, L.; Schilmiller, A.L.; Staswick, P.E.; He, S.Y.; Howe, G.A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 2008, 105, 7100–7105. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.X.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Castells, E.; Casacuberta, J.M. Signalling through kinase-defective domains: The prevalence of atypical receptor-like kinases in plants. J. Exp. Bot. 2007, 58, 3503–3511. [Google Scholar] [CrossRef]
- Haderlein, L.; Jensen, T.L.; Dowbenko, R.E.; Blaylock, A.D. Controlled release urea as a nitrogen source for spring wheat in western Canada: Yield, grain N content, and N use efficiency. Sci. World J. 2001, 1, 114–121. [Google Scholar] [CrossRef]
- Bai, L.; Zhang, G.; Zhou, Y.; Zhang, Z.; Wang, W.; Du, Y.; Wu, Z.; Song, C.P. Plasma membrane-associated proline-rich extension-like receptor kinase 4, a novel regulator of Ca2+ signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J. 2009, 60, 314–327. [Google Scholar] [CrossRef] [PubMed]
- Lehti-Shiu, M.D.; Zou, C.; Hanada, K.; Shiu, S.H. Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 2009, 150, 12–26. [Google Scholar] [CrossRef]
- Den Herder, G.; Yoshida, S.; Antolin-Llovera, M.; Ried, M.K.; Parniske, M. Lotus japonicus E3 ligase SEVEN IN ABSENTIA4 destabilizes the symbiosis receptor-like kinase SYMRK and negatively regulates rhizobial infection. Plant Cell 2012, 24, 1691–1707. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.B.; Liu, C.; Tang, D.Y.; Yan, L.; Wang, D.; Yang, Y.Z.; Gui, J.S.; Zhao, X.Y.; Li, L.G.; Tang, X.D.; et al. The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 2018, 30, 1100–1118. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Z.; Zhao, X.; Liu, L.; Tang, Q.; Fu, J.; Tang, X.; Yang, R.; Lin, J.; Liu, X.; et al. Receptor-like cytoplasmic kinase STK confers salt tolerance in rice. Rice 2023, 16, 21. [Google Scholar] [CrossRef]
- Shiu, S.H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef] [PubMed]
- Tor, M.; Lotze, M.T.; Holton, N. Receptor-mediated signalling in plants: Molecular patterns and programmes. J. Exp. Bot. 2009, 60, 3645–3654. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhu, X.; Peng, G.; Liu, M.; Zhang, S.; Chen, M.; Liao, S.; Wei, X.; Xu, P.; Tan, X.; et al. Methylesterification of cell-wall pectin controls the diurnal flower-opening times in rice. Mol. Plant 2022, 15, 956–972. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Luo, X.; Zhu, L. Cytological analysis and genetic control of rice anther development. J. Genet. Genom. 2011, 38, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y.; Duan, E.; Qi, Q.; Zhou, K.; Lin, Q.; Wang, D.; Wang, Y.; Long, W.; Zhao, Z.; et al. OPEN GLUME1: A key enzyme reducing the precursor of JA, participates in carbohydrate transport of lodicules during anthesis in rice. Plant Cell Rep. 2018, 37, 329–346. [Google Scholar] [CrossRef]
- Sosso, D.; Luo, D.; Li, Q.B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; McCarty, D.R.; Chourey, P.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef] [PubMed]
- Streubel, J.; Pesce, C.; Hutin, M.; Koebnik, R.; Boch, J.; Szurek, B. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 2013, 200, 808–819. [Google Scholar] [CrossRef]
- Yang, J.; Luo, D.; Yang, B.; Frommer, W.B.; Eom, J.S. SWEET 11 and 15 as key players in seed filling in rice. New Phytol. 2018, 218, 604–615. [Google Scholar] [CrossRef]
- Chen, J.; Xu, Y.; Fei, K.; Wang, R.; He, J.; Fu, L.; Shao, S.; Li, K.; Zhu, K.; Zhang, W.; et al. Physiological mechanism underlying the effect of high temperature during anthesis on spikelet-opening of photo-thermo-sensitive genic male sterile rice lines. Sci. Rep. 2020, 10, 2210. [Google Scholar] [CrossRef] [PubMed]
- Gómez, J.F.; Talle, B.; Wilson, Z.A. Anther and pollen development: A conserved developmental pathway. J. Integr. Plant Biol. 2015, 57, 876–891. [Google Scholar] [CrossRef]
- Xiao, Y.; Chen, Y.; Charnikhova, T.; Mulder, P.P.J.; Heijmans, J.; Hoogenboom, A.; Agalou, A.; Michel, C.; Morel, J.B.; Dreni, L.; et al. OsJAR1 is required for JA-regulated floret opening and anther dehiscence in rice. Plant Mol. Biol. 2014, 86, 19–33. [Google Scholar] [PubMed]
- Zhang, D.; Yang, L. Specification of tapetum and microsporocyte cells within the anther. Curr. Opin. Plant Biol. 2014, 17, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Gou, Y.; Heng, Y.; Ding, W.; Xu, C.; Tan, Q.; Li, Y.; Fang, Y.; Li, X.; Zhou, D.; Zhu, X.; et al. Natural variation in OsMYB8 confers diurnal floret opening time divergence between indica and japonica subspecies. Nat. Commun. 2024, 15, 2262. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhu, X.; Huang, Z.; Chen, M.; Xu, P.; Liao, S.; Zhao, Y.; Gao, Y.; He, J.; Luo, Y.; et al. Controlling diurnal flower-opening time by manipulating the jasmonate pathway accelerates development of indica–japonica hybrid rice breeding. Plant Biotechnol. J. 2024, 22, 2267–2281. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Gou, Y.; Li, Y.; Li, J.; Fang, Y.; Liu, X.; Zhu, X.; Ye, R.; Heng, Y.; Wang, H.; et al. A jasmonate-mediated regulatory network modulates diurnal floret opening time in rice. New Phytol. 2024, 244, 176–191. [Google Scholar] [CrossRef] [PubMed]
- Hibara, K.; Isono, M.; Mimura, M.; Sentoku, N.; Kojima, M.; Sakakibara, H.; Kitomi, Y.; Yoshikawa, T.; Itoh, J.I.; Nagato, Y. Jasmonate regulates juvenile-adult phase transition in rice. Development 2016, 143, 3407–3416. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef]
- Sanders, P.M.; Lee, P.Y.; Biesgen, C.; Boone, J.D.; Beals, T.P.; Weiler, E.W.; Goldberg, R.B. The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 2000, 12, 1041–1061. [Google Scholar] [CrossRef]
- Stintzi, A.; Browse, J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 10625–10630. [Google Scholar] [CrossRef] [PubMed]
- Adewusi, K.M.; Showemimo, F.A.; Nassir, A.L.; Olagunju, S.O.; Porbeni, J.B.O.; Amira, J.O.; Aderinola, A.P. Assessment of 60Co gamma radiation on early phenological stages of two generations of OFADA rice. Agro-Science 2021, 20, 31–37. [Google Scholar] [CrossRef]
- Hiei, Y.; Komari, T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 2008, 3, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Betaglucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Li, K.; Hu, L.; Chen, S.; Gai, Y.; Jiang, X. Fast and simple droplet sampling of sap from plant tissues and capillary microextraction of soluble saccharides for picogram-scale quantitative determination with GC-MS. J. Agric. Food. Chem. 2010, 58, 9931–9935. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Liu, S.; Tan, Y.; Chao, N.; Tian, X.; Qi, L.; Powell, W.A.; Jiang, X.; Gai, Y. Optimized GC-MS method to simultaneously quantify acetylated aldose, ketose, and alditol for plant tissues based on derivatization in a methyl sulfoxide/1-methylimidazole system. J. Agric. Food. Chem. 2013, 61, 4011–4018. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Guo, Y.; Li, J.; Long, J.; Zhang, B.; Shyr, Y. Steps to ensure accuracy in genotype and SNP calling from Illumina sequencing data. BMC Genom. 2012, 13, S8. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. clusterProfiler: An universal enrichment tool for functional and comparative study. BioRxiv 2018, 1, 256784. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Wei, M.; Tang, Q.; Tang, L.; Fu, J.; Wang, K.; Zhou, Y.; Yang, Y. Receptor-like Kinase GOM1 Regulates Glume-Opening in Rice. Plants 2025, 14, 5. https://doi.org/10.3390/plants14010005
Zhao X, Wei M, Tang Q, Tang L, Fu J, Wang K, Zhou Y, Yang Y. Receptor-like Kinase GOM1 Regulates Glume-Opening in Rice. Plants. 2025; 14(1):5. https://doi.org/10.3390/plants14010005
Chicago/Turabian StyleZhao, Xinhui, Mengyi Wei, Qianying Tang, Lei Tang, Jun Fu, Kai Wang, Yanbiao Zhou, and Yuanzhu Yang. 2025. "Receptor-like Kinase GOM1 Regulates Glume-Opening in Rice" Plants 14, no. 1: 5. https://doi.org/10.3390/plants14010005
APA StyleZhao, X., Wei, M., Tang, Q., Tang, L., Fu, J., Wang, K., Zhou, Y., & Yang, Y. (2025). Receptor-like Kinase GOM1 Regulates Glume-Opening in Rice. Plants, 14(1), 5. https://doi.org/10.3390/plants14010005





