Mechanism of Action of Fusarium oxysporum CCS043 Utilizing Allelochemicals for Rhizosphere Colonization and Enhanced Infection Activity in Rehmannia glutinosa
Abstract
1. Introduction
2. Results
2.1. Whole-Genome Sequencing, De Novo Assembly, and Annotation of F. oxysporum CCS043 from the Rhizosphere of R. glutinosa
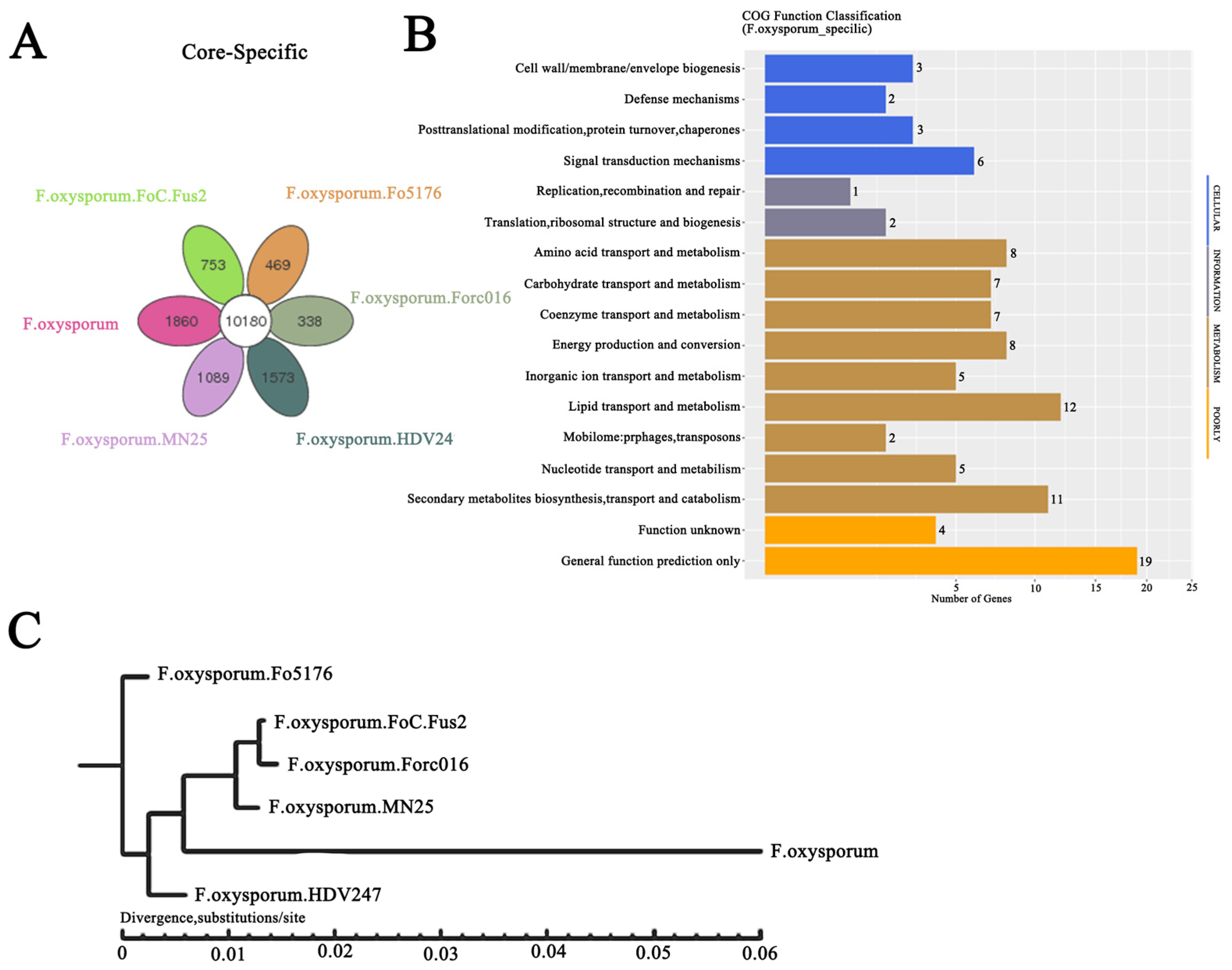
2.2. Evolution of F. oxysporum CCS043 in the Rhizosphere of R. glutinosa
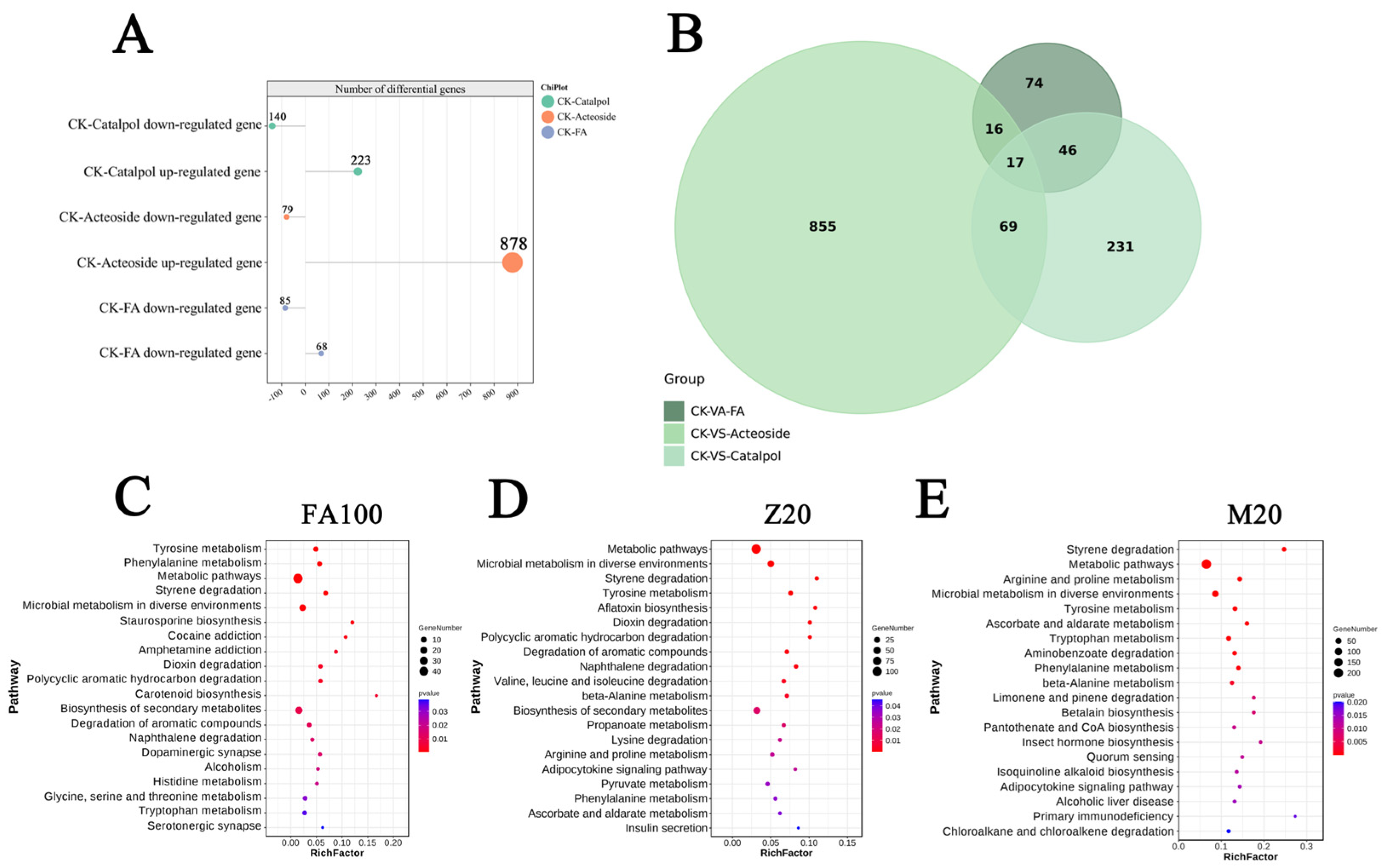
2.3. The Allelopathic Effects of Key Components of Root Exudates from Continuous Cropping of R. glutinosa on F. oxysporum CCS043
2.4. Analysis of Gene Differential Expression Patterns and Metabolic Pathway Enrichment in F. oxysporum CCS043
2.5. Screening and Identification of Effector Proteins in F. oxysporum CCS043 from the Rhizosphere of R. glutinosa
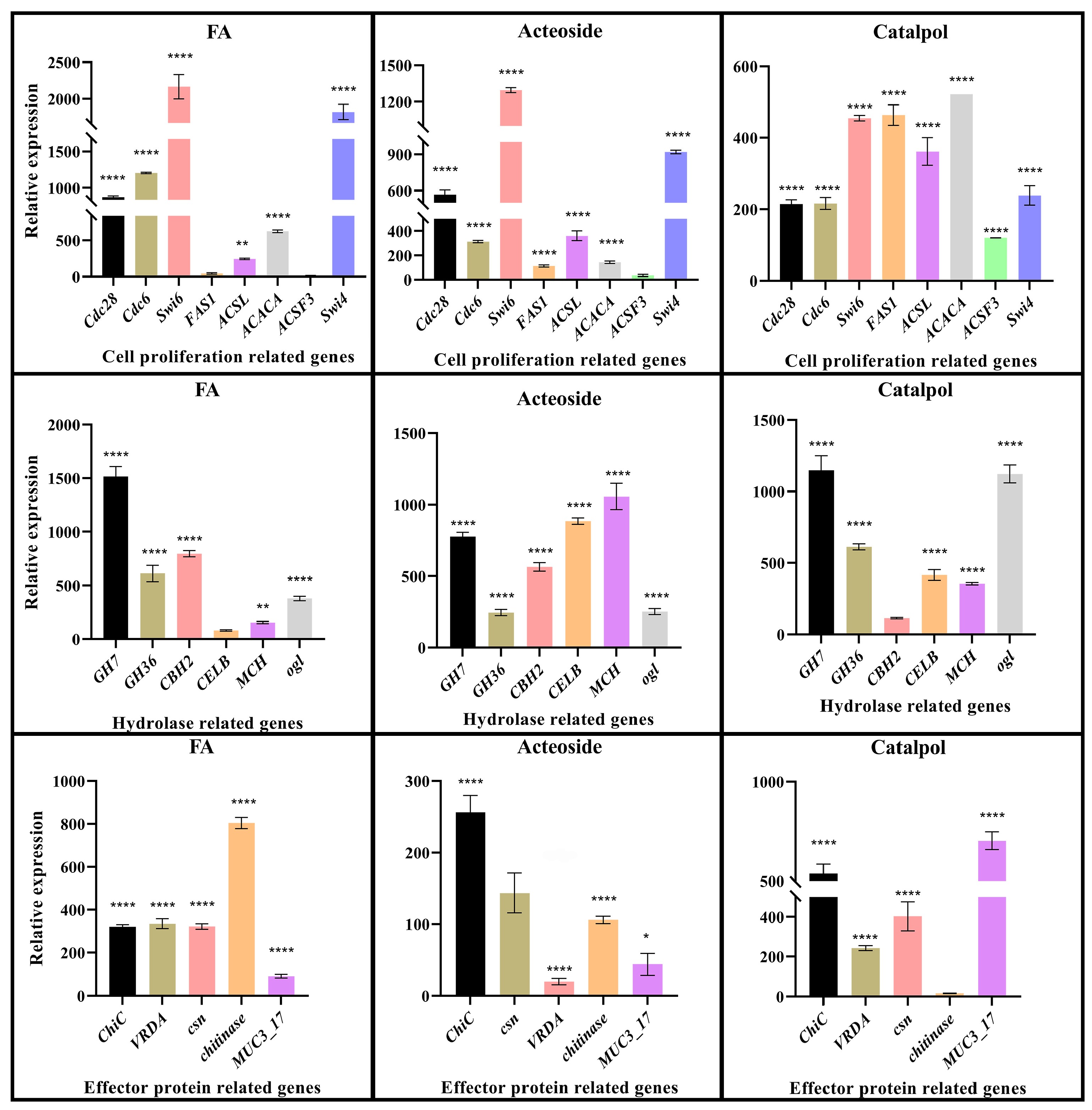
2.6. qRT-PCR Validation of the Key Genes in F. oxysporum CCS043’s Response to Root Exudates
2.7. The Detection of Infection Effects of F. oxysporum CCS043’s Effector Factors on R. glutinosa
3. Discussion
3.1. The Co-Evolutionary Relationship Between Continuous R. glutinosa Cultivation and F. oxysporum CCS043
3.2. The Ten Effector Proteins of F. oxysporum CCS043 from the Rhizosphere of R. glutinosa Identified
3.3. Root Exudates of Continuously Cultivated R. glutinosa Enhance F. oxysporum CCS043 Infection in the Rhizosphere
3.4. F. oxysporum CCS043 Mainly Proliferates and Grows Through Amino Acid Metabolism
3.5. The Molecular Mechanism of R. glutinosa Root Exudates Promoting F. oxysporum CCS043’s Proliferation
4. Materials and Methods
4.1. Determination of the Physiological Indicators of F. oxysporum CCS043
4.2. The RNA-Seq Data Analysis
4.3. RT-qPCR Data Analysis
4.4. Genome Assembly
4.5. Phylogenetic Analysis
4.6. Screening of the Effector Proteins in F. oxysporum CCS043
4.7. ORF Cloning of the Effector Factors in Full Length
4.8. Agrobacterium GV3101 Transformation and Infection
4.9. In Vitro Infection of the R. glutinosa Leaves
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, F.Q.; Zhi, J.Y.; Zang, Z.Y.; Wang, L.N.; Suo, Y.F.; Xie, C.X.; Li, M.J.; Zhang, B.; Du, J.F.; Gu, L.; et al. Transcriptome Analysis of Salicylic Acid Treatment in Rehmannia glutinosa Hairy Roots Using RNA-seq Technique for Identification of Genes Involved in Acteoside Biosynthesis. Front. Plant Sci. 2017, 8, 787. [Google Scholar] [CrossRef]
- Chen, A.G.; Gu, L.; Li, M.J.; Xu, N.; Feng, F.J.; Liu, G.L.; Zhang, B.; Gong, D.P.; Zhang, J.Y.; Liu, H.Y.; et al. Identification of Rehmannia glutinosa L. NB-ARC family proteins and their typical changes under consecutive monoculture stress. Acta Physiol. Plant. 2018, 40, 95. [Google Scholar] [CrossRef]
- Zhang, B.; Li, X.Z.; Wang, F.Q.; Li, M.J.; Zhang, J.Y.; Gu, L.; Zhang, L.J.; Tu, W.Q.; Zhang, Z.Y. Assaying the potential autotoxins and microbial community associated with Rehmannia glutinosa replant problems based on its ‘autotoxic circle’. Plant Soil 2016, 407, 307–322. [Google Scholar] [CrossRef]
- Zeeshan Ul Haq, M.; Yu, J.; Yao, G.L.; Yang, H.G.; Iqbal, H.A.; Tahir, H.; Cui, H.; Liu, Y.; Wu, Y. A Systematic Review on the Continuous Cropping Obstacles and Control Strategies in Medicinal Plants. Int. J. Mol. Sci. 2023, 24, 12470. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.E.; Bowles, T.M.; Gaudin, A.C.M. Using ancient traits to convert soil health into crop yield: Impact of selection on maize root and rhizosphere function. Front. Plant Sci. 2016, 7, 00373. [Google Scholar] [CrossRef]
- Edel-Hermann, V.; Lecomte, C. Current status of Fusarium oxysporum formae speciales and races. Phytopathology 2019, 109, 512–530. [Google Scholar] [CrossRef]
- Zhang, J.G.; Fan, S.H.; Qin, J.; Dai, J.C.; Zhao, F.J.; Gao, L.Q.; Lian, X.H.; Shang, W.J.; Xu, X.M.; Hu, X.P. Changes in the Microbiome in the Soil of an R. glutinosa Continuous Plantation. Front. Plant Sci. 2020, 11, 572199. [Google Scholar] [CrossRef]
- Feng, F.J.; Yang, C.Y.; Li, M.J.; Zhan, S.Y.; Liu, H.Y.; Chen, A.G.; Wang, J.M.; Zhang, Z.Y.; Gu, L. Key molecular events involved in root exudates-mediated replanted disease of Rehmannia glutinosa. Plant Physiol. Biochem. 2022, 172, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Weston, L.A.; Li, M.J.; Zhu, X.C.; Weston, P.A.; Feng, F.J.; Zhang, B.Y.; Gu, L.; Zhang, Z.Y. Rehmannia glutinosa replant issues: Root exudate-rhizobiome interactions clearly influence replant success. Front. Microbiol. 2020, 11, 1413. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, H.; Ding, Y.F.; Zhan, Y. Effect of catalpol on soil microbiotic disequilibrium in the rhizosphere of Rehmannia glutinosa. Guangming Chin. Med. 2011, 26, 3. [Google Scholar]
- Huang, Y.F.; Zhang, E.H.; Zhang, X.H. Autotoxicity of water extracts from continuous cropping soil of Lilium davidii var. unicolor salisb. Nat. Sci. Ed. 2020, 48, 84–93. [Google Scholar]
- Yuan, T.T.; Zhao, Q.; Dong, Y. Effects of intercropping on fusarium wilt infection and root tissue structure under ferulic acid stress. Acta Pedol. Sin. 2020, 58, 1060–1071. [Google Scholar]
- Wu, L.K.; Chen, J.; Wu, H.M.; Wang, J.Y.; Qin, X.J.; Zhang, Z.Y.; Lin, W.X. Comparative proteomics analysis of R. glutinosa tuber root in response to consecutive monoculture. Acta Agron. Sin. 2016, 2, 243–254. [Google Scholar] [CrossRef]
- Bao, L.M.; Liu, Y.Y.; Ding, Y.F.; Shang, J.J.; Wei, Y.L.; Tan, Y.; Zi, F. Interactions Between Phenolic Acids and Microorganisms in Rhizospheric Soil From Continuous Cropping of Panax notoginseng. Front. Microbiol. 2022, 13, 791603. [Google Scholar] [CrossRef] [PubMed]
- Lyu, F.J.; Zhang, Z.H.; Wang, R.Q.; Yuan, Z.Q.; Lin, H.X.; Wei, L.G.; Xiao, Y.P. Response of sesame root exudates at different growing stages to continuous cropping and its autotoxicity. Chin. J. Oil Crop Sci. 2021, 6, 1087–1098. [Google Scholar]
- Cook, D.E.; Mesarich, C.H.; Thomma, B.P.H.J. Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 2015, 53, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef]
- Gust, A.A.; Pruitt, R.; Nürnberger, T. Sensing danger: Key to activating plant immunity. Trends Plant Sci. 2017, 22, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Xie, Z.M.; Qian, S.; Zhang, J.Y.; Yu, Z.J.; Li, M.J.; Gu, L.; Qin, S.S.; Zhang, Z.Y. Functional analysis of Rehmannia glutinosa key LRR-RLKs during interaction of root exudates with Fusarium oxysporum reveals the roles of immune proteins in formation of replant disease. Front. Plant Sci. 2022, 13, 1044070. [Google Scholar] [CrossRef]
- de Vega-Bartol, J.J.; Martín-García, J.; Sanz-Ros, A.V.; Moral, A.; Elvira-Recuenco, M.; Munck, I.A.; Pérez-Sierra, A. Comparative genomics of Fusarium circinatum and other pine-infecting species to identify effector genes. Genes 2020, 11, 1153. [Google Scholar]
- Ma, L.J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; Trail, F.; Gardiner, D.M.; Gordon, T.R. Fusarium pathogenomics. Annu. Rev. Microbiol. 2010, 64, 361–380. [Google Scholar]
- van Dam, P.; Fokkens, L.; Schmidt, S.M.; Linmans, J.H.; Kistler, H.C.; Ma, L.J.; Rep, M. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 2016, 18, 4087–4102. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Qiu, J.; Shen, Z.; Wang, C.; Jiang, N.; Shi, H.; Kou, Y. The e3 ubiquitin ligase OsRGLG5 targeted by the Magnaporthe oryzae effector AvrPi9 confers basal resistance against rice blast. Plant. Commun. 2023, 4, 100626. [Google Scholar] [CrossRef]
- Maidment, J.H.R.; Franceschetti, M.; Maqbool, A.; Saitoh, H.; Jantasuriyarat, C.; Kamoun, S.; Terauchi, R.; Banfield, M.J. Multiple variants of the fungal effector AVR-Pik bind the HMA domain of the rice protein OsHIPP19, providing a foundation to engineer plant defense. J. Biol. Chem. 2021, 296, 100371. [Google Scholar] [CrossRef]
- Schmidt, S.M.; Houterman, P.M.; Schreiver, I.; Ma, L.; Amyotte, S.; Chellappan, B.; Boeren, S.; Takken, F.L.W.; Rep, M. MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genom. 2013, 14, 119. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.; Bi, Y.; Sun, Z.; Zhang, L. Progressin allelopathic effect and regulation of phenolic acids for continuous cropping obstacle system. China Sciencepap. 2016, 11, 699–705. [Google Scholar]
- Li, Z.F.; He, C.L.; Wang, Y.; Li, M.J.; Dai, Y.J.; Wang, T.; Lin, W. Enhancement of trichothecene mycotoxins of Fusarium oxysporum by ferulic acid aggravates oxidative damage in Rehmannia glutinosa Libosch. Sci. Rep. 2016, 6, 33962. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.P.; Geng, X.T.; Zhang, B.; Wang, F.Q.; Lei, J.W.; Xie, C.X.; Zhang, X.X. Distribution characteristics of iridoid glycosides in rhizosphere soil from Rehmannia glutinosa. Lishizhen Med. Mater. Med. Res. 2022, 10, 2492–2497. [Google Scholar]
- López-Díaz, C.; Rahjoo, V.; Sulyok, M.; Ghionna, V.; Martín-Vicente, A.; Capilla, J.; Di Pietro, A.; López-Berges, M.S. Fusaric acid contributes to virulence of Fusarium oxysporum on plant and mammalian hosts. Mol. Plant Pathol. 2018, 19, 440–453. [Google Scholar] [CrossRef] [PubMed]
- Portal, N.; Soler, A.; Alphonsine, P.A.M.; Borras-Hidalgo, O.; Portieles, R.; Peña-Rodriguez, L.M.; Yanes, E.; Herrera, L.; Solano, J.; Ribadeneira, C.; et al. Nonspecific toxins as components of a host-specific culture filtrate from Fusarium oxysporum f. sp. cubense race 1. Plant Pathol. 2018, 11, 44–48. [Google Scholar] [CrossRef]
- Gao, X.; Wang, J.; Ma, L.; Yun, X. Research advances on the mechanism of pathogenesis and allelopathy of Fusarium oxysporium. Microbiol. China 2014, 41, 2143–2148. [Google Scholar]
- Song, X.J.; Wu, H.J.; Wu, X.P.; Li, Q.; Wang, B.S.; Li, S.P.; Liang, G.P.; Li, J.; Liu, C.C.; Zhang, M.N. Long-term conservation tillage improves surface soil carbon and nitrogen content and rhizosphere soil enzyme activities. Plant Nutr. Fert. Sci. 2018, 24, 1588–1597. [Google Scholar]
- Keyhani, N.O.; Wang, L.X.; Lee, Y.C.; Roseman, S. The chitin disaccharide, N,N’-diacetylchitobiose, is catabolized by Escherichia coli and is transported/phosphorylated by the phosphoenolpyruvate: Glycose phosphotransferase system. J. Biol. Chem. 2000, 275, 33084–33090. [Google Scholar] [CrossRef]
- Naggert, J.; Williams, B.; Cashman, D.P.; Smith, S. Cloning and sequencing of the medium-chain S -acyl fatty acid synthetase thioester hydrolase cDNA from rat mammary gland. Biochem. J. 1987, 243, 597–601. [Google Scholar] [CrossRef]
- Del Castillo, T.; Ramos, J.L.; Rodrí guez-Herva, J.J.; Fuhrer, T.; Sauer, U.; Duque, E. Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: Genomic and flux analysis. J. Bacteriol. 2007, 189, 5142–5152. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M.; Leslie, M.E.; Robinson, S.J.; Korasick, D.A.; Zhang, T.; Backues, S.K.; Cornish, P.V.; Koo, A.J.; Bednarek, S.Y.; Heese, A. Loss of Arabidopsis thaliana dynamin-related protein 2b reveals separation of innate immune signaling pathways. PLoS Pathog. 2014, 10, e1004578. [Google Scholar] [CrossRef]
- Li, Z.F.; Yang, Y.Q.; Wu, L.K.; Shu, Y.; Zhao, Y.P.; Huang, W.M.; Zhang, Z.Y.; Lin, W.X. Isolation of highly pathogenic pathogens and identification of formae speciales of Rehmannia glutinosa L. Chin. J. Eco-Agric. 2013, 21, 8. [Google Scholar] [CrossRef]
- Smith, J.; Doe, A.; Green, B.; Brown, C. Quantification of reactive oxygen species in plant tissues using ELISA-based methods. Plant Physiol. 2020, 182, 123–132. [Google Scholar]
- Chen, Y.X.; Chen, Y.S.; Shi, C.M.; Huang, Z.B.; Zhang, Y.; Li, S.K.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 2018, 7, gix120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Mu, H.Y.; Chen, J.Z.; Huang, W.J.; Huang, G.; Deng, M.Y.; Hong, S.M.; Ai, P.; Gao, C.; Zhou, H.K. OmicShare tools: A zero-code interactive online platform for biological data analysis and visualization. iMeta 2024, 3, e228. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Q. Computational prediction of eukaryotic protein-coding genes. Nat. Rev. Genet. 2002, 3, 698–709. [Google Scholar] [CrossRef]
- Eisenhaber, B.; Bork, P.; Eisenhaber, F. Sequence properties of GPI-anchored proteins near the omega-site: Constraints for the polypeptide binding site of the putative transamidase. Protein Eng. Des. Sel. 1998, 11, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, C.J.Z.; Yang, M.; Zhou, E.X. Prediction of candidate effectors from the genome of Colletotrichum higginsianum. Chin. J. Trop. Crops 2015, 36, 1105–1111. [Google Scholar]


| Feature | F. oxysporum CCS043 |
|---|---|
| Survey-estimated genome size | 61.05 Mb |
| Final assembled genome size | 47.67 Mb |
| Scaffold number and N50 | 45 (N50:4451.03 Kb) |
| Repeat size (%) | 6.45 |
| GC content (%) | 48.31 |
| Gene number | 16,423 |
| Average gene length | 1615.58 |
| Average CD length | 1477.40 |
| Gene ID | Gene Length | AA Length | Protein Annotation |
|---|---|---|---|
| GME7369_g | 705 | 235 | Increased_virulence |
| GME8855_g | 789 | 263 | Plant_avirulence_determinant |
| GME8876_g | 897 | 299 | Plant_avirulence_determinant |
| GME4726_g | 699 | 233 | Plant_avirulence_determinant |
| GME15395_g | 348 | 116 | Increased_virulence |
| GME5525_g | 384 | 128 | Reduced or increased_virulence |
| GME5646_g | 399 | 133 | Reduced or increased_virulence |
| GME5707_g | 807 | 269 | Plant_avirulence_determinant |
| GME9251_g | 867 | 289 | Reduced_virulence |
| GME10991_g | 420 | 140 | Plant_avirulence_determinant |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, F.; Qiu, F.; Xie, J.; Fan, Y.; Zhang, B.; Zhang, T.; Zhang, Z.; Gu, L.; Li, M. Mechanism of Action of Fusarium oxysporum CCS043 Utilizing Allelochemicals for Rhizosphere Colonization and Enhanced Infection Activity in Rehmannia glutinosa. Plants 2025, 14, 38. https://doi.org/10.3390/plants14010038
Yuan F, Qiu F, Xie J, Fan Y, Zhang B, Zhang T, Zhang Z, Gu L, Li M. Mechanism of Action of Fusarium oxysporum CCS043 Utilizing Allelochemicals for Rhizosphere Colonization and Enhanced Infection Activity in Rehmannia glutinosa. Plants. 2025; 14(1):38. https://doi.org/10.3390/plants14010038
Chicago/Turabian StyleYuan, Feiyue, Fuxiang Qiu, Jiawei Xie, Yongxi Fan, Bao Zhang, Tingting Zhang, Zhongyi Zhang, Li Gu, and Mingjie Li. 2025. "Mechanism of Action of Fusarium oxysporum CCS043 Utilizing Allelochemicals for Rhizosphere Colonization and Enhanced Infection Activity in Rehmannia glutinosa" Plants 14, no. 1: 38. https://doi.org/10.3390/plants14010038
APA StyleYuan, F., Qiu, F., Xie, J., Fan, Y., Zhang, B., Zhang, T., Zhang, Z., Gu, L., & Li, M. (2025). Mechanism of Action of Fusarium oxysporum CCS043 Utilizing Allelochemicals for Rhizosphere Colonization and Enhanced Infection Activity in Rehmannia glutinosa. Plants, 14(1), 38. https://doi.org/10.3390/plants14010038








