Characterization of MADS-Box Gene Family in Isatis indigotica and Functional Study of IiAP1 in Regulating Floral Transition and Formation
Abstract
1. Introduction
2. Results
2.1. Identification of the IiMADS Genes in I. indigotica
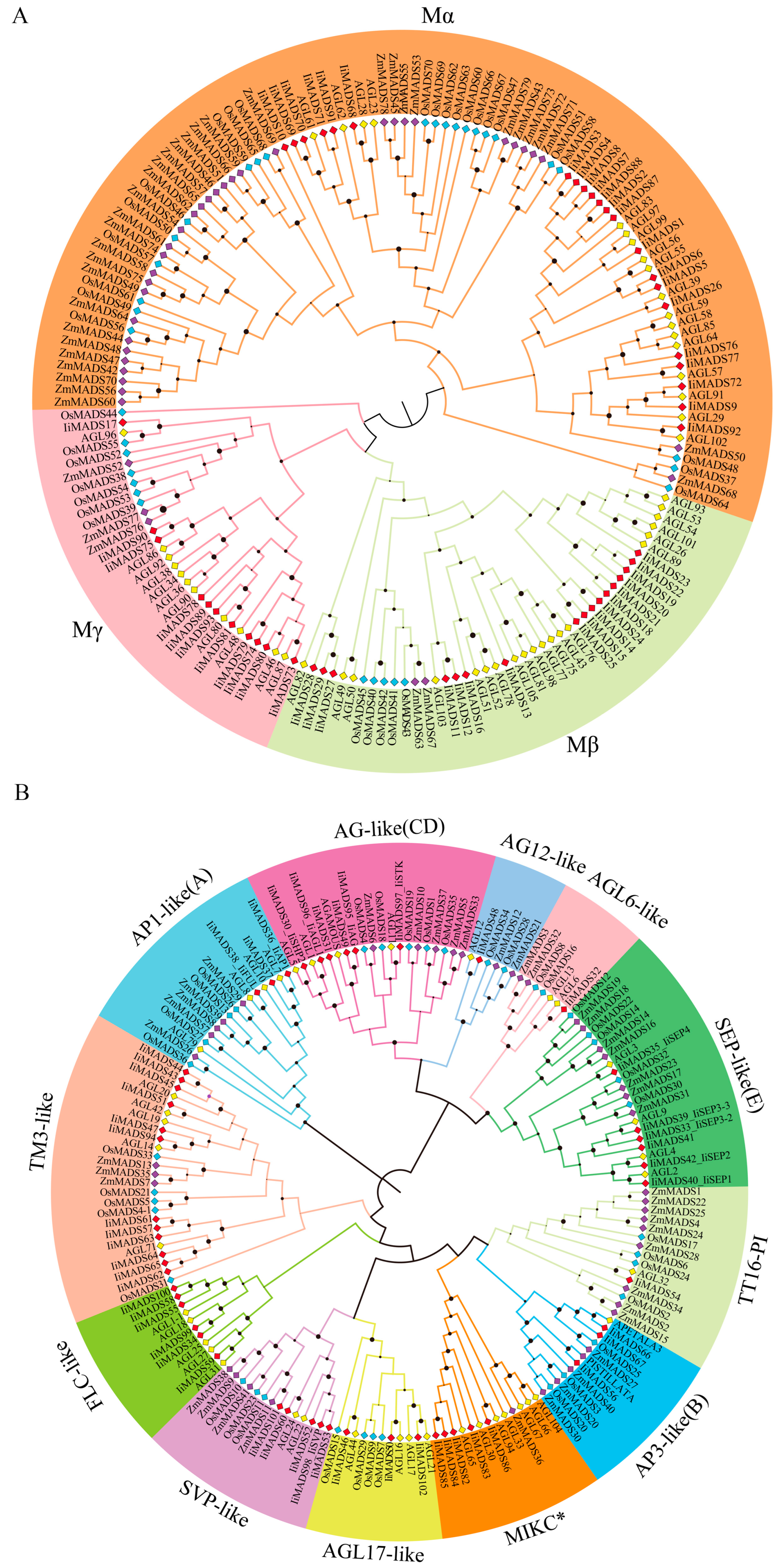
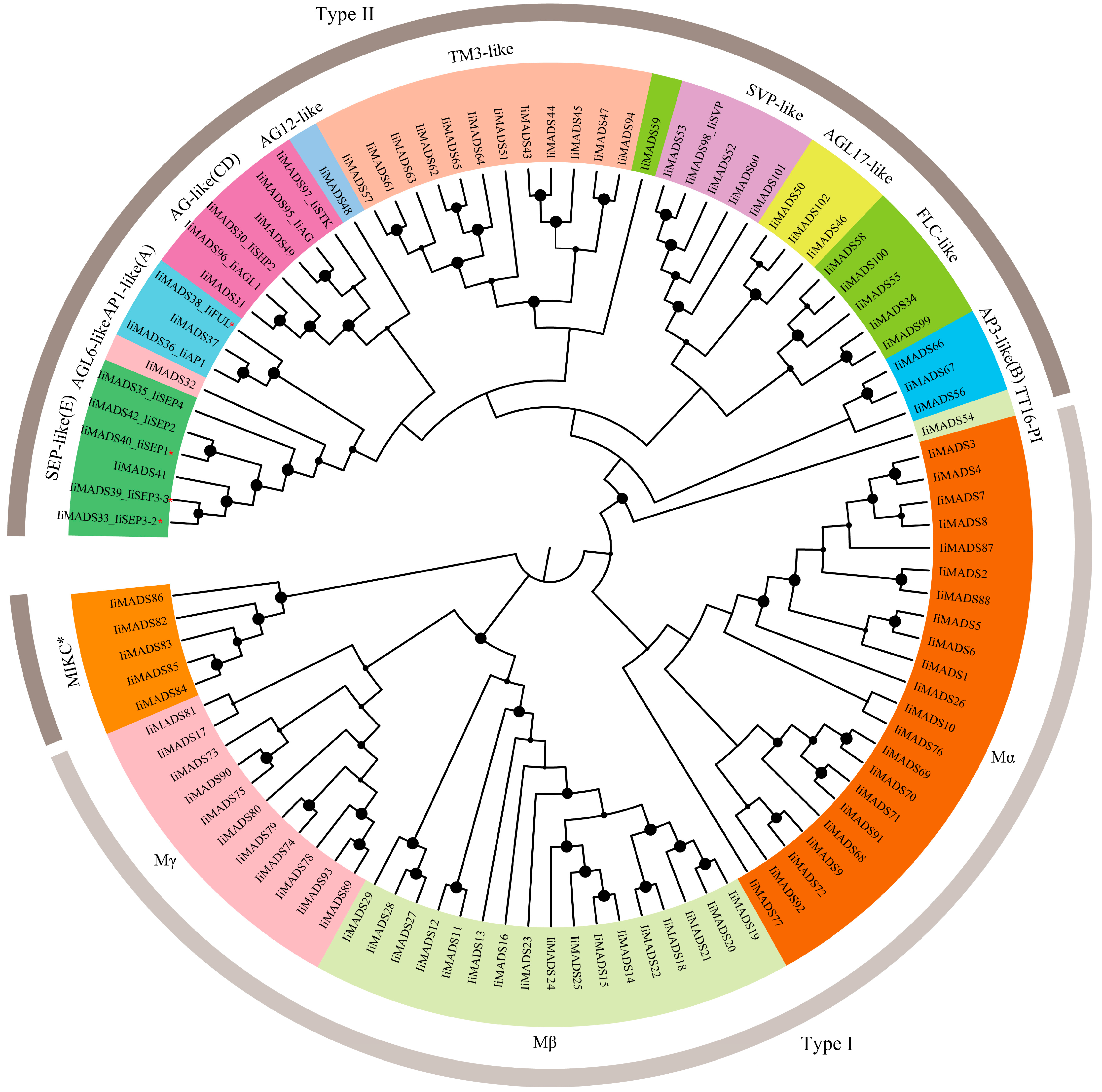
2.2. Phylogenetic Investigation of IiMADS Genes
2.3. Analysis of Gene Structure, Conserved Motifs, and Cis-Acting Elements in the IiMADS Genes
2.4. Chromosomal Distribution and Collinearity Analysis of IiMADS Genes
2.5. Protein Network Interaction of IiMADS Genes
2.6. Isolation of IiAP1 from I. indigotica
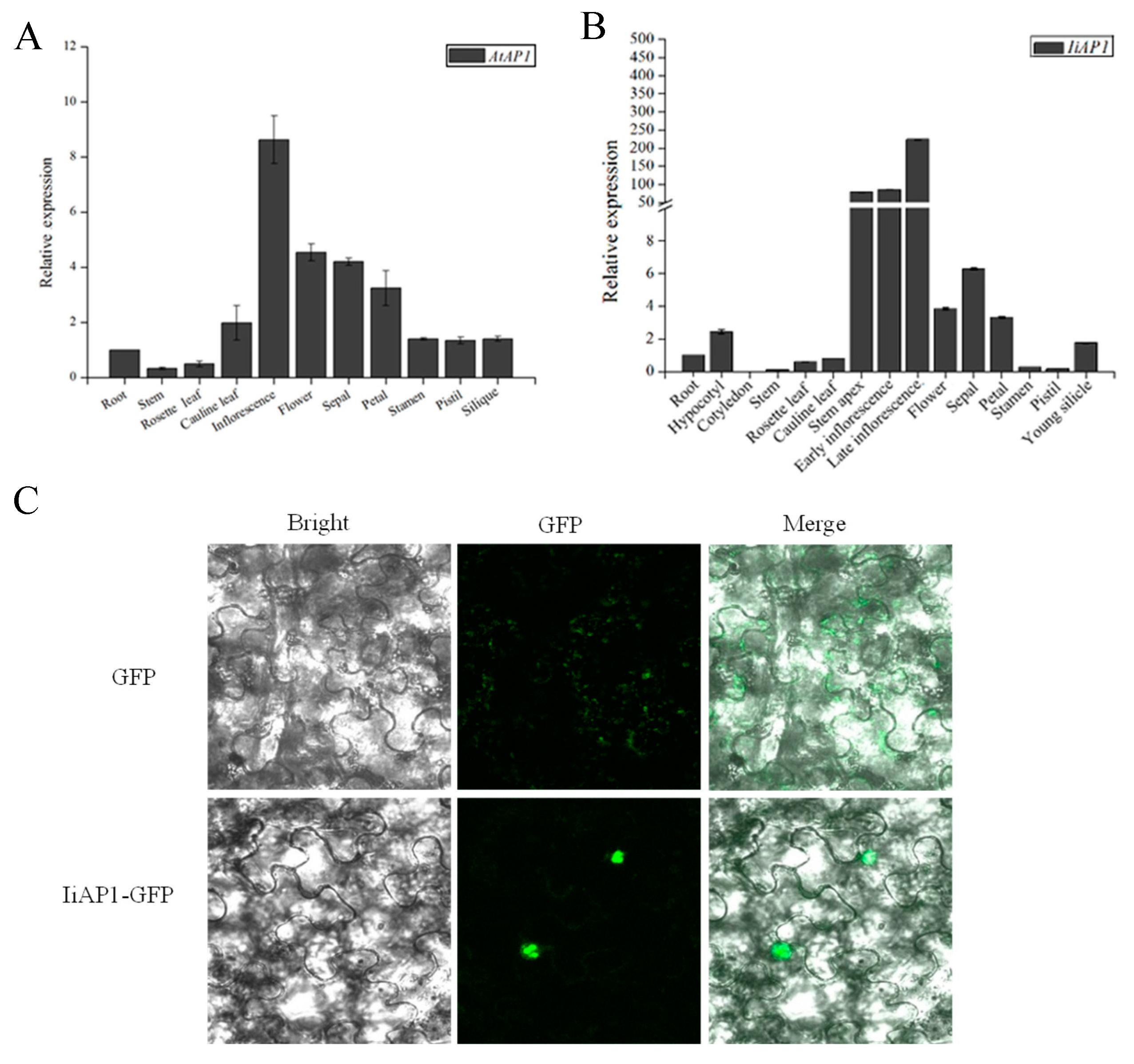
2.7. Expression Analysis of IiAP1 and Nuclear Localization of the Encoding Product
2.8. Ectopic Expression of IiAP1 in Arabidopsis
3. Discussion
3.1. Identified 102 MADS-Box Genes from the Transcriptome of I. indigotica
3.2. Type II MADS-Box Genes Belong to Well-Defined Subfamilies
3.3. Ectopic Expression of IiAP1 in Arabidopsis Promotes Early Flowering
3.4. Ectopic Expression of IiAP1 in Arabidopsis Altered Flower Morphology
4. Materials and Methods
4.1. Plant Materials, RNA Extraction, and cDNA Synthesis
4.2. Identification and Sequence Analysis of MADS Genes in I. indigotica
4.3. Phylogenetic Investigation and Categorization of the IiMADS Genes
4.4. Exploration of Gene Structure, Conserved Motifs, and Cis-Acting Elements in the IiMADS Genes
4.5. Chromosomal Location and Collinearity Analysis of the IiMADS Genes
4.6. Protein–protein Interaction Network of IiMADS Genes
4.7. Analyses of the IiAP1 Expression Patterns Using qRT-PCR
4.8. Subcellular Localization of IiAP1 Protein
4.9. Obtaining Transgenic Arabidopsis Plants Ectopic Expression of IiAP1
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Coen:, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, L.; Thieffry, D.; Alvarez-Buylla, E.R. Genetic control of flower morphogenesis in Arabidopsis thaliana: A logical analysis. Bioinformatics 1999, 15, 593–606. [Google Scholar] [CrossRef]
- Theißen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Favaro, R.; Pinyopich, A.; Battaglia, R.; Kooiker, M.; Borghi, L.; Ditta, G.; Yanofsky, M.F.; Kater, M.M.; Colombo, L. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 2003, 15, 2603–2611. [Google Scholar] [CrossRef]
- Su, C.L.; Chen, W.C.; Lee, A.Y.; Chen, C.Y.; Chang, Y.C.; Chao, Y.; Shih, M.C. A modified ABCDE model of flowering in orchids based on gene expression profiling studies of the moth orchid Phalaenopsis Aphrodite. PLoS ONE 2013, 8, e80462. [Google Scholar] [CrossRef]
- Theissen, G.; Saedler, H. Plant biology: Floral quartets. Nature 2001, 409, 469–471. [Google Scholar] [CrossRef]
- Schilling, S.; Pan, S.; Kennedy, A.; Melzer, R. MADS-box genes and crop domestication: The jack of all traits. J. Exp. Bot. 2018, 69, 1447–1469. [Google Scholar] [CrossRef]
- Yan, W.; Chen, D.; Kaufmann, K. Molecular mechanisms of floral organ specification by MADS domain proteins. Curr. Opin. Plant Biol. 2016, 29, 154–162. [Google Scholar] [CrossRef]
- Gramzow, L.; Ritz, M.S.; Theißen, G. On the origin of MADS-domain transcription factors. Trends Genet. 2010, 26, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; Ribas de Pouplana, L.; Martínez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef]
- Henschel, K.; Kofuji, R.; Hasebe, M.; Saedler, H.; Münster, T.; Theißen, G. Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 2002, 19, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Parenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Shore, P.; Sharrocks, A.D. The MADS-box family of transcription factors. Eur. J. Biochem. 1995, 229, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Meyerowitz, E.M. MADS domain proteins in plant development. Biol. Chem. 1997, 378, 1079–1102. [Google Scholar] [PubMed]
- Yang, Y.Z.; Fanning, L.; Jack, T. The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 2003, 33, 47–59. [Google Scholar] [CrossRef]
- Yang, Y.Z.; Jack, T. Defining subdomains of the K domain important for protein–protein interactions of plant MADS proteins. Plant Mol. Biol. 2004, 55, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Melzer, R.; Theißen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K. MADS-box gene family in rice: Genome-wide identiffcation, organization and expression proffling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef]
- Zhao, D.; Chen, Z.; Xu, L.; Zhang, L.J.; Zou, Q. Genome-Wide analysis of the MADS-Box gene family in maize: Gene structure, evolution, and relationships. Genes 2021, 12, 1956. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.Y.; Cao, D.D.; Damaris, R.N.; Yang, P.F. Genome-wide identification of MADS-box gene family in sacred lotus (Nelumbo nucifera) identifies a SEPALLATA homolog gene involved in floral development. BMC Plant Biol. 2020, 20, 497. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020, 225, 511–529. [Google Scholar] [CrossRef] [PubMed]
- Raza, Q.; Riaz, A.; Atif, R.M.; Hussain, B.; Rana, I.A.; Ali, Z.; Budak, H.; Alaraidh, I.A. Genome-Wide Diversity of MADS-Box Genes in Bread Wheat is Associated with its Rapid Global Adaptability. Front. Genet. 2022, 12, 818880. [Google Scholar] [CrossRef] [PubMed]
- Won, S.Y.; Jung, J.A.; Kim, J.S. Genome-wide analysis of the MADS-Box gene family in Chrysanthemum. Comput. Biol. Chem. 2021, 90, 107424. [Google Scholar] [CrossRef]
- Zhou, P.Y.; Qu, Y.S.; Wang, Z.W.; Huang, B.; Wen, Q.; Xin, Y.; Ni, Z.X.; Xu, L.A. Gene Structural Specificity and Expression of MADS-Box Gene Family in Camellia chekiangoleosa. Int. J. Mol. Sci. 2023, 24, 3434. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Qi, X.W.; Wan, Y.; Chen, Z.Q.; Fang, H.L.; Liang, C.Y. Genome-wide analysis of the MADS-box gene family in Lonicera japonica and a proposed floral organ identity model. BMC Genom. 2023, 24, 447. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.F.; Zhou, Z.X.; Zhu, L.; Gu, Y.Y.; Guo, B.J.; Lv, C.; Zhu, J.; Xu, R.G. Genome-wide analysis of the MADS-box gene family involved in salt and waterlogging tolerance in barley (Hordeum vulgare L.). Front. Plant Sci. 2023, 14, 1178065. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.X.; Huang, Q.Y.; Shen, Z.L.; Baron, G.C.; Li, X.Y.; Lu, X.Y.; Li, Y.Q.; Chen, W.R.; Xu, L.S.; Lv, J.C.; et al. Genome-Wide Identification and Analysis of the MADS-Box Transcription Factor Genes in Blueberry (Vaccinium spp.) and Their Expression Pattern during Fruit Ripening. Plants 2023, 12, 1424. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.L.; Chang, Y.S. Studies on the antinociceptive, anti-inflammatory and antipyretic effects of Isatis indigotica root. Phytomedicine 2002, 9, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.Y.; Meng, Q.; Tan, X.M.; Yang, L.; Zhang, K.L.; Xu, Z.Q. Functional identification of the different regions in B-class floral homeotic MADS-box proteins IiAP3 and IiPI from Isatis indigotica. Physiol. Plant 2022, 174, e13713. [Google Scholar] [CrossRef]
- Yang, L.; Qiao, X.; He, H.E.; Yi, W.W.; Gao, Y.N.; Tan, X.M.; Cheng, H.; Hou, X.F.; Ma, Y.Y.; Wang, H.L.; et al. IiAGL6 participates in the regulation of stamen development and pollen formation in Isatis indigotica. Plant Sci. 2024, 340, 111974. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.Q.; Li, D.Z.; Zhang, L.; Li, Q.; Yao, J.W.; Ma, Z.; Huang, X.; Xu, Z.Q. Ectopic expression of IiFUL isolated from Isatis indigotica could change the reproductive growth of Arabidopsis thaliana. Plant Physiol. Biochem. 2017, 121, 140–152. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.Q.; Pu, Z.Q.; Zhang, L.; Lu, M.X.; Zhu, Y.; Hao, C.Y.; Xu, Z.Q. A SEPALLATA1-like gene of Isatis indigotica Fort. regulates flowering time and specifies floral organs. Gene 2019, 713, 143974. [Google Scholar] [CrossRef]
- Ma, Y.Q.; Pu, Z.Q.; Tan, X.M.; Meng, Q.; Zhang, K.L.; Yang, L.; Ma, Y.Y.; Huang, X.; Xu, Z.Q. SEPALLATA--like genes of Isatis indigotica can affect the architecture of the inflorescences and the development of the floral organs. PeerJ 2022, 10, e13034. [Google Scholar] [CrossRef]
- Ma, Y.Q.; Li, Q.; Cheng, H.; Hou, X.F.; Tan, X.M.; Meng, Q.; Huang, X.; Chang, W.; Yang, L.; Xu, Z.Q. Alternative splicing variants of IiSEP3 in Isatis indigotica are involved in floral transition and flower development. Plant Physiol Biochem. 2024, 216, 109153. [Google Scholar] [CrossRef]
- Pu, Z.Q.; Ma, Y.Y.; Lu, M.X.; Ma, Y.Q.; Xu, Z.Q. Cloning of a SEPALLATA4-like gene (IiSEP4) in Isatis indigotica Fortune and characterization of its function in Arabidopsis thaliana. Plant Physiol. Biochem. 2020, 154, 229–237. [Google Scholar] [CrossRef]
- Lu, M.X.; Li, D.Z.; Pu, Z.Q.; Ma, Y.Q.; Huang, X.; Xu, Z.Q. Ectopic expression of IiSHP2 from Isatis indigotica Fortune, a PLE-lineage MADS-box gene, influences leaf, floral organ and silique morphology in Arabidopsis thaliana. Physiol. Mol. Biol. Plants 2020, 26, 379–389. [Google Scholar] [CrossRef]
- Meng, Q.; Hou, X.F.; Cheng, H.; Tan, X.M.; Pu, Z.Q.; Xu, Z.Q. IiSVP of Isatis indigotica can reduce the size and repress the development of floral organs. Plant Cell Rep. 2023, 42, 561–574. [Google Scholar] [CrossRef]
- Wang, P.; Wang, S.B.; Chen, Y.; Xu, X.M.; Guang, X.M.; Zhang, Y.H. Genome-wide Analysis of the MADS-Box Gene Family in Watermelon. Comput. Biol. Chem. 2019, 80, 341–350. [Google Scholar] [CrossRef]
- Lakhwani, D.; Vikarm, D.Y.; Singh, S.; Pandey, A.; Kumar, T.P.; Hasan, A.M. Genome wide identification of MADS box gene family in Musa balbisiana and their divergence during evolution. Gene 2022, 836, 146666. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Wu, H.; Yang, Q.; Huang, L.; Hu, Q.; Ma, T.; Li, Z.; Liu, J. A chromosome-scale genome assembly of Isatis indigotica, an important medicinal plant used in traditional Chinese medicine: An Isatis genome. Hortic. Res. 2020, 7, 18. [Google Scholar] [CrossRef]
- Daminato, M.; Masiero, S.; Resentini, F.; Lovisetto, A.; Casadoro, G. Characterization of TM8, a MADS-box gene expressed in tomato flowers. BMC Plant Biol. 2014, 14, 319. [Google Scholar] [CrossRef]
- Wei, L.; Sun, L.Q.; Zhang, C.Y.; Tang, X.Q.; Wang, F.Q.; Wang, K.C.; Wang, J. Genome-Wide Characterization of the Isatis indigotica MADS-box Family and Role of IiSVP in Flowering. Russ. J. Plant Physiol. 2023, 70, 69. [Google Scholar] [CrossRef]
- Castro, J.C.; Castro, C.G.; Cobos, M. Genetic and biochemical strategies for regulation of L-ascorbic acid biosynthesis in plants through the L-galactose pathway. Front. Plant Sci. 2023, 14, 1099829. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Cudjoe, D.K.; Kamruzzaman, M.; Siddique, S.; Fiorani, F.; Léon, J.; Naz, A.A. Abscisic acid-responsive element binding transcription factors contribute to proline synthesis and stress adaptation in Arabidopsis. J. Plant Physiol. 2021, 261, 153414. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Blazquez, M.A.; Weigel, D. Integration of floral inductive signals in Arabidopsis. Nature 2000, 404, 889–892. [Google Scholar] [CrossRef]
- Lin, M.; Yan, J.W.; Ali, M.M.; Wang, S.J.; Tian, S.N.; Chen, F.X.; Lin, Z.M. Isolation and Functional Characterization of a Green-Tissue Promoter in Japonica Rice (Oryza sativa subsp. Japonica). Biology 2022, 11, 1092. [Google Scholar] [CrossRef] [PubMed]
- Ohta, T. Gene conversion and evolution of gene families: An overview. Genes 2010, 1, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef] [PubMed]
- Michaels, S.D.; Amasino, R.M. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 2001, 13, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Nardeli, S.M.; Artico, S.; Aoyagi, G.M.; de Moura, S.M.; da Franca, S.T.; Grossi-de-Sa, M.F.; Romanel, E.; Alves-Ferreira, M. Genome-wide analysis of the MADS-box gene family in polyploid cotton (Gossypium hirsutum) and in its diploid parental species (Gossypium arboreum and Gossypium raimondii). Plant Physiol. Biochem. 2018, 127, 169–184. [Google Scholar] [CrossRef]
- Lin, C.S.; Hsu, C.T.; Liao, D.C.; Chang, W.J.; Chou, M.L.; Huang, Y.T.; Chen, J.J.; Ko, S.S.; Chan, M.T.; Shih, M.C. Transcriptome-wide analysis of the MADS-box gene family in the orchid Erycina pusilla. Plant Biotechnol. J. 2016, 14, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Ferrándiz, C.; Gu, Q.; Martienssen, R.; Yanofsky, M.F. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 2000, 127, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Wigge, P.A.; Kim, M.C.; Jaeger, K.E.; Busch, W.; Schmid, M.; Lohmann, J.U.; Weigel, D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 2005, 309, 1056–1059. [Google Scholar] [CrossRef]
- Mandel, M.A.; Yanofsky, M.F. A gene triggering flower formation in Arabidopsis. Nature 1995, 377, 522–524. [Google Scholar] [CrossRef]
- Chen, M.K.; Lin, I.C.; Yang, C.H. Functional analysis of three lily (Lilium longiflorum) APETALA1-like MADS box genes in regulating floral transition and formation. Plant Cell Physiol. 2008, 49, 704–717. [Google Scholar] [CrossRef] [PubMed]
- Ning, K.; Han, Y.Y.; Chen, Z.J.; Luo, C.; Wang, S.L.; Zhang, W.J.; Li, L.; Zhang, X.L.; Fan, S.X.; Wang, Q. Genome-wide analysis of MADS-box family genes during flower development in lettuce. Plant Cell Environ. 2019, 42, 1868–1881. [Google Scholar] [CrossRef]
- Liu, Z.X.; Fei, Y.; Zhang, K.B.; Fang, Z.W. Ectopic expression of a Fagopyrum esculentum APETALA1 ortholog only rescues sepal development in Arabidopsis ap1 mutant. Int. J. Mol. Sci. 2019, 20, 2021. [Google Scholar] [CrossRef]
- Han, Y.; Tang, A.Y.; Yu, J.Y.; Cheng, T.R.; Wang, J.; Yang, W.R.; Pan, H.T.; Zhang, Q.X. RcAP1, a Homolog of APETALA1, is Associated with Flower Bud Differentiation and Floral Organ Morphogenesis in Rosa chinensis. Int. J. Mol. Sci. 2019, 20, 3557. [Google Scholar] [CrossRef]
- Sawettalake, N.; Bunnag, S.; Wang, Y.; Shen, L.; Yu, H. DOAP1 Promotes Flowering in the Orchid Dendrobium Chao Praya Smile. Front. Plant Sci. 2017, 8, 400. [Google Scholar] [CrossRef]
- Hu, T.Q.; Li, X.H.; Du, L.R.; Manuela, D.; Xu, M.L. LEAFY and APETALA1 down-regulate ZINC FINGER PROTEIN 1 and 8 to release their repression on class B and C floral homeotic genes. Proc. Natl. Acad. Sci. USA 2023, 120, e2221181120. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Ito, T.; Wellmer, F.; Meyerowitz, E.M. Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat Genet. 2004, 36, 157–161. [Google Scholar] [CrossRef]
- Kotoda, N.; Wada, M.; Kusaba, S.; Kano-Murakami, Y.; Masuda, T.; Soejima, J. Overexpression of MdMADS5, an APETALA1-like gene of apple, causes early flowering in transgenic Arabidopsis. Plant Sci. 2002, 162, 679–687. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.Q.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic. Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Lu, Y.C. BLASTP-ACC: Parallel Architecture and Hardware Accelerator Design for BLAST-Based Protein Sequence Alignment. IEEE Trans. Biomed. Circuits. Syst. 2019, 13, 1771–1782. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic. Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Abdullah-Zawawi, M.R.; Ahmad-Nizammuddin, N.F.; Govender, N.; Harun, S.; Mohd-Assaad, N.; Mohamed-Hussein, Z.A. Comparative genome-wide analysis of WRKY, MADS-box and MYB transcription factor families in Arabidopsis and rice. Sci. Rep. 2021, 11, 19678. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput. Biol. 2008, 4, e1000069. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.N.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic. Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.H.; Lercher, M.J.; Hu, S.N.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic. Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Doncheva, N.T.; Morris, J.H.; Holze, H.; Kirsch, R.; Nastou, K.C.; Cuesta-Astroz, Y.; Rattei, T.; Szklarczyk, D.; von Mering, C.; Jensen, L.J. Cytoscape stringApp 2.0: Analysis and visualization of heterogeneous biological networks. J. Proteome Res. 2023, 22, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]






| Multiple-Copy Gene Pairs | Synonymous Substitution Rate (Ka) | Nonsynonymous Substitution Rate (Ks) | Ka/Ks |
|---|---|---|---|
| IiMADS91/IiMADS69 | 0.0921623 | 0.502525 | 0.183398 |
| IiMADS36/IiMADS38 | 0.0265616 | 0.401908 | 0.0660888 |
| IiMADS90/ IiMADS42 | 0.0194865 | 0.387274 | 0.050317 |
| IiMADS40/ IiMADS89 | 0.0672956 | 0.511771 | 0.131496 |
| IiMADS64/ IiMADS63 | 0.037241 | 0.266845 | 0.139561 |
| IiMADS20/ IiMADS21 | 0.0850036 | 0.452594 | 0.187814 |
| IiMADS45/ IiMADS47 | 0.0603465 | 0.288191 | 0.209397 |
| IiMADS30/ IiMADS96 | 0.0776146 | 0.39195 | 0.198022 |
| IiMADS5/ IiMADS6 | 0.0439878 | 0.195654 | 0.224824 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Lan, Y.; Li, J.; Long, H.; Zhou, Y.; Li, Z.; Miao, M.; Zhong, J.; Wang, H.; Chang, W.; et al. Characterization of MADS-Box Gene Family in Isatis indigotica and Functional Study of IiAP1 in Regulating Floral Transition and Formation. Plants 2025, 14, 129. https://doi.org/10.3390/plants14010129
Ma Y, Lan Y, Li J, Long H, Zhou Y, Li Z, Miao M, Zhong J, Wang H, Chang W, et al. Characterization of MADS-Box Gene Family in Isatis indigotica and Functional Study of IiAP1 in Regulating Floral Transition and Formation. Plants. 2025; 14(1):129. https://doi.org/10.3390/plants14010129
Chicago/Turabian StyleMa, Yanqin, Yanhong Lan, Ju Li, Haicheng Long, Yujie Zhou, Zhi Li, Mingjun Miao, Jian Zhong, Haie Wang, Wei Chang, and et al. 2025. "Characterization of MADS-Box Gene Family in Isatis indigotica and Functional Study of IiAP1 in Regulating Floral Transition and Formation" Plants 14, no. 1: 129. https://doi.org/10.3390/plants14010129
APA StyleMa, Y., Lan, Y., Li, J., Long, H., Zhou, Y., Li, Z., Miao, M., Zhong, J., Wang, H., Chang, W., Xu, Z., & Yang, L. (2025). Characterization of MADS-Box Gene Family in Isatis indigotica and Functional Study of IiAP1 in Regulating Floral Transition and Formation. Plants, 14(1), 129. https://doi.org/10.3390/plants14010129







