Mapping and Omics Integration: Towards Precise Rice Disease Resistance Breeding
Abstract
1. Introduction
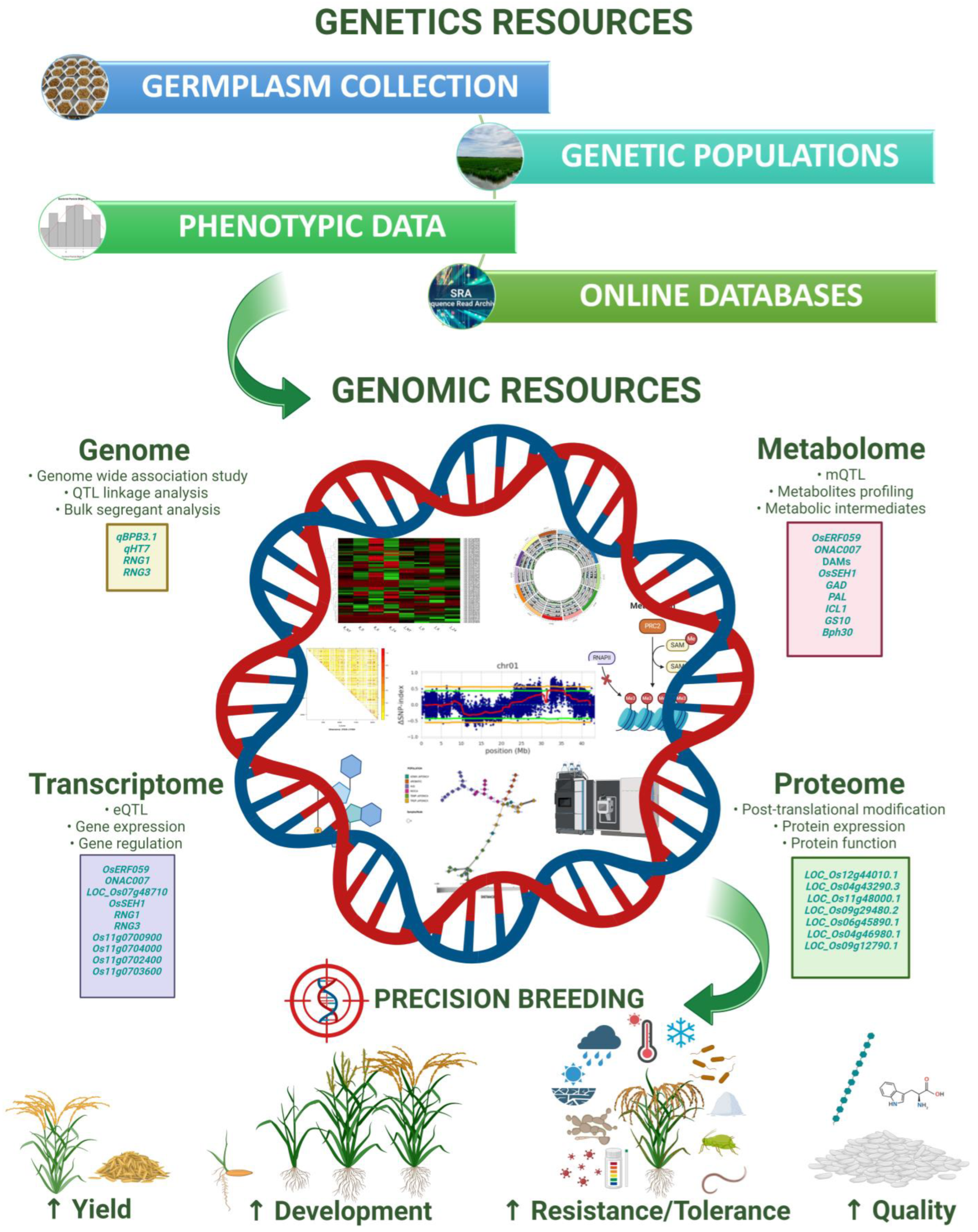
2. Omics for Decoding Resistance Mechanisms
2.1. Harnessing Genomics for Enhancing Rice Disease Resistance
2.2. Harnessing Transcriptomics to Safeguard Rice against Disease
2.3. Harnessing Proteomics for Fortifying Rice against Disease
2.4. Harnessing Metabolomics for Bolstering Rice Disease Resistance
3. Integrative Omics: Bridging the Layers
3.1. Integrative Studies in Rice for Disease Resistance
3.2. Integrative Studies in Rice for Tolerance to Abiotic Stresses
3.3. Navigating the Path to Precision by Combinining QTL Mapping and Omics
| Abiotic/Biotic Factors | QTL/Loci/Genes (Gene Products) | Methods | Critical Information | Reference |
|---|---|---|---|---|
| Drought tolerance | OsERF059 (ethylene response factor 59) and ONAC007 (NAC domain-containing protein 7) | Transcriptomics and metabolomics |
| Lu et al. (2022) [55] |
| Heat tolerance | qHT7/LOC_Os07g48710 (VQ motif-containing protein 30) | GWAS and transcriptomics |
| Li et al. (2023) [56] |
| Arsenic toxicity | DEGs and DAMs ion transporters, ROS, etc. | Transcriptomics and metabolomics |
| Ma et al. (2023) [57] |
| Cold tolerance | OsSEH1 (nucleoporin SEH1) | Transcriptomics and metabolomics |
| Gu et al. (2023) [58] |
| Blast resistance (Magnaporthe grisea) | RNG1 (Zinc finger protein with B-box-domain) and RNG3 (Dehydrogenase) | GWAS and transcriptomics |
| Xu et al. (2023) [47] |
| Blast resistance (Magnaporthe grisea) | Os11g0700900 (glycoside hydrolase), Os11g0704000 (SelT selenoprotein family), Os11g0702400 (zinc finger, C2H2-type domain containing protein), and Os11g0703600 (hypothetical protein) | BSA, QTL-mapping, and transcriptomics |
| Tan et al. (2022) [48] |
| Sheath blight resistance (Rhizoctonia solani) | LOC_Os12g44010.1 (purple acid phosphatase 10b), LOC_Os04g43290.3 (actin-related protein (ARP) C2 subunit), LOC_Os11g48000.1 (EPF zinc-finger), LOC_Os09g29480.2 (2-aminoethanethiol dioxygenase), LOC_Os06g45890.1 (MYB-like transcription factor), LOC_Os04g46980.1 (cis-zeatin-O-glucosyltransferase), and LOC_Os09g12790.1 (potassium channel protein) | Proteomics and transcriptomics |
| Prathi et al. (2018) [49] |
| Bacterial blight resistance (Xanthomonas oryzae pv oryzae) | GAD (Glutamate decarboxylase), PAL (Phenylalanine ammonia-lyase), ICL1 (Isocitrate lyase), and GS10 (Glutathione-S-transferase) | Transcriptomics and metabolomics |
| Sana et al. (2010) [41] |
| Brown plant hopper resistance (Tungro virus) | Bph30 (Leucine rich repeat (LRR) family protein) | Transcriptomics and metabolomics |
| Shi et al. (2023) [50] |
| Bacterial panicle blight resistance (Burkholderia glumae) | qBPB3.1 | QTL-mapping and QTL-seq |
| Ontoy et al. (2023) [20] |
4. Challenges and Gaps: Exploring the Intersections
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Singh, P.K.; Nag, A.; Arya, P.; Kapoor, R.; Singh, A.; Jaswal, R.; Sharma, T.R. Prospects of Understanding the Molecular Biology of Disease Resistance in Rice. Int. J. Mol. Sci. 2018, 19, 1141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, C.; Si, J.; Han, Z.; Chen, D. Action Mechanisms of Effectors in Plant-Pathogen Interaction. Int. J. Mol. Sci. 2022, 23, 6758. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Yang, L. Pathogen-triggered changes in plant development: Virulence strategies or host defense mechanism? Front. Microbiol. 2023, 14, 1122947. [Google Scholar] [CrossRef] [PubMed]
- Mapuranga, J.; Zhang, N.; Zhang, L.; Chang, J.; Yang, W. Infection Strategies and Pathogenicity of Biotrophic Plant Fungal Pathogens. Front. Microbiol. 2022, 13, 799396. [Google Scholar] [CrossRef]
- Pilet-Nayel, M.-L.; Moury, B.; Caffier, V.; Montarry, J.; Kerlan, M.-C.; Fournet, S.; Durel, C.-E.; Delourme, R. Quantitative Resistance to Plant Pathogens in Pyramiding Strategies for Durable Crop Protection. Front. Plant Sci. 2017, 8, 1838. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhu, Y.; Shi, H.; Qiu, J.; Ding, X.; Kou, Y. Recent Progress in Rice Broad-Spectrum Disease Resistance. Int. J. Mol. Sci. 2021, 22, 11658. [Google Scholar] [CrossRef] [PubMed]
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Powder, K.E. Quantitative Trait Loci (QTL) Mapping. In Methods in Molecular Biology; Humana: New York, NY, USA, 2020; pp. 211–229. [Google Scholar] [CrossRef]
- Zeng, Z.B. QTL Mapping. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: San Diego, MA, USA, 2001; pp. 8–12. [Google Scholar]
- Mohan, M.; Nair, S.; Bhagwat, A.; Krishna, T.G.; Yano, M.; Bhatia, C.R.; Sasaki, T. Genome mapping, molecular markers and marker-assisted selection in crop plants. Mol. Breed. 1997, 3, 87–103. [Google Scholar] [CrossRef]
- Mulualem, T.; Bekeko, Z. Advances in Quantitative Trait Loci, Mapping and Importance of Markers Assisted Selection in Plant Breeding Research. Int. J. Plant Breed. Genet. 2016, 10, 58–68. [Google Scholar] [CrossRef][Green Version]
- Miles, C.; Wayne, M. Quantitative trait locus (QTL) analysis. Nat. Educ. 2008, 1, 208. [Google Scholar]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Prim. 2021, 1, 59. [Google Scholar] [CrossRef]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef]
- Sattayachiti, W.; Wanchana, S.; Arikit, S.; Nubankoh, P.; Patarapuwadol, S.; Vanavichit, A.; Darwell, C.T.; Toojinda, T. Genome-Wide Association Analysis Identifies Resistance Loci for Bacterial Leaf Streak Resistance in Rice (Oryza sativa L.). Plants 2020, 9, 1673. [Google Scholar] [CrossRef]
- Liu, Q.; Lan, G.; Zhu, Y.; Chen, K.; Shen, C.; Zhao, X.; Zhang, F.; Xu, J.; Li, Z. Genome-Wide Association Study on Resistance to Rice Black-Streaked Dwarf Disease Caused by Rice black-streaked dwarf virus. Plant Dis. 2021, 105, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Majeed, A.; Johar, P.; Raina, A.; Salgotra, R.K.; Feng, X.; Bhat, J.A. Harnessing the potential of bulk segregant analysis sequencing and its related approaches in crop breeding. Front. Genet. 2022, 13, 944501. [Google Scholar] [CrossRef]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Uemura, A.; Utsushi, H.; Tamiru, M.; Takuno, S.; et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef]
- Li, Z.; Chen, X.; Shi, S.; Zhang, H.; Wang, X.; Chen, H.; Li, W.; Li, L. DeepBSA: A deep-learning algorithm improves bulked segregant analysis for dissecting complex traits. Mol. Plant 2022, 15, 1418–1427. [Google Scholar] [CrossRef]
- Ontoy, J.C.; Shrestha, B.; Karki, H.S.; Barphagha, I.; Angira, B.; Famoso, A.; Ham, J.H. Genetic Characterization of the Partial Disease Resistance of Rice to Bacterial Panicle Blight and Sheath Blight by Combined QTL Linkage and QTL-seq Analyses. Plants 2023, 12, 559. [Google Scholar] [CrossRef] [PubMed]
- Riangwong, K.; Aesomnuk, W.; Sonsom, Y.; Siangliw, M.; Unartngam, J.; Toojinda, T.; Wanchana, S.; Arikit, S. QTL-seq Identifies Genomic Regions Associated with Resistance to Dirty Panicle Disease in Rice. Agronomy 2023, 13, 1905. [Google Scholar] [CrossRef]
- Kankanala, P.; Nandety, R.S.; Mysore, K.S. Genomics of Plant Disease Resistance in Legumes. Front. Plant Sci. 2019, 10, 1345. [Google Scholar] [CrossRef]
- Yin, K.; Qiu, J.-L. Genome editing for plant disease resistance: Applications and perspectives. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180322. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, J.; Yeom, S.-I.; Kim, Y.-M.; Seo, E.; Kim, K.-T.; Kim, M.-S.; Lee, J.M.; Cheong, K.; Shin, H.-S.; et al. New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 2017, 18, 210. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Neme, R.; Beh, L.Y.; Chen, X.; Braun, J.; Lu, M.W.; Landweber, L.F. Comparative genomics reveals insight into the evolutionary origin of massively scrambled genomes. eLife 2022, 11, e82979. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Gu, X.; Ding, J.; Yao, L.; Gao, X.; Zhang, M.; Meng, Q.; Wei, S.; Fu, J. Gene expression analysis of resistant and susceptible rice cultivars to sheath blight after inoculation with Rhizoctonia solani. BMC Genom. 2022, 23, 278. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.P.; Liu, H.; Argueso, C.T.; Pereira, A.; Cruz, C.V.; Verdier, V.; Leach, J.E. RNA-Seq analysis reveals insight into enhanced rice Xa7-mediated bacterial blight resistance at high temperature. PLoS ONE 2017, 12, e0187625. [Google Scholar] [CrossRef] [PubMed]
- Stokes, T. Transcriptional responses to plant pathogen interactions. Trends Plant Sci. 2001, 6, 50–51. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, P.; Singh, D.; Mathur, S.; Singh, A.; Ranjan, R. Upcoming progress of transcriptomics studies on plants: An overview. Front. Plant Sci. 2022, 13, 1030890. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, H.; Wang, H.; Xiang, Z.; Wei, S.; Zheng, W. Comparative transcriptome analysis of rice cultivars resistant and susceptible to Rhizoctonia solani AG1-IA. BMC Genom. 2022, 23, 606. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, S.; Liu, K.; Wang, S.; Huang, L.; Guo, L. Proteomics: A powerful tool to study plant responses to biotic stress. Plant Methods 2019, 15, 135. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Gupta, R.; Min, C.W.; Kwon, S.W.; Wang, Y.; Je, B.I.; Kim, Y.-J.; Jeon, J.-S.; Agrawal, G.K.; Rakwal, R.; et al. Proteomics of Rice—Magnaporthe oryzae Interaction: What Have We Learned So Far? Front. Plant Sci. 2019, 10, 1383. [Google Scholar] [CrossRef]
- Yong, Y.; Qiujun, L.; Xinyu, C.; Weifang, L.; Yuwen, F.; Zhengjin, X.; Yuanhua, W.; Xuming, W.; Jie, Z.; Chulang, Y.; et al. Characterization and Proteomic Analysis of Novel Rice Lesion Mimic Mutant with Enhanced Disease Resistance. Rice Sci. 2021, 28, 466–478. [Google Scholar] [CrossRef]
- Gupta, R.; Min, C.W.; Son, S.; Lee, G.H.; Jang, J.W.; Kwon, S.W.; Park, S.R.; Kim, S.T. Comparative proteome profiling of susceptible and resistant rice cultivars identified an arginase involved in rice defense against Xanthomonas oryzae pv. oryzae. Plant Physiol. Biochem. 2022, 171, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Min, C.W.; Park, S.-R.; Kim, S.T. Label-free proteome data of susceptible and resistant rice cultivars in response to Xanthomonas oryzae pv. oryzae inoculation. Data Brief 2022, 41, 107890. [Google Scholar] [CrossRef]
- Tian, D.; Yang, L.; Chen, Z.; Chen, Z.; Wang, F.; Zhou, Y.; Luo, Y.; Yang, L.; Chen, S. Proteomic analysis of the defense response to Magnaporthe oryzae in rice harboring the blast resistance gene Piz-t. Rice 2018, 11, 47. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Wang, D.; Gupta, R.; Kim, S.T.; Wang, Y. A Proteomics Insight into Advancements in the Rice–Microbe Interaction. Plants 2023, 12, 1079. [Google Scholar] [CrossRef]
- Zhang, F.; Huang, L.; Zeng, D.; Cruz, C.V.; Li, Z.; Zhou, Y. Comparative proteomic analysis reveals novel insights into the interaction between rice and Xanthomonas oryzae pv. oryzae. BMC Plant Biol. 2020, 20, 563. [Google Scholar] [CrossRef]
- Sharma, V.; Gupta, P.; Priscilla, K.; Kumar, S.; Hangargi, B.; Veershetty, A.; Ramrao, D.P.; Suresh, S.; Narasanna, R.; Naik, G.R.; et al. Metabolomics Intervention towards Better Understanding of Plant Traits. Cells 2021, 10, 346. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, S.G.; Wu, J.; Huh, H.-H.; Lee, S.-J.; Rakwal, R.; Agrawal, G.K.; Park, Z.-Y.; Kang, K.Y.; Kim, S.T. Secretome analysis of the rice bacterium Xanthomonas oryzae (Xoo) using in vitro and in planta systems. Proteomics 2013, 13, 1901–1912. [Google Scholar] [CrossRef]
- Sana, T.R.; Fischer, S.; Wohlgemuth, G.; Katrekar, A.; Jung, K.-H.; Ronald, P.C.; Fiehn, O. Metabolomic and transcriptomic analysis of the rice response to the bacterial blight pathogen Xanthomonas oryzae pv. oryzae. Metabolomics 2010, 6, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Castro-Moretti, F.R.; Gentzel, I.N.; Mackey, D.; Alonso, A.P. Metabolomics as an Emerging Tool for the Study of Plant-Pathogen Interactions. Metabolites 2020, 10, 52. [Google Scholar] [CrossRef]
- Suharti, W.S.; Nose, A.; Zheng, S.-H. Metabolite profiling of sheath blight disease resistance in rice: In the case of positive ion mode analysis by CE/TOF-MS. Plant Prod. Sci. 2016, 19, 279–290. [Google Scholar] [CrossRef][Green Version]
- Gong, L.; Chen, W.; Gao, Y.; Liu, X.; Zhang, H.; Xu, C.; Yu, S.; Zhang, Q.; Luo, J. Genetic analysis of the metabolome exemplified using a rice population. Proc. Natl. Acad. Sci. USA 2013, 110, 20320–20325. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Bohra, A.; Pandey, A.K.; Pandey, M.K.; Kumar, A. Metabolomics for Plant Improvement: Status and Prospects. Front. Plant Sci. 2017, 8, 1302. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Saand, M.A.; Huang, L.; Abdelaal, W.B.; Zhang, J.; Wu, Y.; Li, J.; Sirohi, M.H.; Wang, F. Applications of Multi-Omics Technologies for Crop Improvement. Front. Plant Sci. 2021, 12, 563953. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Bai, L.; Liu, M.; Liu, Y.; Peng, S.; Hu, P.; Wang, D.; Liu, Q.; Yan, S.; Gao, L.; et al. Identification of two novel rice S genes through combination of association and transcription analyses with gene-editing technology. Plant Biotechnol. J. 2023, 21, 1628–1641. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; He, H.; Chen, W.; Huang, L.; Zhao, D.; Chen, X.; Li, J.; Yang, X. Integrated genetic analysis of leaf blast resistance in upland rice: QTL mapping, bulked segregant analysis and transcriptome sequencing. AoB Plants 2022, 14, plac047. [Google Scholar] [CrossRef]
- Prathi, N.B.; Palit, P.; Madhu, P.; Ramesh, M.; Laha, G.; Balachandran, S.; Madhav, M.S.; Sundaram, R.; Mangrauthia, S.K. Proteomic and transcriptomic approaches to identify resistance and susceptibility related proteins in contrasting rice genotypes infected with fungal pathogen Rhizoctonia solani. Plant Physiol. Biochem. 2018, 130, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Zha, W.; Yu, X.; Wu, Y.; Li, S.; Xu, H.; Li, P.; Li, C.; Liu, K.; Chen, J.; et al. Integrated transcriptomics and metabolomics analysis provide insight into the resistance response of rice against brown planthopper. Front. Plant Sci. 2023, 14, 1213257. [Google Scholar] [CrossRef]
- Roychowdhury, R.; Das, S.P.; Gupta, A.; Parihar, P.; Chandrasekhar, K.; Sarker, U.; Kumar, A.; Ramrao, D.P.; Sudhakar, C. Multi-Omics Pipeline and Omics-Integration Approach to Decipher Plant’s Abiotic Stress Tolerance Responses. Genes 2023, 14, 1281. [Google Scholar] [CrossRef]
- Naik, B.; Kumar, V.; Rizwanuddin, S.; Chauhan, M.; Choudhary, M.; Gupta, A.K.; Kumar, P.; Kumar, V.; Saris, P.E.J.; Rather, M.A.; et al. Genomics, Proteomics, and Metabolomics Approaches to Improve Abiotic Stress Tolerance in Tomato Plant. Int. J. Mol. Sci. 2023, 24, 3025. [Google Scholar] [CrossRef]
- Ullah, M.A.; Abdullah-Zawawi, M.-R.; Zainal-Abidin, R.-A.; Sukiran, N.L.; Uddin, I.; Zainal, Z. A Review of Integrative Omic Approaches for Understanding Rice Salt Response Mechanisms. Plants 2022, 11, 1430. [Google Scholar] [CrossRef]
- Zargar, S.M.; Mir, R.A.; Ebinezer, L.B.; Masi, A.; Hami, A.; Manzoor, M.; Salgotra, R.K.; Sofi, N.R.; Mushtaq, R.; Rohila, J.S.; et al. Physiological and Multi-Omics Approaches for Explaining Drought Stress Tolerance and Supporting Sustainable Production of Rice. Front. Plant Sci. 2022, 12, 803603. [Google Scholar] [CrossRef]
- Lu, S.; Jia, Z.; Meng, X.; Chen, Y.; Wang, S.; Fu, C.; Yang, L.; Zhou, R.; Wang, B.; Cao, Y. Combined Metabolomic and Transcriptomic Analysis Reveals Allantoin Enhances Drought Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 14172. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Jiang, J.; Zhang, G.; Miao, S.; Lu, J.; Qian, Y.; Zhao, X.; Wang, W.; Qiu, X.; Zhang, F.; et al. Integrating GWAS and transcriptomics to identify candidate genes conferring heat tolerance in rice. Front. Plant Sci. 2023, 13, 1102938. [Google Scholar] [CrossRef]
- Ma, L.; Zeng, J.; Zhang, R.Q.; Wang, L.; Zhang, F.; Zhao, X.; Yuan, Y.; Li, L. Integrated transcriptomic and metabolomic analysis the variation of rice cultivars response to arsenite stress. Environ. Technol. Innov. 2023, 31, 103207. [Google Scholar] [CrossRef]
- Gu, S.; Zhuang, J.; Zhang, Z.; Chen, W.; Xu, H.; Zhao, M.; Ma, D. Multi-omics approach reveals the contribution of OsSEH1 to rice cold tolerance. Front. Plant Sci. 2023, 13, 1110724. [Google Scholar] [CrossRef]
- Iqbal, Z.; Iqbal, M.S.; Khan, M.I.R.; Ansari, M.I. Toward Integrated Multi-Omics Intervention: Rice Trait Improvement and Stress Management. Front. Plant Sci. 2021, 12, 741419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhang, C.; Yu, C.; Dong, J.; Hu, J. Integration of multi-omics technologies for crop improvement: Status and prospects. Front. Bioinform. 2022, 2, 1027457. [Google Scholar] [CrossRef]
- Dai, L.; Li, P.; Li, Q.; Leng, Y.; Zeng, D.; Qian, Q. Integrated Multi-Omics Perspective to Strengthen the Understanding of Salt Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 5236. [Google Scholar] [CrossRef] [PubMed]
- Zaghum, M.J.; Ali, K.; Teng, S. Integrated Genetic and Omics Approaches for the Regulation of Nutritional Activities in Rice (Oryza sativa L.). Agriculture 2022, 12, 1757. [Google Scholar] [CrossRef]
- Geng, L.; Zhang, W.; Zou, T.; Du, Q.; Ma, X.; Cui, D.; Han, B.; Zhang, Q.; Han, L. Integrating linkage mapping and comparative transcriptome analysis for discovering candidate genes associated with salt tolerance in rice. Front. Plant Sci. 2023, 14, 1065334. [Google Scholar] [CrossRef] [PubMed]
- McCouch, S.R.; Wright, M.H.; Tung, C.-W.; Maron, L.G.; McNally, K.L.; Fitzgerald, M.; Singh, N.; DeClerck, G.; Agosto-Perez, F.; Korniliev, P.; et al. Open access resources for genome-wide association mapping in rice. Nat. Commun. 2016, 7, 10532. [Google Scholar] [CrossRef] [PubMed]
- Shew, A.M.; Durand-Morat, A.; Nalley, L.L.; Zhou, X.-G.; Rojas, C.; Thoma, G. Warming increases Bacterial Panicle Blight (Burkholderia glumae) occurrences and impacts on USA rice production. PLoS ONE 2019, 14, e0219199. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ontoy, J.C.; Ham, J.H. Mapping and Omics Integration: Towards Precise Rice Disease Resistance Breeding. Plants 2024, 13, 1205. https://doi.org/10.3390/plants13091205
Ontoy JC, Ham JH. Mapping and Omics Integration: Towards Precise Rice Disease Resistance Breeding. Plants. 2024; 13(9):1205. https://doi.org/10.3390/plants13091205
Chicago/Turabian StyleOntoy, John Christian, and Jong Hyun Ham. 2024. "Mapping and Omics Integration: Towards Precise Rice Disease Resistance Breeding" Plants 13, no. 9: 1205. https://doi.org/10.3390/plants13091205
APA StyleOntoy, J. C., & Ham, J. H. (2024). Mapping and Omics Integration: Towards Precise Rice Disease Resistance Breeding. Plants, 13(9), 1205. https://doi.org/10.3390/plants13091205






