Adoption of the 2A Ribosomal Skip Principle to Track Assembled Virions of Pepper Mild Mottle Virus in Nicotiana benthamiana
Abstract
1. Introduction
2. Results
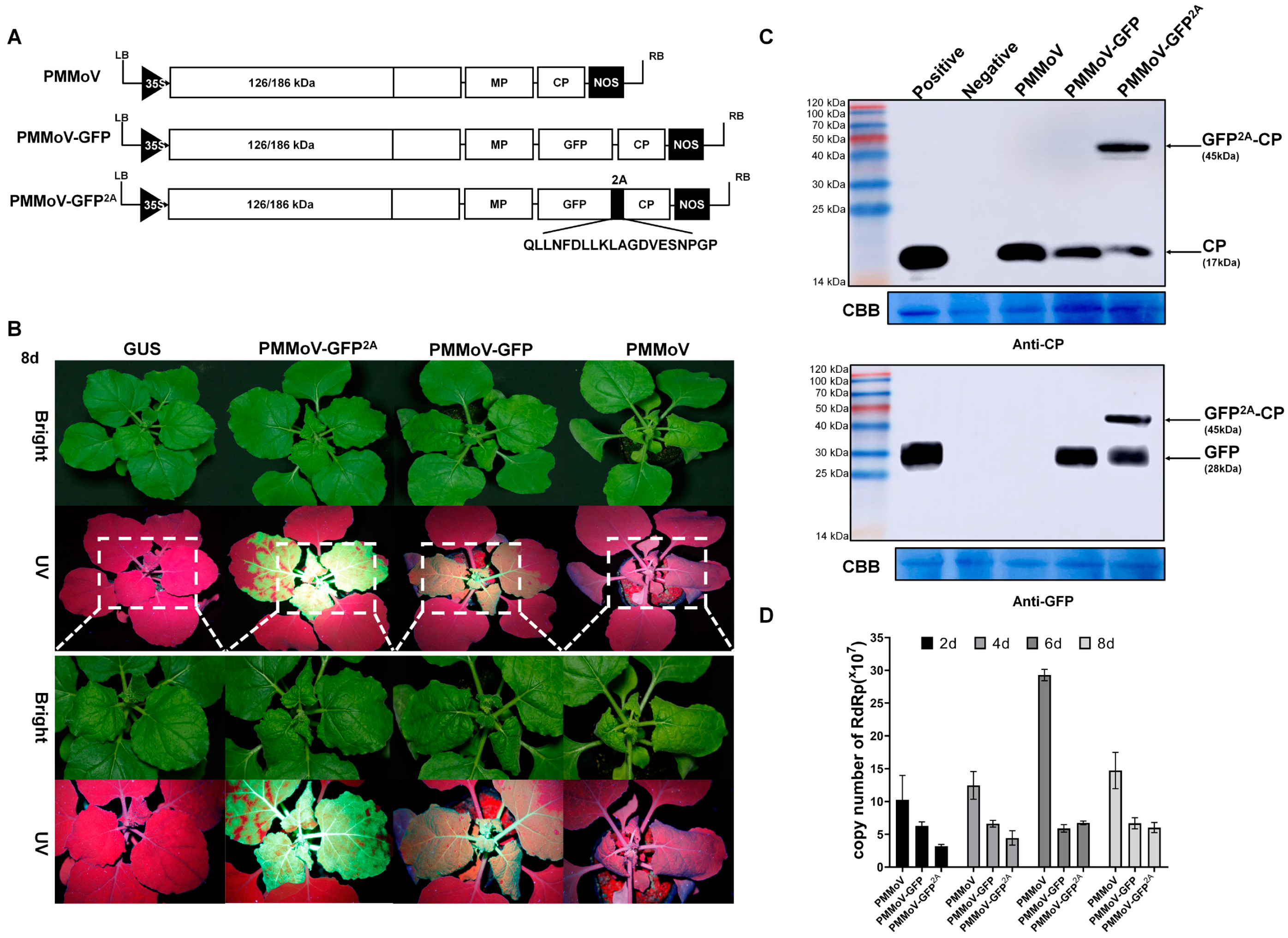
2.1. The Constructed PMMoV-GFP2A Vector Is Capable of Infecting Plants under Normal Conditions
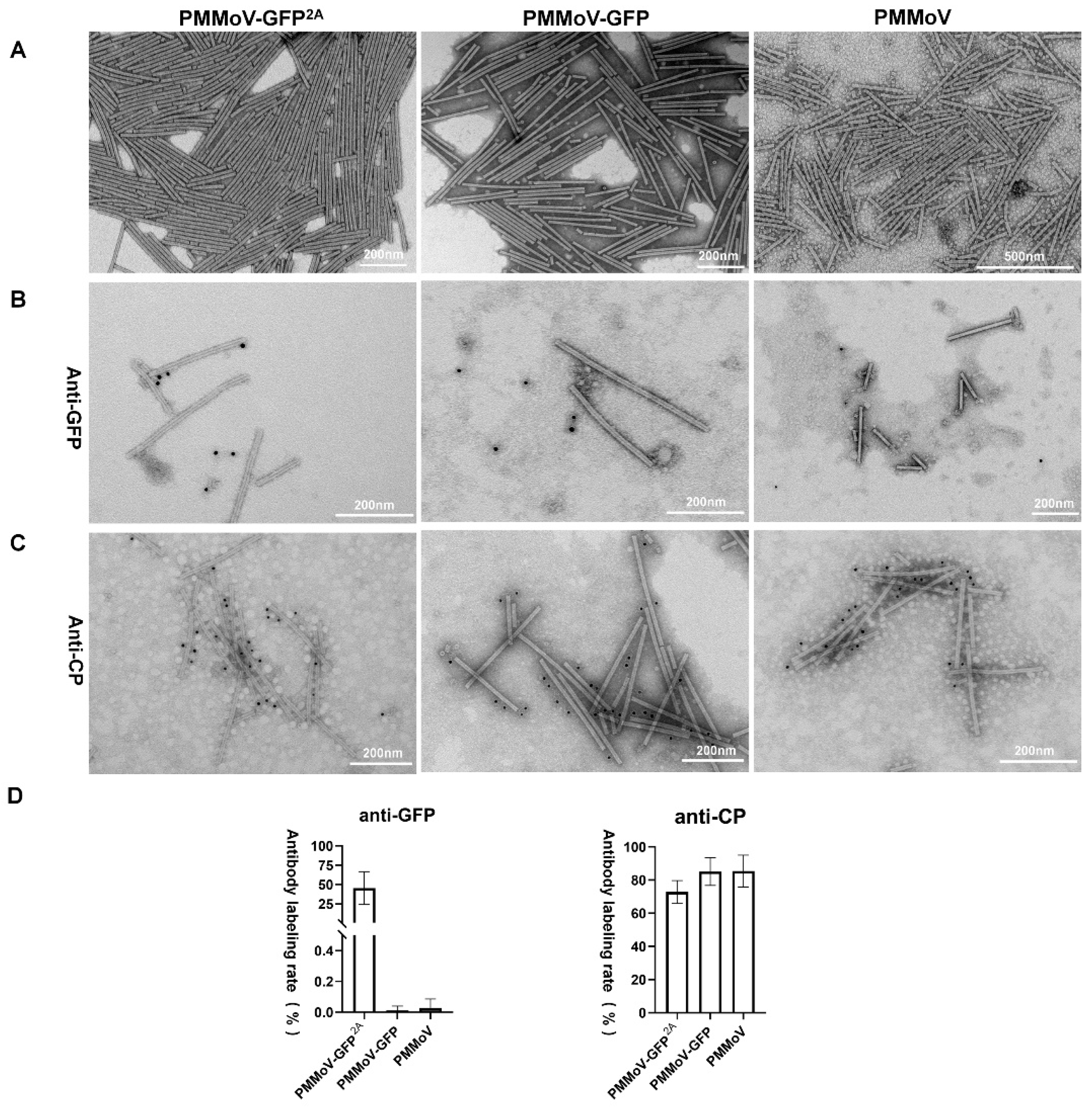
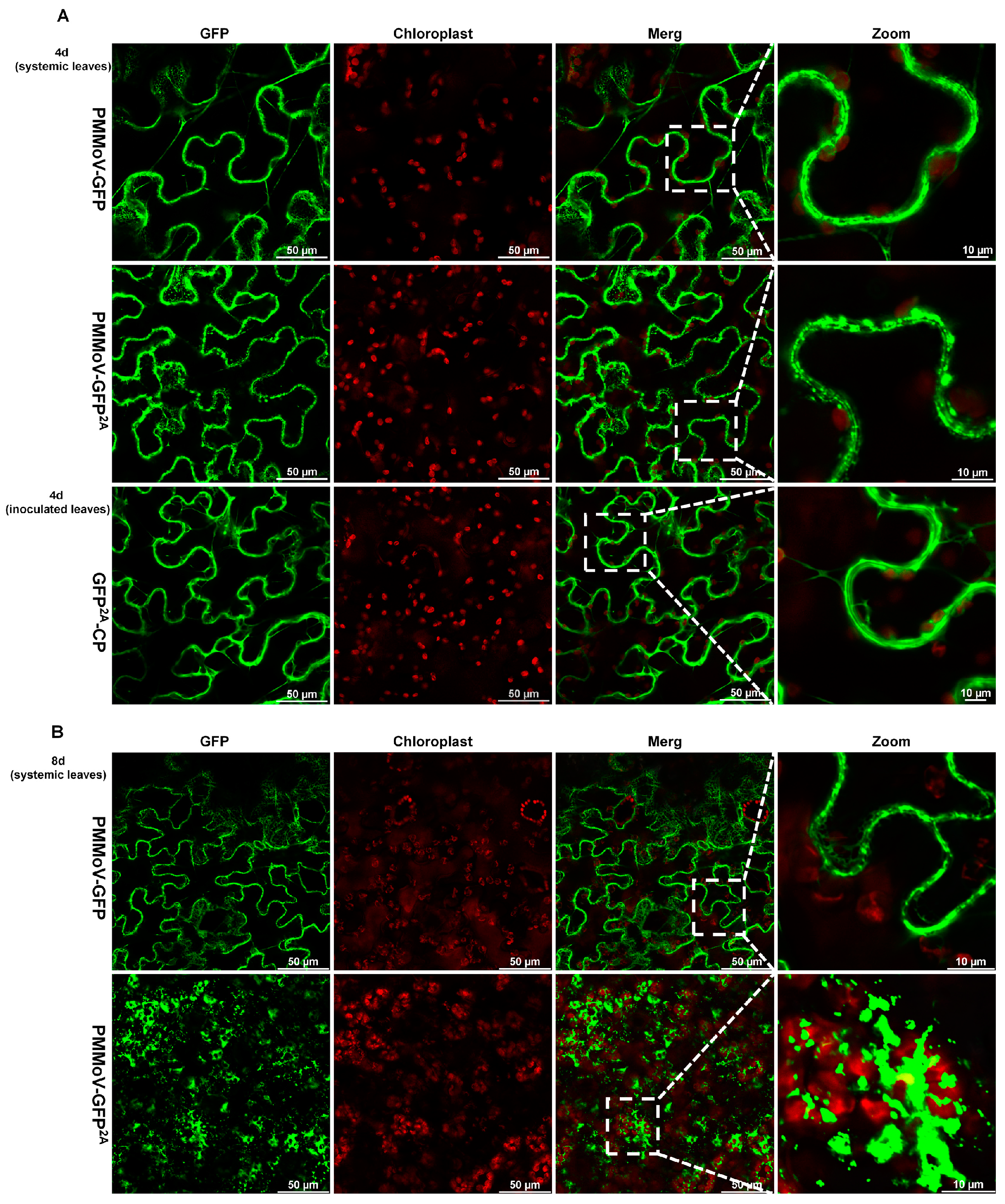
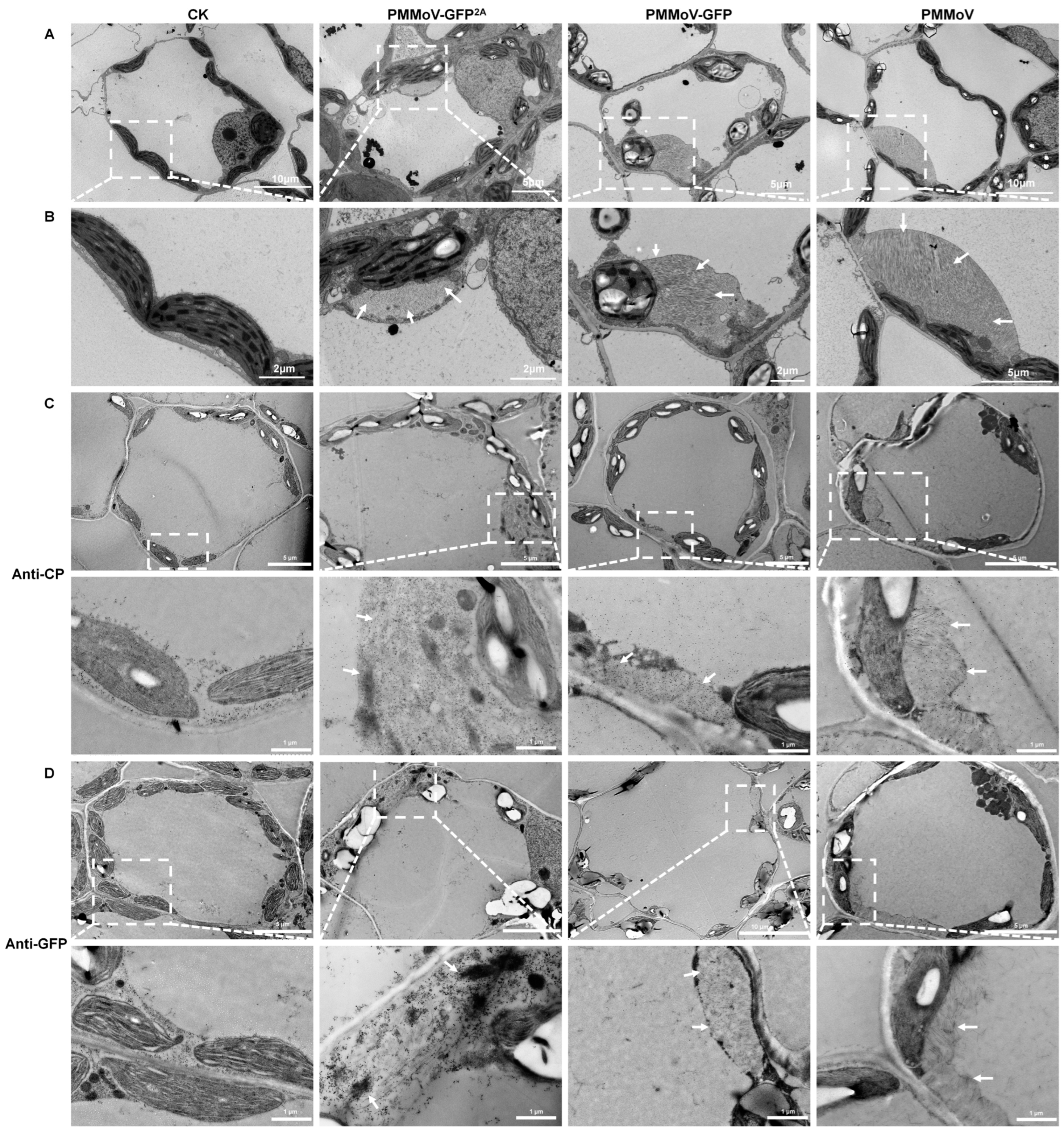
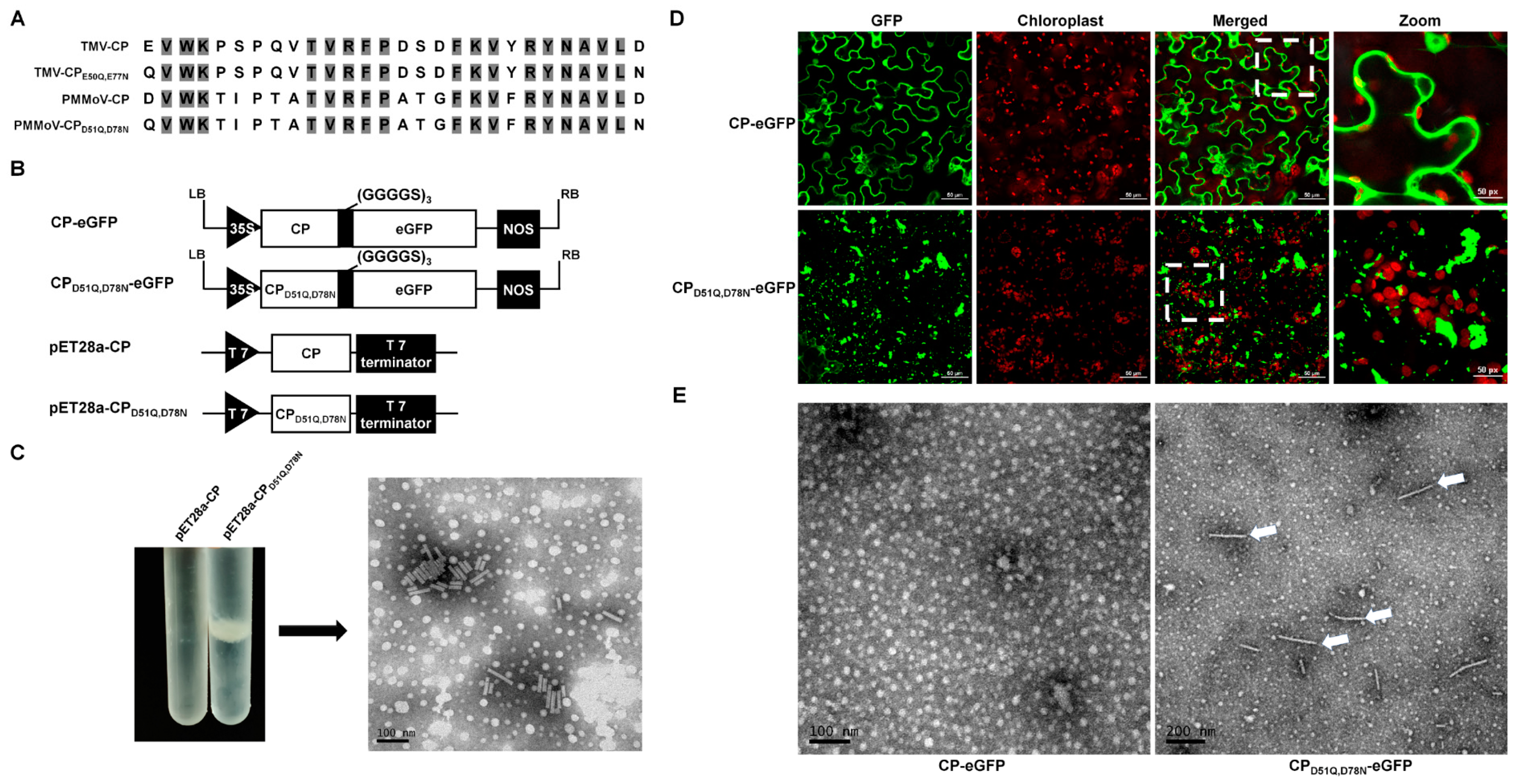
2.2. The Virus Particles Produced by PMMoV-GFP2A Incorporate GFP
2.3. PMMoV-GFP2A Can Track the Dynamic Changes of Virus Coat Protein by Tracking GFP
2.4. Virions or Virion Aggregation Can Form Fluorescent Plaques
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Plant Infection and Fluorescence Detection
4.3. Western Blot
4.4. Quantitative Real-Time PCR
4.5. Virus Preparation and Electron Microscopy
4.6. Ultrathin Sectioning
4.7. Immunocolloidal Gold Technique
4.8. Immuno Electron Microscopy
4.9. Virus-like Particle Purification
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, Z.; Zhang, D.; Wang, X.; Zhang, X.; Wen, Z.; Zhang, Q.; Li, D.; Dinesh-Kumar, S.P.; Zhang, Y. Coat proteins of necroviruses target 14-3-3a to subvert MAPKKKα-mediated antiviral immunity in plants. Nat. Commun. 2022, 13, 716. [Google Scholar] [CrossRef]
- Niu, E.; Ye, C.; Zhao, W.; Kondo, H.; Wu, Y.; Chen, J.; Andika, I.B.; Sun, L. Coat protein of Chinese wheat mosaic virus upregulates and interacts with cytosolic glyceraldehyde-3-phosphate dehydrogenase, a negative regulator of plant autophagy, to promote virus infection. J. Integr. Plant Biol. 2022, 64, 1631–1645. [Google Scholar] [CrossRef]
- Callaway, A.; Giesman-Cookmeyer, D.; Gillock, E.T.; Sit, T.L.; Lommel, S.A. The multifunctional capsid proteins of plant RNA viruses. Annu. Rev. Phytopathol. 2001, 39, 419–460. [Google Scholar] [CrossRef] [PubMed]
- Herranz, M.C.; Pallas, V.; Aparicio, F. Multifunctional roles for the N-terminal basic motif of Alfalfa mosaic virus coat protein: Nucleolar/cytoplasmic shuttling, modulation of RNA-binding activity, and virion formation. Mol. Plant Microbe Interact. 2012, 25, 1093–1103. [Google Scholar] [CrossRef]
- Vaira, A.M.; Lim, H.S.; Bauchan, G.; Gulbronson, C.J.; Miozzi, L.; Vinals, N.; Natilla, A.; Hammond, J. The interaction of Lolium latent virus major coat protein with ankyrin repeat protein NbANKr redirects it to chloroplasts and modulates virus infection. J. Gen. Virol. 2018, 99, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Ishikawa, M. Replication of Tobamovirus RNA. Annu. Rev. Phytopathol. 2016, 54, 55–78. [Google Scholar] [CrossRef] [PubMed]
- Alonso, E.; García-Luque, I.; de la Cruz, A.; Wicke, B.; Avila-Rincón, M.J.; Serra, M.T.; Castresana, C.; Díaz-Ruíz, J.R. Nucleotide sequence of the genomic RNA of pepper mild mottle virus, a resistance-breaking tobamovirus in pepper. J. Gen. Virol. 1991, 72 Pt 12, 2875–2884. [Google Scholar] [CrossRef] [PubMed]
- Makarov, V.V.; Kalinina, N.O. Structure and Noncanonical Activities of Coat Proteins of Helical Plant Viruses. Biochemistry 2016, 81, 1–18. [Google Scholar] [CrossRef]
- Culver, J.N.; Dawson, W.O.; Plonk, K.; Stubbs, G. Site-directed mutagenesis confirms the involvement of carboxylate groups in the disassembly of tobacco mosaic virus. Virology 1995, 206, 724–730. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Han, K.; Zheng, H.; Ji, M.; Cui, W.; Hu, S.; Peng, J.; Zhao, J.; Lu, Y.; Lin, L.; Liu, Y.; et al. A single amino acid in coat protein of Pepper mild mottle virus determines its subcellular localization and the chlorosis symptom on leaves of pepper. J. Gen. Virol. 2020, 101, 565–570. [Google Scholar] [CrossRef]
- Weber, P.H.; Bujarski, J.J. Multiple functions of capsid proteins in (+) stranded RNA viruses during plant-virus interactions. Virus Res. 2015, 196, 140–149. [Google Scholar] [CrossRef]
- Peng, J.; Shi, B.; Zheng, H.; Lu, Y.; Lin, L.; Jiang, T.; Chen, J.; Yan, F. Detection of pepper mild mottle virus in pepper sauce in China. Arch. Virol. 2015, 160, 2079–2082. [Google Scholar] [CrossRef]
- Rosario, K.; Symonds, E.M.; Sinigalliano, C.; Stewart, J.; Breitbart, M. Pepper mild mottle virus as an indicator of fecal pollution. Appl. Environ. Microbiol. 2009, 75, 7261–7267. [Google Scholar] [CrossRef]
- Zheng, K.; Zhang, R.; Wan, Q.; Zhang, G.; Lu, Y.; Zheng, H.; Yan, F.; Peng, J.; Wu, J. Pepper mild mottle virus can infect and traffick within Nicotiana benthamiana plants in non-virion forms. Virology 2023, 587, 109881. [Google Scholar] [CrossRef]
- Tsuda, S.; Kirita, M.; Watanabe, Y. Characterization of a pepper mild mottle tobamovirus strain capable of overcoming the L3 gene-mediated resistance, distinct from the resistance-breaking Italian isolate. Mol. Plant Microbe Interact. 1998, 11, 327–331. [Google Scholar] [CrossRef]
- Han, K.; Zheng, H.; Yan, D.; Zhou, H.; Jia, Z.; Zhai, Y.; Wu, J.; Lu, Y.; Wu, G.; Rao, S.; et al. Pepper mild mottle virus coat protein interacts with pepper chloroplast outer envelope membrane protein OMP24 to inhibit antiviral immunity in plants. Hortic. Res. 2023, 10, uhad046. [Google Scholar] [CrossRef]
- Asurmendi, S.; Berg, R.H.; Koo, J.C.; Beachy, R.N. Coat protein regulates formation of replication complexes during tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA 2004, 101, 1415–1420. [Google Scholar] [CrossRef] [PubMed]
- Shivprasad, S.; Pogue, G.P.; Lewandowski, D.J.; Hidalgo, J.; Donson, J.; Grill, L.K.; Dawson, W.O. Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 1999, 255, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Vasques, R.M.; Lacorte, C.; da Luz, L.L.; Aranda, M.A.; Nagata, T. Development of a new tobamovirus-based viral vector for protein expression in plants. Mol. Biol. Rep. 2019, 46, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Xiao, C.; Han, K.; Peng, J.; Lin, L.; Lu, Y.; Xie, L.; Wu, X.; Xu, P.; Li, G.; et al. Development of an agroinoculation system for full-length and GFP-tagged cDNA clones of cucumber green mottle mosaic virus. Arch. Virol. 2015, 160, 2867–2872. [Google Scholar] [CrossRef] [PubMed]
- Shahgolzari, M.; Pazhouhandeh, M.; Milani, M.; Yari Khosroushahi, A.; Fiering, S. Plant viral nanoparticles for packaging and in vivo delivery of bioactive cargos. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol 2020, 12, e1629. [Google Scholar] [CrossRef]
- Conti, G.; Rodriguez, M.C.; Venturuzzi, A.L.; Asurmendi, S. Modulation of host plant immunity by Tobamovirus proteins. Ann. Bot. 2017, 119, 737–747. [Google Scholar] [CrossRef]
- Röder, J.; Fischer, R.; Commandeur, U. Adoption of the 2A Ribosomal Skip Principle to Tobacco Mosaic Virus for Peptide Display. Front. Plant Sci. 2017, 8, 1125. [Google Scholar] [CrossRef]
- Werner, S.; Marillonnet, S.; Hause, G.; Klimyuk, V.; Gleba, Y. Immunoabsorbent nanoparticles based on a tobamovirus displaying protein A. Proc. Natl. Acad. Sci. USA 2006, 103, 17678–17683. [Google Scholar] [CrossRef]
- Brown, A.D.; Naves, L.; Wang, X.; Ghodssi, R.; Culver, J.N. Carboxylate-directed in vivo assembly of virus-like nanorods and tubes for the display of functional peptides and residues. Biomacromolecules 2013, 14, 3123–3129. [Google Scholar] [CrossRef]
- Lu, P.; Feng, M.G. Bifunctional enhancement of a beta-glucanase-xylanase fusion enzyme by optimization of peptide linkers. Appl. Microbiol. Biotechnol. 2008, 79, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, M.L.L.; Luke, G.; Mehrotra, A.; Li, X.; Hughes, L.E.; Gani, D.; Ryan, M.D. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal ‘skip’. J. Gen. Virol. 2001, 82, 1013–1025. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Jiang, L.; Zhou, Z.; Fan, J.; Zhang, Q.; Zhu, H.; Han, Q.; Xu, Z. Expression of foot-and-mouth disease virus epitopes in tobacco by a tobacco mosaic virus-based vector. Vaccine 2003, 21, 4390–4398. [Google Scholar] [CrossRef]
- Larkin, E.J.; Brown, A.D.; Culver, J.N. Fabrication of Tobacco Mosaic Virus-Like Nanorods for Peptide Display. Methods Mol. Biol. 2018, 1776, 51–60. [Google Scholar] [CrossRef]
- Dawson, W.O.; Lehto, K.M. Regulation of tobamovirus gene expression. Adv. Virus Res. 1990, 38, 307–342. [Google Scholar] [CrossRef] [PubMed]
- Grdzelishvili, V.Z.; Chapman, S.N.; Dawson, W.O.; Lewandowski, D.J. Mapping of the Tobacco mosaic virus movement protein and coat protein subgenomic RNA promoters in vivo. Virology 2000, 275, 177–192. [Google Scholar] [CrossRef]
- Dawson, W.O.; Lewandowski, D.J.; Hilf, M.E.; Bubrick, P.; Raffo, A.J.; Shaw, J.J.; Grantham, G.L.; Desjardins, P.R. A tobacco mosaic virus-hybrid expresses and loses an added gene. Virology 1989, 172, 285–292. [Google Scholar] [CrossRef]
- Rabindran, S.; Dawson, W.O. Assessment of recombinants that arise from the use of a TMV-based transient expression vector. Virology 2001, 284, 182–189. [Google Scholar] [CrossRef][Green Version]
- Creager, A.N.; Scholthof, K.B.; Citovsky, V.; Scholthof, H.B. Tobacco mosaic virus. Pioneering research for a century. Plant Cell 1999, 11, 301–308. [Google Scholar] [CrossRef]
- Cruz, S.S.; Chapman, S.; Roberts, A.G.; Roberts, I.M.; Prior, D.A.; Oparka, K.J. Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc. Natl. Acad. Sci. USA 1996, 93, 6286–6290. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Dickmeis, C.; Fischer, R.; Commandeur, U.; Steinmetz, N.F. In Planta Production of Fluorescent Filamentous Plant Virus-Based Nanoparticles. Methods Mol. Biol. 2018, 1776, 61–84. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.; Kwon, S.J.; Kim, M.H.; Choe, S.; Kwak, H.R.; Kim, M.K.; Jung, C.; Seo, J.K. A Plant Virus-Based Vector System for Gene Function Studies in Pepper. Plant. Physiol. 2019, 181, 867–880. [Google Scholar] [CrossRef]
- Gopinath, K.; Wellink, J.; Porta, C.; Taylor, K.M.; Lomonossoff, G.P.; van Kammen, A. Engineering cowpea mosaic virus RNA-2 into a vector to express heterologous proteins in plants. Virology 2000, 267, 159–173. [Google Scholar] [CrossRef]
- Li, Q.; Li, W.; Yin, W.; Guo, J.; Zhang, Z.P.; Zeng, D.; Zhang, X.; Wu, Y.; Zhang, X.E.; Cui, Z. Single-Particle Tracking of Human Immunodeficiency Virus Type 1 Productive Entry into Human Primary Macrophages. ACS Nano 2017, 11, 3890–3903. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Yin, Y.; Hua, M.; Wang, S.; Mo, X.; Yuan, E.; Zheng, H.; Lin, L.; Chen, H.; Lu, Y.; et al. Pod pepper vein yellows virus, a new recombinant polerovirus infecting Capsicum frutescens in Yunnan province, China. Virol. J. 2021, 18, 42. [Google Scholar] [CrossRef]
| Target | Name | Sequence (5′-3′) |
|---|---|---|
| PMMoV-GFP | Vec-PMMoV-GFP f | ATGGCTTACACAGTTTCCAGTG |
| Vec-PMMoV-GFP r | GATTAAGCACAGTATAAACG | |
| PMMoV-GFP-1 f | CGTTTATACTGTGCTTAATC | |
| PMMoV-GFP-1 r | TTAAAACGAAGAAGACTCGGCG | |
| PMMoV-GFP-2 f | CGCCGAGTCTTCTTCGTTTTAACTATGAAGACTAATCTTTTTC | |
| PMMoV-GFP-2 r | CACTGGAAACTGTGTAAGCCATAGTTAAAACGTACTCGATGAC | |
| PMMoV-GFP2A | PMMoV-GFP2A-2 f | GCCGAGTCTTCTTCGTTTTAACTATGAAGACTAATCTTTTTCTC |
| PMMoV-GFP2A-2 r | CACTGGAAACTGTGTAAGCCATGGGCCCAGGGTTGGACTCGAC | |
| GFP2A-CP | GFP2A-CP f | TTGGAGAGAACACGGGGGACGAGCTCATGAAGACTAATCTTTTTCTC |
| GFP2A-CP r | AACGAAAGCTCTGCAGTCTAGATTAAGGAGTTGTAGCCCAGGTGAG | |
| pET28a-CP/ pET28a-CPD51Q,D78N | 28a-CP f | TTGTTTAACTTTAAGAAGGAGATATACCATGGCTTACACAGTTTCCAGTG |
| 28a-CP r | CAGTGGTGGTGGTGGTGGTGCTCGAGCTAAGGAGTTGTAGCCCAGGTG | |
| CP-eGFP/ CPD51Q,D78N-eGFP | CP-eGFP-1 f | TTGGAGAGAACACGGGGGACGAGCTCATGGCTTACACAGTTTCC |
| CP-eGFP-1 r | CCCGAGCCACCGCCACCGGTACCAGGAGTTGTAGCCCAGGTGAGTCC | |
| CP-eGFP-2 f | ACCGGTGGCGGTGGCTCGGGCGGTGGTGGGTCGGGTGGCGGTGGCTCG GGATCCATGGTGAGCAAGGGCGAGGA | |
| CP-eGFP-2 r | GAACGAAAGCTCTGCAGTCTAGATTACTTGTACAGCTCGTCCAT | |
| Real-time qPCR | RdRp f | AAGAAAGGAGGTTATGGTCAGC |
| RdRp r | CGGCAGTAATCTCATCCCAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, M.; Yin, Y.; Tian, Y.; Lei, J.; Lin, L.; Wu, J.; Lu, Y.; Zheng, H.; Yan, F.; Wang, J.; et al. Adoption of the 2A Ribosomal Skip Principle to Track Assembled Virions of Pepper Mild Mottle Virus in Nicotiana benthamiana. Plants 2024, 13, 928. https://doi.org/10.3390/plants13070928
Jiao M, Yin Y, Tian Y, Lei J, Lin L, Wu J, Lu Y, Zheng H, Yan F, Wang J, et al. Adoption of the 2A Ribosomal Skip Principle to Track Assembled Virions of Pepper Mild Mottle Virus in Nicotiana benthamiana. Plants. 2024; 13(7):928. https://doi.org/10.3390/plants13070928
Chicago/Turabian StyleJiao, Mengting, Yueyan Yin, Yanzhen Tian, Jianing Lei, Lin Lin, Jian Wu, Yuwen Lu, Hongying Zheng, Fei Yan, Jianguang Wang, and et al. 2024. "Adoption of the 2A Ribosomal Skip Principle to Track Assembled Virions of Pepper Mild Mottle Virus in Nicotiana benthamiana" Plants 13, no. 7: 928. https://doi.org/10.3390/plants13070928
APA StyleJiao, M., Yin, Y., Tian, Y., Lei, J., Lin, L., Wu, J., Lu, Y., Zheng, H., Yan, F., Wang, J., & Peng, J. (2024). Adoption of the 2A Ribosomal Skip Principle to Track Assembled Virions of Pepper Mild Mottle Virus in Nicotiana benthamiana. Plants, 13(7), 928. https://doi.org/10.3390/plants13070928





