Genotype-Dependent Response of Root Microbiota and Leaf Metabolism in Olive Seedlings Subjected to Drought Stress
Abstract
1. Introduction
2. Results
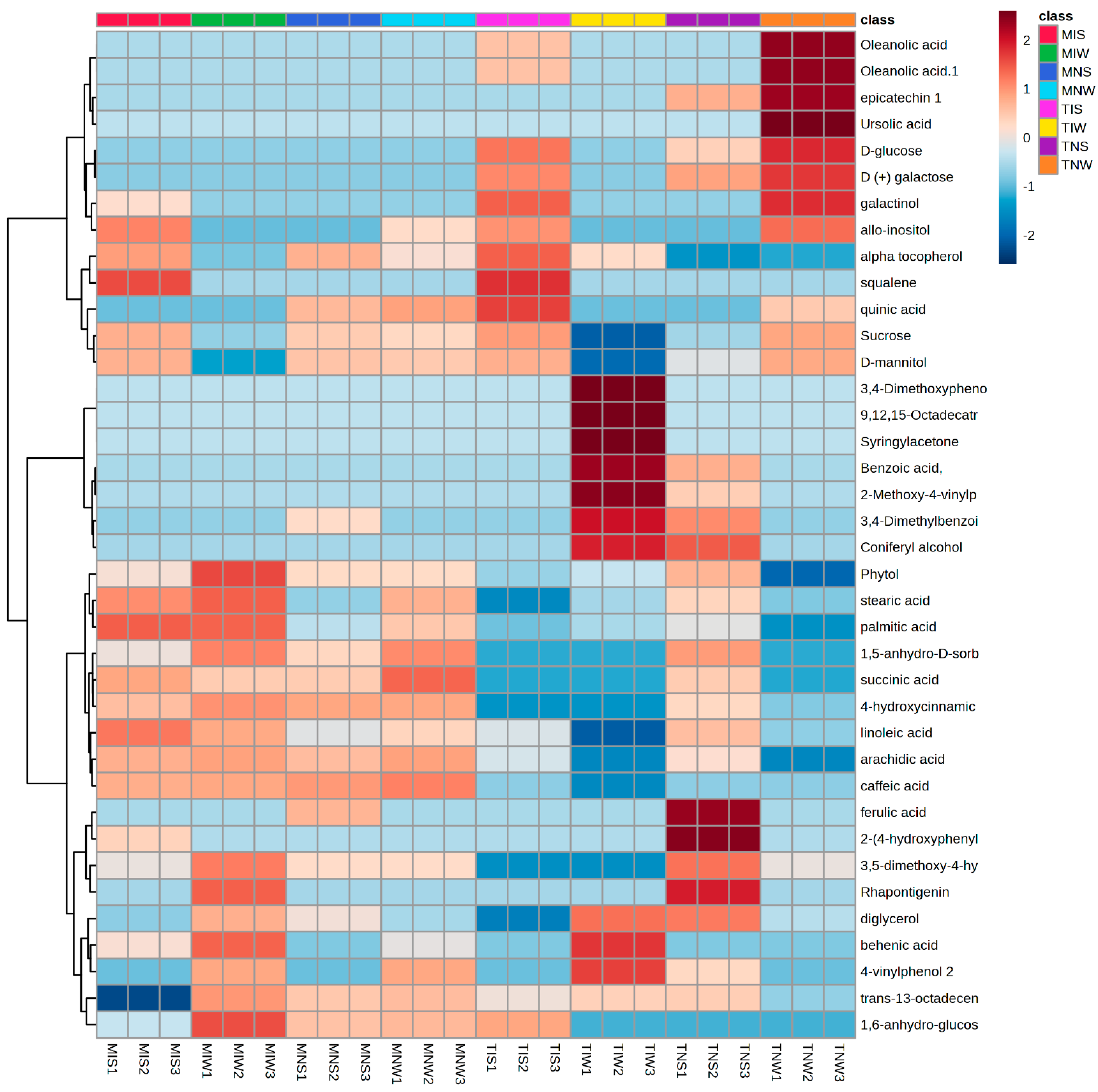
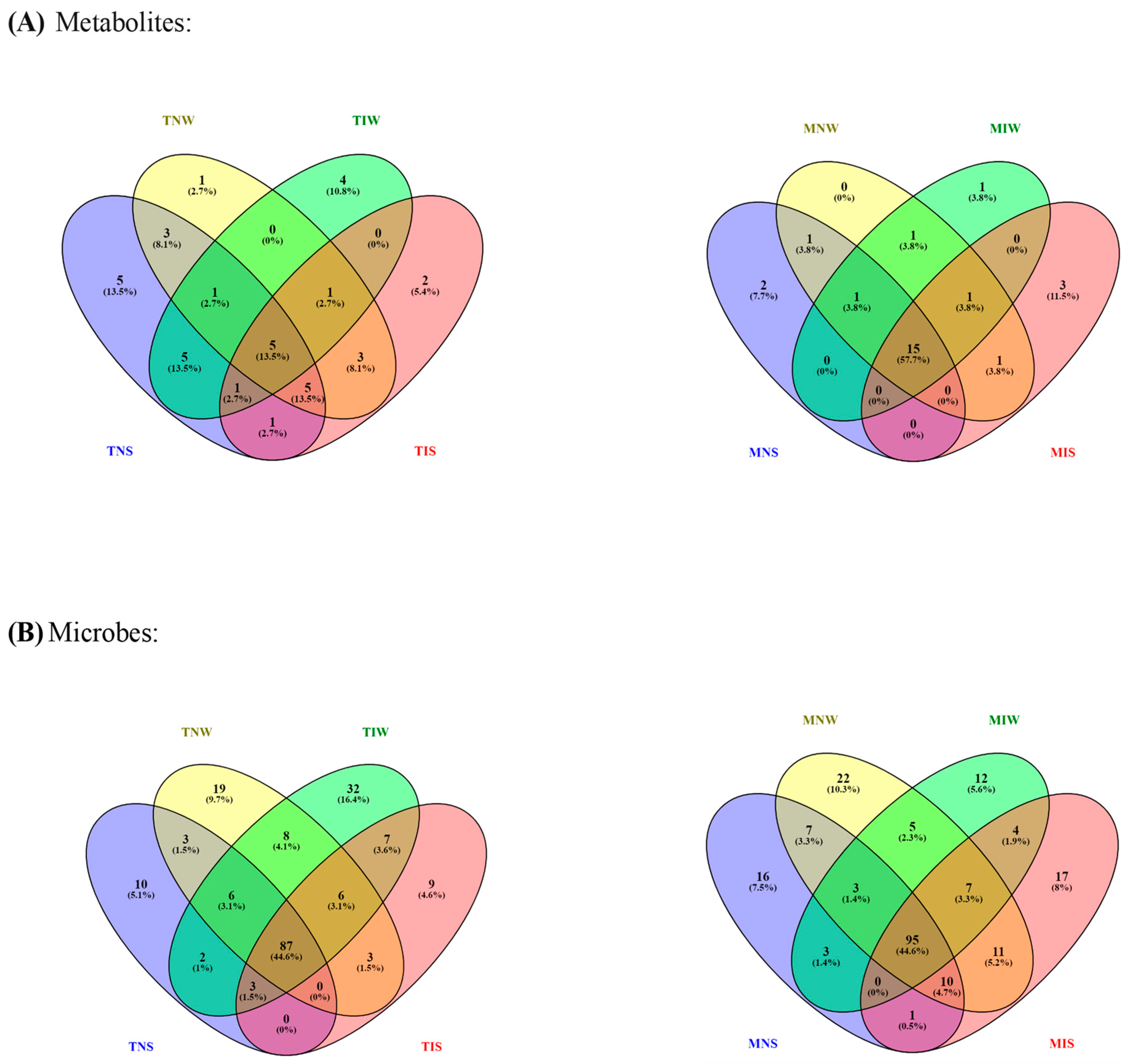
2.1. Assignment of Significant Metabolic Biomarkers Related to Stress and Inoculation
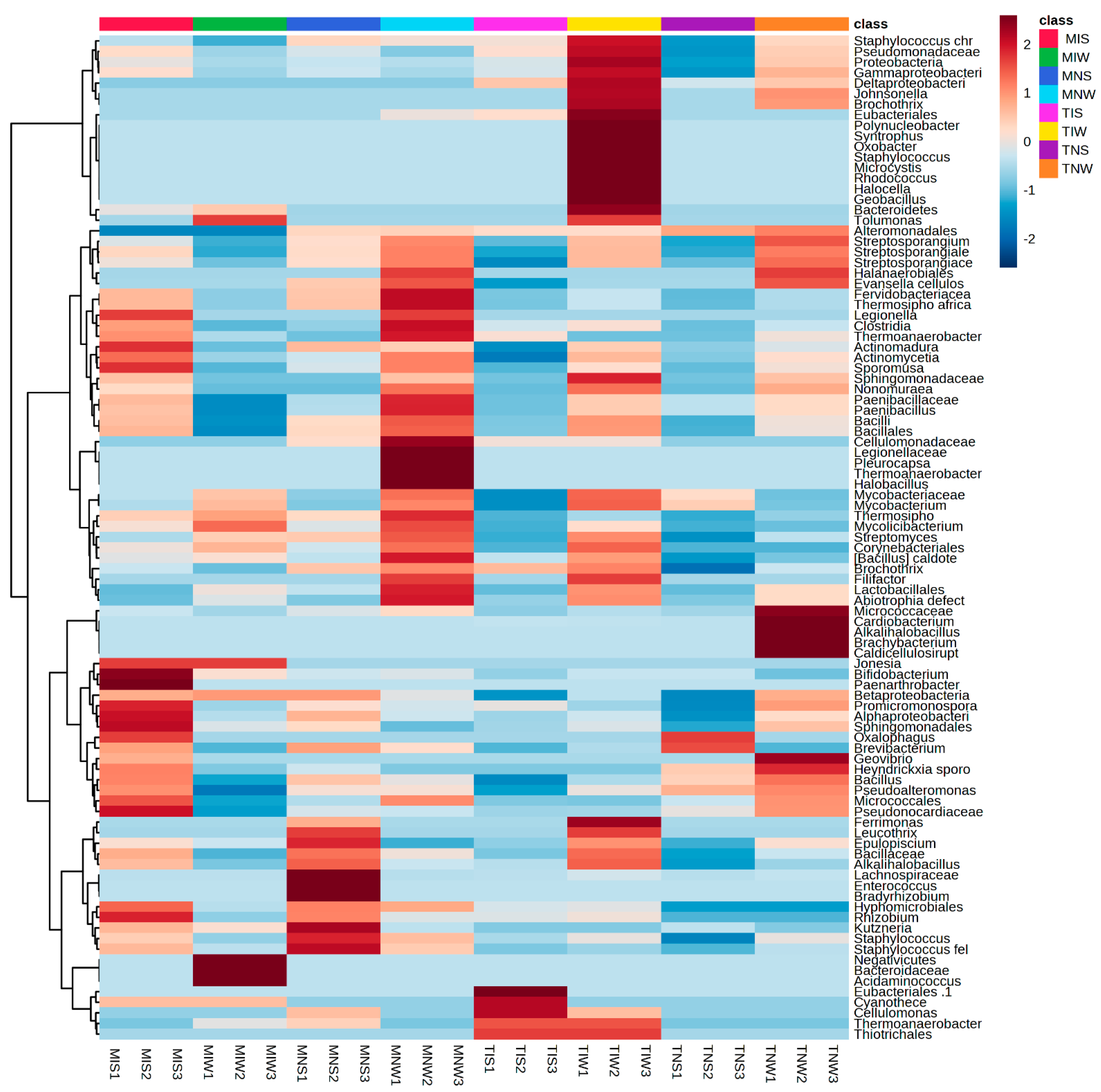
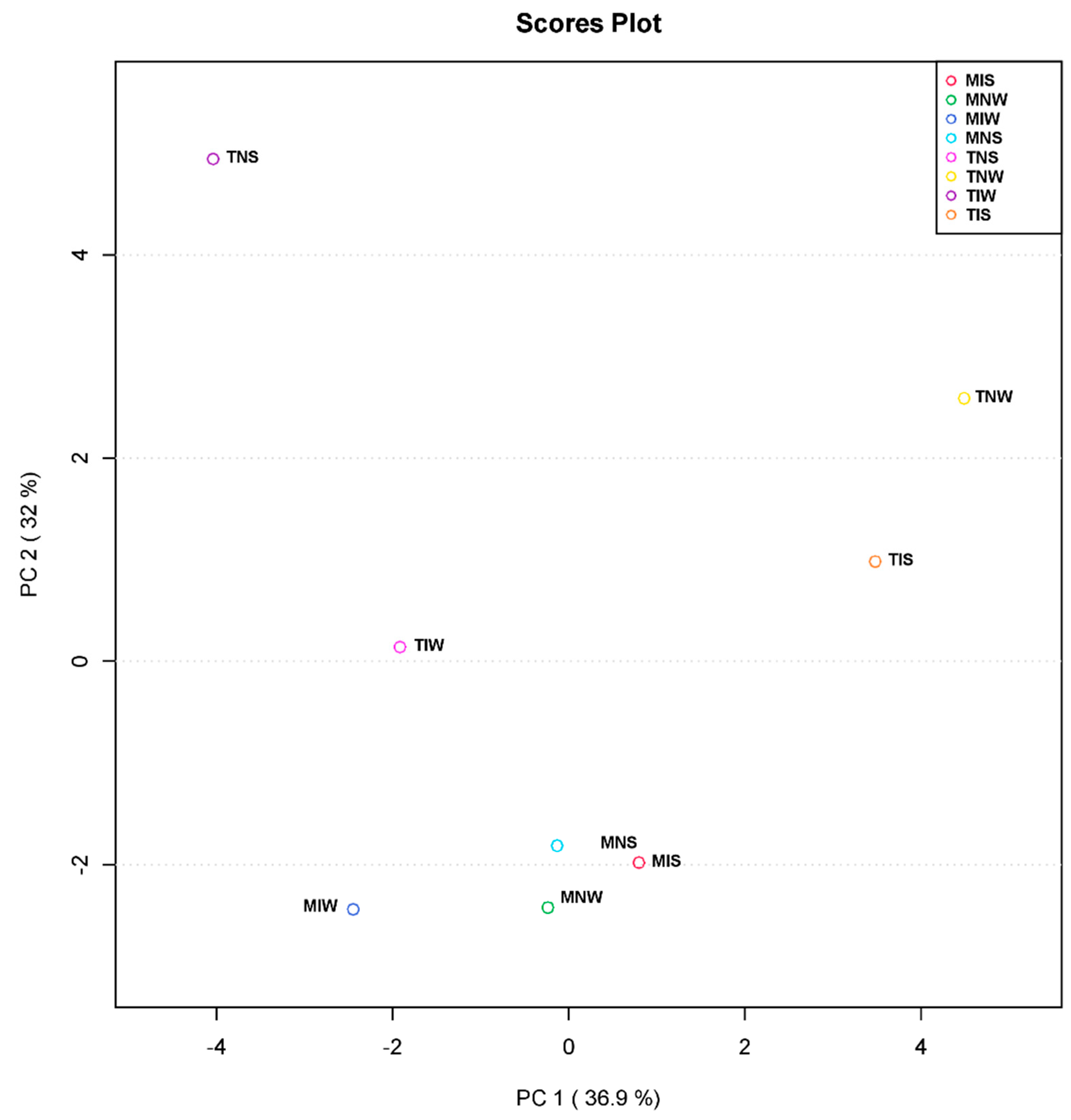
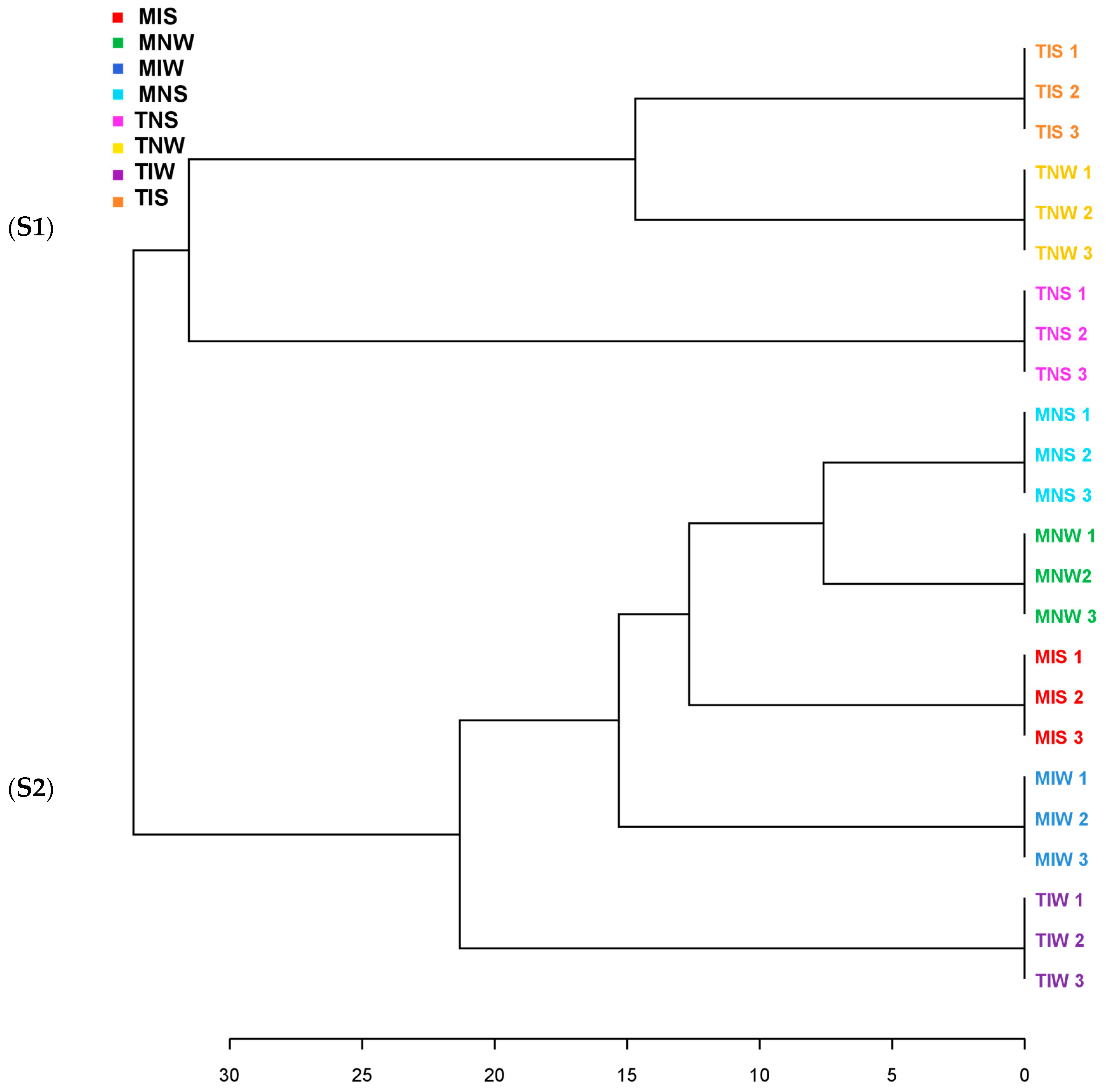
2.2. Matching Olive-Genotype-Dependent Response to Drought and PGPR Application
2.3. Metabolic Profiling of PGPR-Induced Changes in Olive Tissues
2.4. Soil Characterization
3. Materials and Methods
3.1. Soil Sampling
3.2. Physicochemical Analysis
3.3. Experimental Design
3.4. Untargeted Metabolomics
3.5. GC-TOF/MS Data Processing
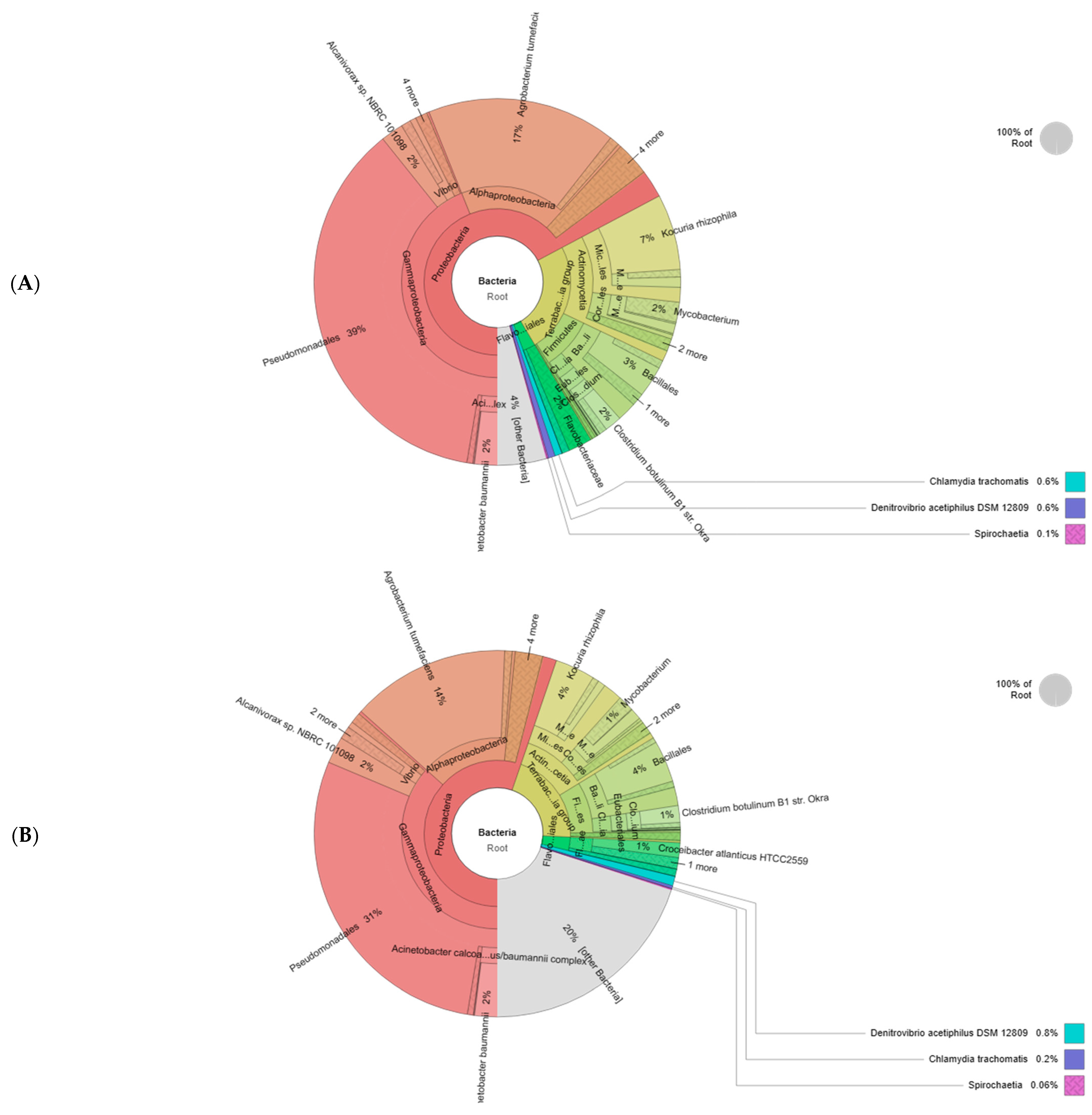
3.6. Microbial Sequencing and Bioinformatic Analysis
3.7. Statistics
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Morgado, R.; Ribeiro, P.F.; Santos, J.L.; Rego, F.; Beja, P.; Moreira, F. Drivers of irrigated olive grove expansion in Mediterranean landscapes and associated biodiversity impacts. Landsc. Urban Plan. 2022, 225, 104429. [Google Scholar] [CrossRef]
- Brito, C.; Dinis, L.T.; Moutinho-Pereira, J.; Correia, C.M. Drought stress effects and olive tree acclimation under a changing climate. Plants 2019, 8, 232. [Google Scholar] [CrossRef] [PubMed]
- Sofo, A.; Manfreda, S.; Fiorentino, M.; Dichio, B.; Xiloyannis, C. The olive tree: A paradigm for drought tolerance in Mediterranean climates. Hydrol. Earth Syst. Sci. 2008, 12, 293–301. [Google Scholar] [CrossRef]
- Ozdemir, Y. Effects of climate change on olive cultivation and table olive and olive oil quality. Sci. Pap. Ser. B Hortic. 2016, 60, 65–69. [Google Scholar]
- Tsiourtis, N.X.; Engineer, S.W. Drought Management Plans for the Mediterranean Region; Report of the Water Engineer Water Development Department; European Commission: Nicosia, Cyprus, 2001. [Google Scholar]
- Parry, M.A.J.; Flexas, J.; Medrano, H. Prospects for crop production under drought: Research priorities and future directions. Ann. Appl. Biol. 2005, 147, 211–226. [Google Scholar] [CrossRef]
- Rolli, E.; Marasco, R.; Vigani, G.; Ettoumi, B.; Mapelli, F.; Deangelis, M.L.; Gandolfi, C.; Casati, E.; Previtali, F.; Gerbino, R.; et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ. Microbiol. 2015, 17, 316–331. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, A.; Schmid, M.; Tuinen, D.V.; Berg, G. Plant-driven selection of microbes. Plant Soil Springer 2009, 321, 235–257. [Google Scholar] [CrossRef]
- Thoms, D.; Liang, Y.; Haney, C.H. Maintaining symbiotic homeostasis: How do plants engage with beneficial microorganisms while at the same time restricting pathogens? Mol. Plant-Microbe Interact. 2021, 34, 462–469. [Google Scholar] [CrossRef]
- Andreote, F.D.; e Silva, M.D.C.P. Microbial communities associated with plants: Learning from nature to apply it in agriculture. Curr. Opin. Microbiol. 2017, 37, 29–34. [Google Scholar] [CrossRef]
- Koskella, B.; Bergelson, J. The study of host–microbiome (co)evolution across levels of selection. Philos. Trans. R. Soc. B 2020, 375, 20190604. [Google Scholar] [CrossRef]
- Gao, C.; Ren, X.; Mason, A.S.; Liu, H.; Xiao, M.; Li, J.; Fu, D. Horizontal gene transfer in plants. Funct. Integr. Genom. 2014, 14, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Sudheer, S.; Usmani, Z.; Rani, R.; Gupta, P. Deciphering the omics of plant-microbe interaction: Perspectives and new insights. Curr. Genom. 2020, 21, 343–362. [Google Scholar] [CrossRef] [PubMed]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 39. [Google Scholar] [CrossRef] [PubMed]
- Adeniji, A.A.; Babalola, O.O. Metabolomic applications for understanding complex tripartite plant-microbes interactions: Strategies and perspectives. Biotechnol. Rep. 2020, 25, e00425. [Google Scholar] [CrossRef]
- Min, X.; Lin, X.; Ndayambaza, B.; Wang, Y.; Liu, W. Coordinated mechanisms of leaves and roots in response to drought stress underlying full-length transcriptome profiling in Vicia sativa L. BMC Plant Biol. 2020, 20, 165. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Bano, A.; Rahman, M.A.; Guo, J.; Kang, Z.; Babar, M.A. Comparative physiological and metabolic analysis reveals a complex mechanism involved in drought tolerance in chickpea (Cicer arietinum L.) induced by PGPR and PGRs. Sci. Rep. 2019, 9, 2097. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.P.; Goodman, R.M. Host variation for interactions with beneficial plant-associated microbes. Annu. Rev. Phytopathol. 1999, 37, 473–491. [Google Scholar] [CrossRef]
- Gontia-Mishra, I.; Sapre, S.; Deshmukh, R.; Sikdar, S.; Tiwari, S. Microbe-mediated drought tolerance in plants: Current developments and future challenges. In Plant Microbiomes for Sustainable Agriculture; Springer: Cham, Switzerland, 2020; pp. 351–379. [Google Scholar]
- Baker, B.; Zambryski, P.; Staskawicz, B.; Dinesh-Kumar, S.P. Signaling in plant-microbe interactions. Science 1997, 276, 726–733. [Google Scholar] [CrossRef]
- Verma, P.; Yadav, A.N.; Kumar, V.; Singh, D.P.; Saxena, A.K. Beneficial plant-microbe interactions: Biodiversity of microbes from diverse extreme environments and its impact for crop improvement. In Plant-Microbe Interactions in Agro-Ecological Perspectives: Volume 2: Microbial Interactions and Agro-Ecological Impacts; Springer: Singapore, 2017; pp. 543–580. [Google Scholar]
- Ramirez, K.S.; Snoek, L.B.; Koorem, K.; Geisen, S.; Bloem, L.J.; Ten Hooven, F.; Kostenko, O.; Krigas, N.; Manrubia, M.; Caković, D.; et al. Range-expansion effects on the belowground plant microbiome. Nat. Ecol. Evol. 2019, 3, 604–611. [Google Scholar] [CrossRef]
- Biggs, E.; Taylor, M.W.; Middleton, D.M. Beyond the theory: From holobiont concept to microbiome engineering. Environ. Microbiol. 2023, 25, 832–835. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, C.; Pieltain, F. Analyse Chimique des Sols Méthodes Choisies; Editions Tec et Doc/Lavoisier: Paris, France, 2003; 408p. [Google Scholar]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Bansal, M. Variation of gene expression in plants is influenced by gene architecture and structural properties of promoters. PLoS ONE 2019, 14, e0212678. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Zhao, Q.; Chen, L.; Yao, X.; Zhang, W.; Zhang, B.; Xie, F. Effect of drought stress on sugar metabolism in leaves and roots of soybean seedlings. Plant Physiol. Biochem. 2020, 146, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bertolli, S.C.; Vítolo, H.F.; Souza, G.M. Network connectome analysis as a tool to understand homeostasis of plants under environmental changes. Plants 2013, 2, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Marček, T.; Hamow, K.A.; Végh, B.; Janda, T.; Darko, E. Metabolic response to drought in six winter wheat genotypes. PLoS ONE 2019, 14, e0212411. [Google Scholar] [CrossRef] [PubMed]
- Adams, W.W.; Stewart, J.J.; Demmig-Adams, B. Photosynthetic modulation in response to plant activity and environment. In The Leaf: A Platform for Performing Photosynthesis; Springer: Cham, Switzerland, 2018; pp. 493–563. [Google Scholar]
- Kang, Z.; Babar, M.A.; Khan, N.; Guo, J.; Khan, J.; Islam, S.; Shrestha, S.; Shahi, D. Comparative metabolomic profiling in the roots and leaves in contrasting genotypes reveals complex mechanisms involved in post-anthesis drought tolerance in wheat. PLoS ONE 2019, 14, e0213502. [Google Scholar] [CrossRef] [PubMed]
- Akšić, M.F.; Tosti, T.; Sredojević, M.; Milivojević, J.; Meland, M.; Natić, M. Comparison of sugar profile between leaves and fruits of blueberry and strawberry cultivars grown in organic and integrated production system. Plants 2019, 8, 205. [Google Scholar] [CrossRef]
- Ozturk, M.; Turkyilmaz Unal, B.; García-Caparrós, P.; Khursheed, A.; Gul, A.; Hasanuzzaman, M. Osmoregulation and its actions during the drought stress in plants. Physiol. Plant. 2021, 172, 1321–1335. [Google Scholar] [CrossRef]
- Lloyd, D.H.; Viac, J.; Werling, D.; Rème, C.A.; Gatto, H. Role of sugars in surface microbe–host interactions and immune reaction modulation. Vet. Dermatol. 2007, 18, 197–204. [Google Scholar] [CrossRef]
- Jha, Y.; Subramanian, R.B. Effect of root-associated bacteria on soluble sugar metabolism in plant under environmental stress. In Plant Metabolites and Regulation under Environmental Stress; Academic Press: Cambridge, MA, USA, 2018; pp. 231–240. [Google Scholar]
- Hadley, N.F. Lipid water barriers in biological systems. Prog. Lipid Res. 1989, 28, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Janda, M.; Planchais, S.; Djafi, N.; Martinec, J.; Burketova, L.; Valentova, O.; Zachowski, A.; Ruelland, E. Phosphoglycerolipids are master players in plant hormone signal transduction. Plant Cell Rep. 2013, 32, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Martinoia, E.; Massonneau, A.; Frangne, N. Transport processes of solutes across the vacuolar membrane of higher plants. Plant Cell Physiol. 2000, 41, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Macabuhay, A.; Arsova, B.; Walker, R.; Johnson, A.; Watt, M.; Roessner, U. Modulators or facilitators? Roles of lipids in plant root–microbe interactions. Trends Plant Sci. 2022, 27, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Song, J.B.; Huang, R.K.; Guo, M.J.; Zhou, Q.; Guo, R.; Zhang, S.Y.; Yao, J.W.; Bai, Y.N.; Huang, X. Lipids associated with plant-bacteria interaction identified using a metabolomics approach in an Arabidopsis thaliana model. PeerJ 2022, 10, e13293. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Crawford, P.A. Coordinated regulation of the metabolome and lipidome at the host-microbial interface. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2010, 1801, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Vaio, C. Biostimulants improve plant growth and bioactive compounds of young olive trees under abiotic stress conditions. Agriculture 2022, 12, 227. [Google Scholar]
- Kumar, S.; Bhushan, B.; Wakchaure, G.C.; Meena, K.K.; Kumar, M.; Meena, N.L.; Rane, J. Plant phenolics under water-deficit conditions: Biosynthesis, accumulation, and physiological roles in water stress alleviation. Plant Phenolics Sustain. Agric. 2020, 1, 451–465. [Google Scholar]
- Varela, M.C.; Arslan, I.; Reginato, M.A.; Cenzano, A.M.; Luna, M.V. Phenolic compounds as indicators of drought resistance in shrubs from Patagonian shrublands (Argentina). Plant Physiol. Biochem. 2016, 104, 81–91. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, N. Characterization of olive cultivars for drought tolerance potential under rainfed conditions of Himachal Pradesh. Indian J. Agric. Res. 2016, 50, 440–445. [Google Scholar]
- Da Sois, L.; Mencuccini, M.; Castells, E.; Sanchez-Martinez, P.; Martínez-Vilalta, J. How are physiological responses to drought modulated by water relations and leaf economics’ traits in woody plants? Agric. Water Manag. 2024, 291, 108613. [Google Scholar] [CrossRef]
- Dias, M.C.; Pinto, D.C.; Figueiredo, C.; Santos, C.; Silva, A.M. Phenolic and lipophilic metabolite adjustments in Olea europaea (olive) trees during drought stress and recovery. Phytochemistry 2021, 185, 112695. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, L.P.; Chen, X.; Clarke, J.; Salmeron, J.; Nguyen, H.T. RNAi-mediated disruption of squalene synthase improves drought tolerance and yield in rice. J. Exp. Bot. 2012, 63, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Zafar, N.; Akram, N.A.; Fatima, K.; Noreen, S.; Akram, M.S.; Umer, S.; Ashraf, M.; Alsahli, A.A.; Mansoor, S. Drought-induced changes in plant-yield interlinked biochemistry of cauliflower (Brassica oleracea L. var. botrytis) by exogenously applied alpha-tocopherol. J. King Saud Univ.-Sci. 2024, 36, 103028. [Google Scholar] [CrossRef]
- McLaughlin, S.; Zhalnina, K.; Kosina, S.; Northen, T.R.; Sasse, J. The core metabolome and root exudation dynamics of three phylogenetically distinct plant species. Nat. Commun. 2023, 14, 1649. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zeng, X.; Xu, Q.; Mei, X.; Yuan, H.; Jiabu, D.; Sang, Z.; Nyima, T. Metabolite profiling in two contrasting Tibetan hulless barley cultivars revealed the core salt-responsive metabolome and key salt-tolerance biomarkers. AoB Plants 2019, 11, plz021. [Google Scholar] [CrossRef] [PubMed]
- Chialva, M.; De Rose, S.; Novero, M.; Lanfranco, L.; Bonfante, P. Plant genotype and seasonality drive fine changes in olive root microbiota. Curr. Plant Biol. 2021, 28, 100219. [Google Scholar] [CrossRef]
- Brencic, A.; Winans, S.C. Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol. Mol. Biol. Rev. 2005, 69, 155–194. [Google Scholar] [CrossRef]
- Mathur, P.; Roy, S. Insights into the plant responses to drought and decoding the potential of root associated microbiome for inducing drought tolerance. Physiol. Plant. 2021, 172, 1016–1029. [Google Scholar] [CrossRef]
- Finkel, O.M.; Castrillo, G.; Paredes, S.H.; González, I.S.; Dangl, J.L. Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 2017, 38, 155–163. [Google Scholar] [CrossRef]
- Shelby, N.; Duncan, R.P.; Van Der Putten, W.H.; McGinn, K.J.; Weser, C.; Hulme, P.E. Plant mutualisms with rhizosphere microbiota in introduced versus native ranges. J. Ecol. 2016, 104, 1259–1270. [Google Scholar] [CrossRef]
- Deng, S.; Caddell, D.F.; Xu, G.; Dahlen, L.; Washington, L.; Yang, J.; Coleman-Derr, D. Genome wide association study reveals plant loci controlling heritability of the rhizosphere microbiome. ISME J. 2021, 15, 3181–3194. [Google Scholar] [CrossRef] [PubMed]
- Tkacz, A.; Cheema, J.; Chandra, G.; Grant, A.; Poole, P.S. Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition. ISME J. 2015, 9, 2349–2359. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Garcia, B.J.; Furches, A.; Tuskan, G.A.; Jacobson, D. Plant host-associated mechanisms for microbial selection. Front. Plant Sci. 2019, 10, 862. [Google Scholar] [CrossRef] [PubMed]
- Ricks, K.D.; Yannarell, A.C. Soil moisture incidentally selects for microbes that facilitate locally adaptive plant response. Proc. R. Soc. B 2023, 290, 20230469. [Google Scholar] [CrossRef] [PubMed]
- Vorholt, J.A.; Vogel, C.; Carlström, C.I.; Müller, D.B. Establishing causality: Opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 2017, 22, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.R.; Chaitanya, K.V.; Vivekanandan, M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J. Plant Physiol. 2004, 161, 1189–1202. [Google Scholar] [CrossRef]
- Younginger, B.S.; Friesen, M.L. Connecting signals and benefits through partner choice in plant–microbe interactions. FEMS Microbiol. Lett. 2019, 366, fnz217. [Google Scholar] [CrossRef]
- Brown, S.P.; Grillo, M.A.; Podowski, J.C.; Heath, K.D. Soil origin and plant genotype structure distinct microbiome compartments in the model legume Medicago truncatula. Microbiome 2020, 8, 139. [Google Scholar] [CrossRef]
- Lau, J.A.; Lennon, J.T. Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc. Natl. Acad. Sci. USA 2012, 109, 14058–14062. [Google Scholar] [CrossRef]
- Chen, J.Z.; Huang, X.L.; Sun, Q.W.; Liu, J.M. Bulk soil microbial reservoir or plant recruitment dominates rhizosphere microbial community assembly: Evidence from the rare, endangered Lauraceae species Cinmaomum migao. Ecol. Indic. 2023, 148, 110071. [Google Scholar] [CrossRef]
- Chinta, Y.D.; Araki, H. Responses of bulk and rhizosphere soil microbiomes to different cover crop inputs and their connection and contribution to soil fertility and plant growth. Pedobiologia 2023, 101, 150907. [Google Scholar] [CrossRef]
- Malard, L.A.; Pearce, D.A. Bacterial colonisation: From airborne dispersal to integration within the soil community. Front. Microbiol. 2022, 13, 782789. [Google Scholar] [CrossRef]
| Sample | C (g/kg) | TN (mg/kg) | Available P (mg/kg) | EC (mS) | pH | H (%) |
|---|---|---|---|---|---|---|
| Rhizosphere | 18.98 ± 1.65 | 85.21 ± 13.42 | 63.34 ± 5.12 | 150.16 ± 2.46 | 7.40 ± 0.5 | 87 ± 2.75 |
| Bulk soil | 5.05 ± 3.79 | 57.10 ± 4.06 | 54.52 ± 12.32 | 90.07 ± 6.25 | 8.16 ± 0.2 | 74 ± 3.32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azri, R.; Lamine, M.; Bensalem-Fnayou, A.; Hamdi, Z.; Mliki, A.; Ruiz-Lozano, J.M.; Aroca, R. Genotype-Dependent Response of Root Microbiota and Leaf Metabolism in Olive Seedlings Subjected to Drought Stress. Plants 2024, 13, 857. https://doi.org/10.3390/plants13060857
Azri R, Lamine M, Bensalem-Fnayou A, Hamdi Z, Mliki A, Ruiz-Lozano JM, Aroca R. Genotype-Dependent Response of Root Microbiota and Leaf Metabolism in Olive Seedlings Subjected to Drought Stress. Plants. 2024; 13(6):857. https://doi.org/10.3390/plants13060857
Chicago/Turabian StyleAzri, Rahma, Myriam Lamine, Asma Bensalem-Fnayou, Zohra Hamdi, Ahmed Mliki, Juan Manuel Ruiz-Lozano, and Ricardo Aroca. 2024. "Genotype-Dependent Response of Root Microbiota and Leaf Metabolism in Olive Seedlings Subjected to Drought Stress" Plants 13, no. 6: 857. https://doi.org/10.3390/plants13060857
APA StyleAzri, R., Lamine, M., Bensalem-Fnayou, A., Hamdi, Z., Mliki, A., Ruiz-Lozano, J. M., & Aroca, R. (2024). Genotype-Dependent Response of Root Microbiota and Leaf Metabolism in Olive Seedlings Subjected to Drought Stress. Plants, 13(6), 857. https://doi.org/10.3390/plants13060857








