1. Introduction
Stigeoclonium is an important primary producer that is widely distributed in freshwater ecosystems around the world. They often attach to inorganic substrates or other large algae and higher aquatic plant surfaces. Some species of
Stigeoclonium have high application prospects in terms of water quality indication, wastewater treatment [
1,
2,
3], and lipid production [
4].
The genus
Stigeoclonium, comprising approximately 80 described species, was initially established by Kützing [
5] and belongs to the order Chaetophorales (Chlorophyceae, Chlorophyta) [
6].
Stigeoclonium was originally defined based on the type species
Stigeoclonium tenue, encompassing algae forming vibrant green tufts with uniseriate branched heterotrichous filaments [
5]. Further delimitation of the genus occurred through the examination of various vegetative traits, such as cellular dimensions, branching degree, the presence/absence of hairs, thallus color, and habitat, resulting in the description of 29 species [
7]. Subsequent taxonomic revisions by different phycologists led to varied concepts and proposed species. Islam [
8] critically reviewed the genus based on 73 specimens worldwide, recognizing 28 species, with only 10 being considered valid. Printz [
9] reported 42 species, but 11 remained not fully understood. These phycologists primarily delimited
Stigeoclonium species based on branching types and the shape and size of cells in the erect system.
Stigeoclonium is a heterotrichous alga, which is composed of an erect system and a prostrate system. Cox and Bold [
10] introduced a species concept, focusing on the morphological features of the prostrate system and considering it more stable than the erect system. This approach identified seven species from 81
Stigeoclonium-like specimens collected in Texas, which belonged to three groups. Francke and Simons [
11] refined these methods, confirming four groups (
S. helveticum group,
S. aestivale group,
S. tenue group, and
S. farctum group) from 150 strains collected in the Netherlands. Simons et al. [
12] further merged groups based on detailed studies on the morphological development of the prostrate system of
Stigeoclonium, with particular emphasis on the types of zoospore germination, resulting in three groups:
S. helveticum,
S. tenue, and
S. farctum, each with a single species.
Despite these efforts, universally recognized taxonomic criteria for
Stigeoclonium remain elusive. Various authors have proposed species and variants based on different concepts. John et al. [
13] focused on the morphology of the erect system, describing nine species. Branco et al. [
14] identified six species in southeastern Brazil by considering the characteristics of both prostrate and erect systems. Skinner and Entwisle [
15] described three species in Australia following the species concept of Simons et al. [
12].
The genus
Stigeoclonium has undergone comprehensive study for nearly two centuries, resulting in numerous species descriptions based on morphology. However, the traditional taxonomic framework established by Kützing [
5] has persisted without significant changes. The genus
Stigeoclonium was considered a well-defined monophyletic genus for a long time until Caisová et al. [
16] first used SSU rDNA to study the phylogeny of
Stigeoclonium, which contained nine molecular sequences of six species of
Stigeoclonium, confirming that
Stigeoclonium was polyphyletic, challenging the previous notion. Notably, species sharing the same name, including the type species
S. tenue, did not form cohesive clusters, adding complexity to the phylogenetic understanding of
Stigeoclonium [
17,
18].
While molecular phylogenetic studies have primarily concentrated on the order Chaetophorales as a whole, investigations specific to Stigeoclonium have been limited. To address these challenges, the present study generated new sequences for 18S rDNA, ITS2, and tufA. A concatenated dataset, combined with morphological data, was employed to (1) re-evaluate morphological characteristics and (2) reassess the broadly defined genus Stigeoclonium, leading to the proposal of a new genus.
2. Results
2.1. Taxonomic Implications
Pseudostigeoclonium B. Wen Liu, Q. Mei Lan, Q. Yu Dai, Huan Zhu et G. Xiang Liu gen. nov.
Diagnosis: Turf-like thallus; cushion-forming; 1–5 cm. Filaments consist of erect and prostrate systems. The erect filaments are unbranched in the actively growing stage or sparsely branched in the older stage. The branches usually alternate, are rarely approximate or opposite, and branches are long, bluntly attenuating, or ending in a colorless hair. Cells of the main filament and branches are cylindrical and either slightly constricted or not. Chloroplasts are single and parietal; pyrenoid 1–3; typically two per cell. The prostrate systems are underdeveloped, attached to the substrate by a sparse, rhizoid-like basal system developed from the lower or middle cells. Sexual reproduction produces biflagellate isogametes, and asexual reproduction produces four-flagellated zoospores. Zoospore germination is erect. The first cross-wall of the attached germling is parallel to the substrate.
Etymology: The genus is named for its morphological similarity to the genus Stigeoclonium.
Type species (designated herein): Pseudostigeoclonium helveticum (Vischer) B. Wen Liu, Q. Yu Dai, Huan Zhu et G. Xiang Liu comb. nov.
Pseudostigeoclonium helveticum (Vischer) B. Wen Liu, Q. Mei Lan, Q. Yu Dai, Huan Zhu et G. Xiang Liu comb. nov.
Synonym: Stigeoclonium helveticum Vischer.
Formaldehyde-fixed material of Pseudostigeoclonium helveticum in this study was deposited in the Freshwater Algal Herbarium (HBI), Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, Hubei Province, China, as specimen no. WDLC201608.
Locality: Heilongjiang province, China; on a rock in a stream; freshwater.
2.2. Morphological Observations
Species of
Stigeoclonium were highly variable in morphology, especially in the erect system. For instance,
Stigeoclonium farctum specimens displayed different morphologies when growing epiphytically on a plastic cup in their natural habitat compared with those cultured in medium. Filaments cultured on solid medium were abundantly branched, whereas those in liquid medium had extremely long, upright filaments with rare branching (
Figure 1).
In this study, three types of zoospore germination were clearly observed (
Figure 2). The genus
Stigeoclonium was grouped into three clades based on zoospore germination types:
The
Stigeoclonim helveticum clade (
Pseudostigeoclonium gen. nov.): The
Stigeoclonim helveticum clade exclusively included the species
Stigeoclonium, primarily including
Stigeoclonium helveticum and
Stigeoclonium sp. This clade was characterized by the strictly erect germination of zoospores (
Figure 2A) and a prostrate system consisting of one
Ulothrix-like holdfast cell or some very short prostrate filaments and/or slender rhizoids growing out from the basal cell of an erect axis [
19,
20].
The
Stigeoclonim tenue clade: The
Stigeoclonim tenue clade included some species of
Stigeoclonium and the genus
Caespitella. This clade was characterized by an approximately prostrate germination of zoospores. The original zoospore cell unipolar germination usually gave rise to a lateral outgrowth at one side to first form the prostrate part, and later, a pointed erect filament began to develop (
Figure 2D). We placed all species with an approximately prostrate germination of zoospores and an irregular and mostly extensive open prostrate system, with or without rhizoids, into this clade.
The
Stigeoclonim farctum clade: The
Stigeoclonim farctum clade comprised some species of
Stigeoclonium and the genus
Fritschiella. This clade was characterized by an approximately prostrate or pseudo-erect germination of zoospores (
Figure 2B,C,E). What was different from the
Stigeoclonim tenue clade was that the original zoospore cell bipolar germination gave rise to a lateral outgrowth on two sides to form the prostrate part, with the exception of the
Fritschiella tuberosa (approximately gregarious on moist soil or silt, including drying rainwater puddles), which showed the original unipolar germination of zoospore cells, and
S. aestivale, which showed the original unipolar germination of zoospore cells, to first form the erect part. We placed all species with an approximately prostrate and pseudo-erect germination of zoospores, a regular compact development of the prostrate system that results in a closed, pseudoparenchymatous disk, an open star-like form, or intermediate forms and a poorly developed erect system, into this clade.
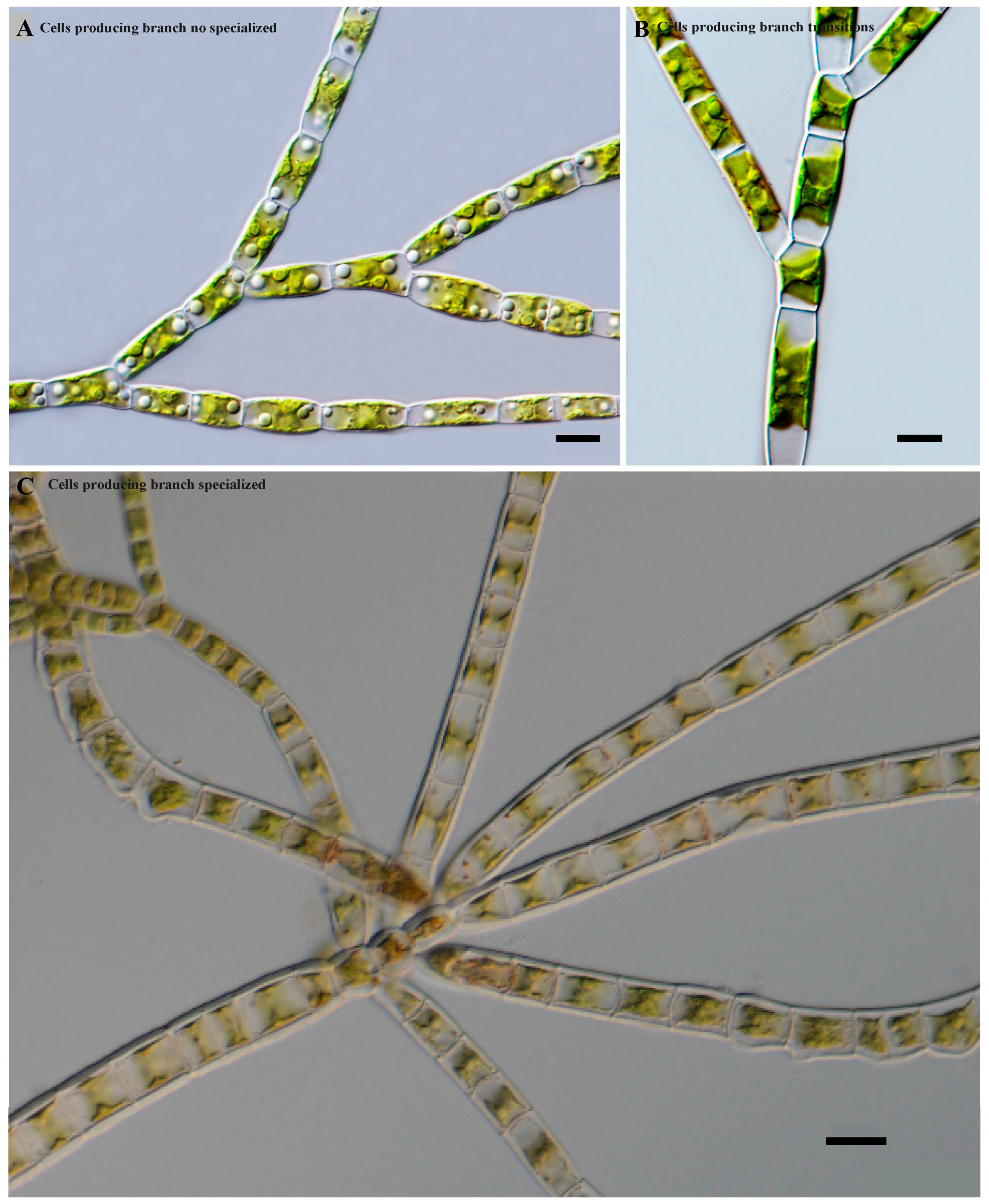
In addition, three cell types of the main axis-producing branch were observed (
Figure 3). Species with a main axis and primary branches were similar, and usually, no specialized or modified cells were present on the main axis producing the branches (
Figure 3A). Species showed transitions in which some differentiation began between the cells of the main axis. Some cells gradually became long and some short, with the latter commonly producing branches (
Figure 3B). Species with a main axis usually consisted of two types of cells. The long cells usually did not produce branches, while the small and short cells usually produced lateral primary branches (
Figure 3C).
2.3. Phylogenetic Analysis of the Stigeoclonium
A total of 135 new sequences, including 18S rDNA, ITS2, and tufA, were generated in this study. The best models were selected for the BI analysis: TrNef + I + G for 18S rDNA, GTR + G for ITS2 rDNA, GTR + G for the first codon position matrix of tufA, and GTR + I + G for the second and third codon positions matrix. The GTRGAMMA model was used for maximum likelihood analysis.
A saturation index analysis (
Iss = 0.386 <
Iss.c = 0.802) showed that the sequence matrix was not saturated and could be used for phylogenetic analysis. The 71-taxa alignment consisted of 2687 positions (18S rDNA + ITS2 +
tufA) (see
Appendix A). A total of 743 sites in these nucleotides were variable, of which 573 sites were parsimoniously informative and 170 sites were singleton sites. The average amount of A, T, C, and G was 24.69%, 26.04%, 20.89%, and 28.38%, respectively, of which A + T (50.73%) was greater than G + C (49.27%). The transition/transversion ratio was 1.61.
The phylogenetic trees created using the Bayesian and ML methods showed similar topologies to previous studies [
21,
22]. The monophyly of the Chaetophorales was strongly supported, including six widely accepted families: Schizomeridaceae, Aphanochaetaceae, Barrancaceae, Uronemataceaea, Fritschiellaceae, and Chaetophoraceae [
19] (
Figure 4).
Stigeoclonium species were dispersed over the family Fritschiellaceae or family Chaetophoraceae and intermixed with the other genera. The family Chaetophoraceae and family Fritschiellaceae diverged into three well-supported sister lineages. The family Chaetophoraceae included the genera
Chaetophora,
Draparnaldia, and the
Stigeoclonium helveticum clade. The
S. helveticum clade showed basal divergence as a sister of the
Chaetophora and
Draparnaldia, while the family Fritschiellaceae comprised the genus
Chaetophoropsis,
S. tenue clade, and
S. farctum clade. The
S. tenue clade showed basal divergence as a sister of the
Chaetophoropsis and
S. farctum clade. The
S. tenue clade and
S. farctum clade, with a similar morphology, were well separated from each other. The three clades of
Stigeoclonium were further split into seven molecular groups. All
Stigeoclonium species were dispersed over these two families and were recovered as independent monophyletic clades (
Stigeoclonium helveticum clade,
S. tenue clade, and
S. farctum clade) with moderate to robust support values (BP/PP, 80/0.85 to 100/1.00). Species of
Stigeoclonium with zoospore germination of an erect type (
Stigeoclonium helveticum clade) formed a monophyletic clade in the Chaetophoraceae, with a robust support value (BP/PP, 100/1.00), which obviously branched independently of the
S. tenue clade and
S. farctum clade (within the Fritschiellaceae) with zoospore germination prostrate or the pseudo-erect type in our phylogenetic trees. In addition, multiple sequences with the same species name were phylogenetically distantly related, including the type species of the genus
Stigeoclonium (
S. tenue in bold;
Figure 1) and
Caespitella pascheri (type species of
Caespitella). Two sequences with the same name,
S. tenue, were dispersed over the Fritschiellaceae and the Chaetophoraceae instead of clustering together.
3. Discussion
Based on concatenated datasets and additional samples, we still had to accept the fact that the Stigeoclonium was paraphyletic. Clearly, the morphologically defined genus Stigeoclonium was unnatural and needed re-evaluation. This study obtained a well-resolved molecular phylogeny with moderate to high support values for Stigeoclonium. All species of Stigeoclonium were divided into three clades with robust support, located in either the family Fritschiellaceae or the family Chaetophoraceae. This provided a basis for further discussion on the phylogenetic relationships within the paraphyletic genus Stigeoclonium.
In cases where paraphyly or polyphyly is demonstrated within a genus, a common approach is to designate one clade as “genus sensu stricto” and analyze the morphological characteristics used in the original diagnosis [
16]. Before that, it is necessary for us to re-examine its type species. However, re-examination of the type species revealed two stains of
S. tenue (
S. tenue CCAC 3492B HF920647 and CCAP 477/11A FN824374), which are clearly unrelated and located in Fritschiellaceae and Chaetophoraceae, respectively. Based on a comprehensive sample observation and literature analysis, the zoospore germination type of the type species (
S. tenue) was consistently identified as prostrate [
10,
11,
12]. On the other hand, recent research has provided evidence indicating that taxa within the family Fritschiellaceae exhibit zoospore germination of the prostrate or pseudo-erect type and, in contrast, taxa belonging to Chaetophoraceae display zoospore germination of the erect type [
19]. Consequently, the inference that was drawn was that
S. tenue CCAC 3492B (HF920647), situated in the Fritschiellaceae, was more likely to represent
Stigeoclonium sensu stricto. This conclusion aligned with the understanding that the germination characteristics of zoospores played a crucial role in delineating the taxonomy of
Stigeoclonium.
After identifying
Stigeoclonium sensu stricto, we next tried to find some effective morphological characteristics to revise the genus
Stigeoclonium. The characteristic cell type of the main axis-producing branch was considered the most important morphological characteristic in the traditional taxonomy of
Stigeoclonium by Islam [
8], although it did not reflect the real phylogeny of the genus
Stigeoclonium in this study. Instead, the characteristics of zoospore germination types correlated well with the phylogenetic results. Drawing upon the morphological features of the prostrate system and the germination type of zoospores, various phycologists have categorized the genus
Stigeoclonium into distinct groups [
10,
11,
12]. Despite the fact that none of these grouping schemes matched the phylogenetic results well, the important studies mentioned above [
10,
11,
12] provide a valuable indication that the germination type of zoospores might serve as a dependable morphological characteristic [
20]. This observation is consistent with findings from prior studies [
19], reinforcing the significance of zoospore germination type in understanding the taxonomy of
Stigeoclonium.
The broadly defined genus
Stigeoclonium diverged into three well-supported clades in our phylogenetic analyses using a concatenated dataset, which was initially unexpected. By observing the zoospore germination type of representative species and combining the phylogeny results, the genus
Stigeoclonium could be divided into at least six groups—the
S. helveticum group,
S. aestivale group,
S. variabile group,
S. farctum group,
S. tenue group and
S. pascheri group—and three clades—the
S. tenue clade,
S. farctum clade, and
S. helveticum clade. The life history and reproductive development characteristics of
S. helveticum (
S. helveticum group) have been well studied, providing us with many taxonomic insights [
23]. All species with erect zoospore germination types (
S. helveticum group) of the broadly defined
Stigeoclonium formed a separate, monophyletic clade that was clearly separate from the
S. tenue clade (
Stigeoclonium sensu stricto) and the
S. farctum clade with zoospore germination of the prostrate and pseudo-erect types. Thus, in light of the morphological and molecular differences, we erected a new genus,
Pseudostigeoclonium gen. nov., and amended the
Stigeoclonium. The genus
Pseudostigeoclonium was newly erected due to its unique morphological characters, as follows: the erect filaments are unbranched in the actively growing stage or sparsely branched in older stages; the prostrate systems are underdeveloped, attached to the substrate by a sparse, rhizoid-like basal system developed from the lower or middle cells; zoospore germination is of the erect type. This clearly distinguishes
Pseudostigeoclonium from
Stigeoclonium sensu stricto.
Usually, samples with typical characteristics of
Stigeoclonium are confidently classified within the genus. However, some species of
Stigeoclonium (such as
S. terrestre and
S. pascheri) spun off to create the new genera [
24,
25]. Vischer [
25] discovered and isolated a new
Stigeoclonium-like taxon with cell division that was entirely apical, distinguishing it from the
Stigeoclonium with intercalary cell division and, rather than describing it as a new species of
Stigeoclonium, created the new genus
Caespitella with the type species
C. pascheri. There is a lack of consensus among phycologists regarding this taxonomic approach. Printz [
9] and Bourrelly [
26] recognized this genus based on the morphological characteristics of the erect system. However, Cox and Bold [
10] reincorporated the genus into
Stigeoclonium based on detailed life history observations, which was accepted by Shyam and Sarma [
27]. Caisová et al. [
16] endorsed the establishment of the genus
Caespitella based on the limited samples and molecular data of
Stigeoclonium. In this study, two stains of
Caespitella pascheri (
S. pascheri HB201611 OP236750 and SAG 410-1 FN824387) (type species of
Caespitella) were not independent of the genus
Stigeoclonium; instead, they were dispersed within the
S. pascheri group. Moreover, the key morphological characteristics (zoospore germination types and prostrate system) were found to be more reliable and were not significantly different between
Caespitella pascheri and other
Stigeoclonium. Thus, proposing
Caespitella pascheri as a separate genus raised questions. Vischer [
25] was, at the time, intensively studying both
S. helveticum and
C. pascheri. These two species exhibited significant differences in growth pattern, both in liquid and solid agar, which might have influenced Vischer’s belief that they belonged to different genera. In fact, apical cell division and intercalary cell division have been observed in the same species [
10]. Apical cell division is common in prostrate filaments, while intercalary cell division is common in erect filaments [
10]. The authentic strains of
C. pascheri and
S. helveticum could also be well distinguished on the phylogenetic tree. Considering the crucial evidence presented by Cox and Bold [
10] and the molecular phylogeny in this study, it is suggested that
C. pascheri should be considered a member of
Stigeoclonium.
It is noteworthy that the sequences with the same species name were not clustered into one clade in the genus Stigeoclonium (e.g., S. tenue, S. pascheri), which may be for the following reasons: (1) the presence of cryptic species within the genus Stigeoclonium, where species share similar morphological characteristics but exhibit significant differences in molecular data; (2) insufficient knowledge among phycologists regarding the genus Stigeoclonium, leading to the use of inadequate morphological characteristics for classification and resulting in the definition of multiple species as a single species; (3) the prevalence of morphological plasticity in Stigeoclonium species.
It is also essential to emphasize that the precise identification of most species is challenging, and several specimens exhibit characteristics that are significantly different from known species in the genus
Stigeoclonium, as reported in previous studies [
8,
9,
28]. Consequently, we refrain from drawing conclusions or hastily proposing new species, primarily due to the limited specimens and the phenotypic plasticity [
11,
29,
30].
Currently, reliable morphological characteristics that can be used to distinguish between Stigeoclonium sensu stricto and the S. farctum clade still have not been identified. Although the zoospore germination types and the prostrate system are important morphological features, they appear to be unsuitable for differentiating between the mentioned clades and species within Stigeoclonium. In the future, with an ample supply of specimens, a well-resolved molecular phylogeny, as presented in this study, could be used to re-evaluate morphological characteristics in Stigeoclonium under precisely defined laboratory conditions.