A Novel Sorbitol-Based Flow Cytometry Buffer Is Effective for Genome Size Estimation across a Cypriot Grapevine Collection
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Sample Preparation
2.3. Flow Cytometry
2.4. Statistical Analyses
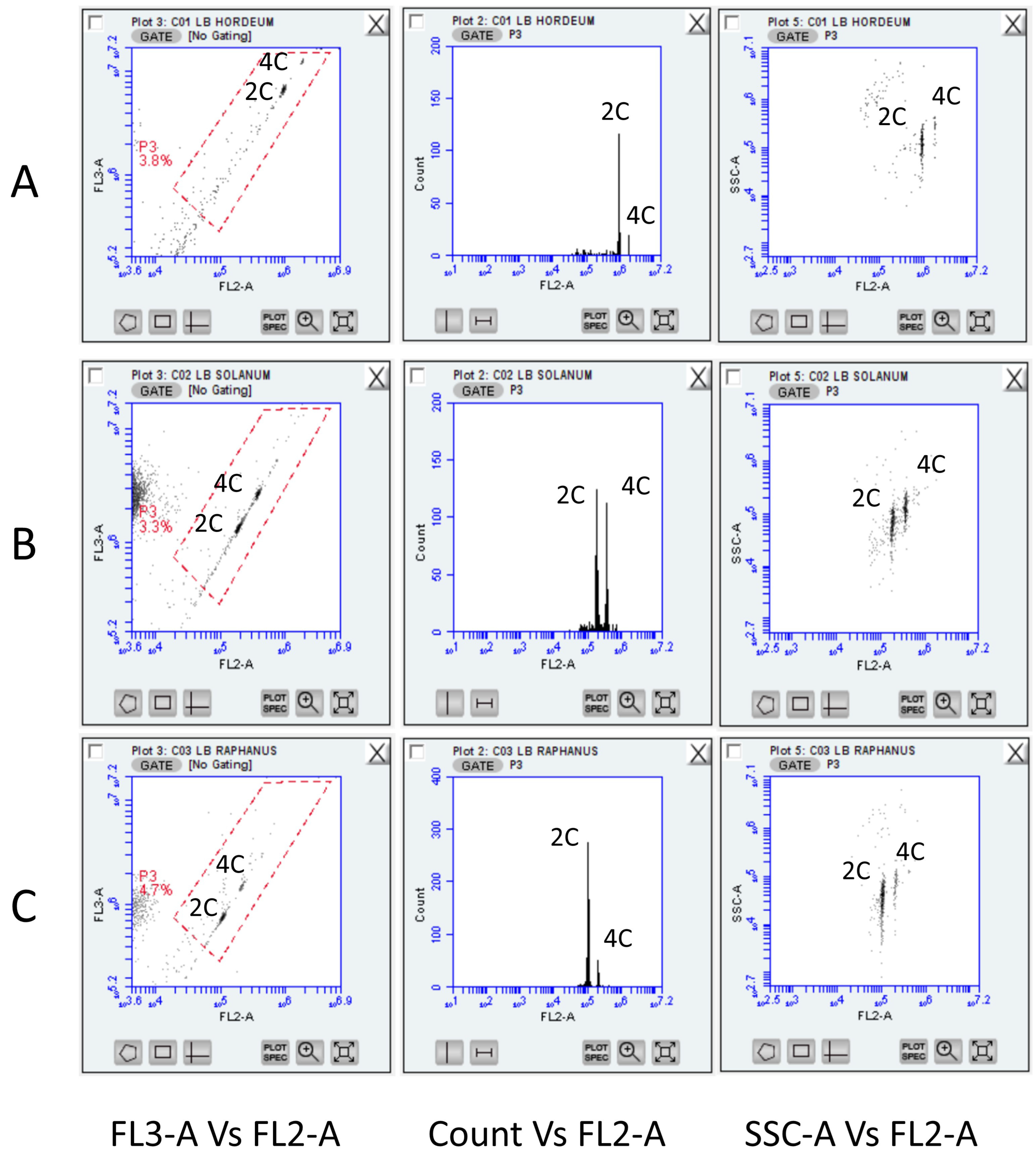
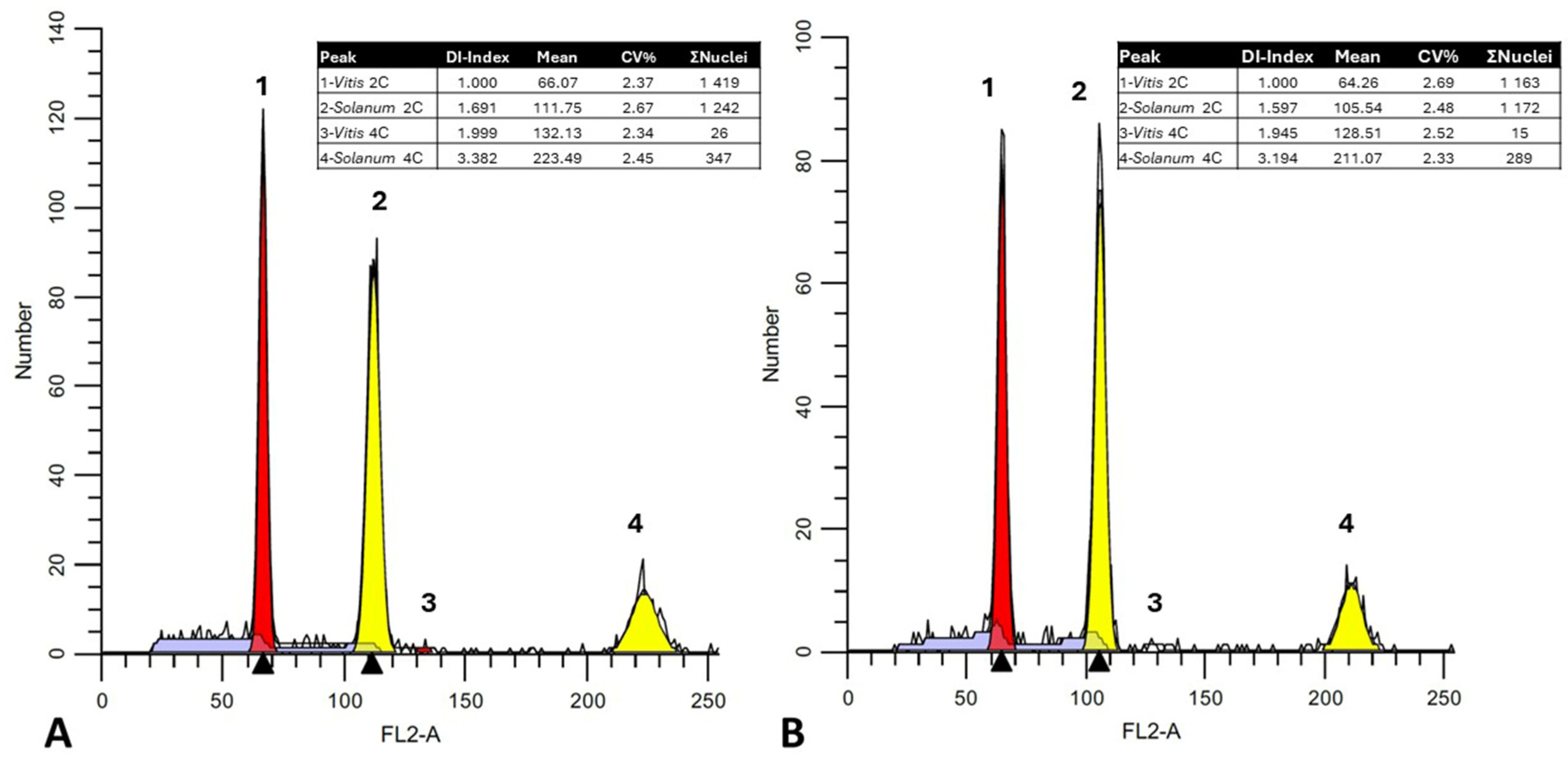
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doležel, J.; Bartoš, J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005, 95, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Galbraith, D.W.; Harkins, K.R.; Maddox, J.M.; Ayres, N.M.; Sharma, D.P.; Firoozabady, E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 1983, 220, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, J.; Rodriguez, E.; Doležel, J.; Santos, C. Two new nuclear isolation buffers for plant DNA flow cytometry: A test with 37 species. Ann. Bot. 2007, 100, 875–888. [Google Scholar] [CrossRef] [PubMed]
- Sadhu, A.; Bhadra, S.; Bandyopadhyay, M. Novel nuclei isolation buffer for flow cytometric genome size estimation of Zingiberaceae: A comparison with common isolation buffers. Ann. Bot. 2016, 118, 1057–1070. [Google Scholar] [CrossRef]
- Gambetta, J.M.; Romat, V.; Schmidtke, L.M.; Holzapfel, B.P. Secondary Metabolites Coordinately Protect Grapes from Excessive Light and Sunburn Damage during Development. Biomolecules 2022, 12, 42. [Google Scholar] [CrossRef]
- Muñoz, P.; Munné-Bosch, S. Photo-Oxidative Stress during Leaf, Flower and Fruit Development. Plant Physiol. 2018, 176, 1004–1014. [Google Scholar] [CrossRef]
- Goufo, P.; Singh, R.K.; Cortez, I. A Reference List of Phenolic Compounds (Including Stilbenes) in Grapevine (Vitis vinifera L.). Antioxidants 2020, 9, 398. [Google Scholar] [CrossRef]
- Ky, I.; Le Floch, A.; Zeng, L.; Pechamat, L.; Jourdes, M.; Teissedre, P.-L. Tannins; Caballero, B., Finglas, P.M., Toldrá, F., Eds.; Academic Press: Oxford, UK, 2016; pp. 247–255. ISBN 978-0-12-384953-3. [Google Scholar]
- Boulet, J.C.; Abi-Habib, E.; Carrillo, S.; Roi, S.; Veran, F.; Verbaere, A.; Meudec, E.; Rattier, A.; Ducasse, M.A.; Jørgensen, B.; et al. Focus on the relationships between the cell wall composition in the extraction of anthocyanins and tannins from grape berries. Food Chem. 2023, 406, 135023. [Google Scholar] [CrossRef]
- Loureiro, J.; Rodriguez, E.; Doležel, J.; Santos, C. Flow cytometric and microscopic analysis of the effect of tannic acid on plant nuclei and estimation of DNA content. Ann. Bot. 2006, 98, 515–527. [Google Scholar] [CrossRef]
- Noirot, M.; Barre, P.; Louarn, J.; Duperray, C.; Hamon, S. Nucleus-cytosol interactions—A source of stoichiometric error in flow cytometric estimation of nuclear DNA content in plants. Ann. Bot. 2000, 86, 309–316. [Google Scholar] [CrossRef]
- Noirot, M.; Barre, P.; Duperray, C.; Louarn, J.; Hamon, S. Effects of caffeine and chlorogenic acid on propidium iodide accessibility to DNA: Consequences on genome size evaluation in coffee tree. Ann. Bot. 2003, 92, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Noirot, M.; Barre, P.; Duperray, C.; Hamon, S.; De Kochko, A. Investigation on the causes of stoichiometric error in genome size estimation using heat experiments: Consequences on data interpretation. Ann. Bot. 2005, 95, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Price, H.J.; Hodnett, G.; Johnston, J.S. Sunflower (Helianthus annuus) leaves contain compounds that reduce nuclear propidium iodide fluorescence. Ann. Bot. 2000, 86, 929–934. [Google Scholar] [CrossRef]
- Walker, D.J.; Moñino, I.; Correal, E. Genome size in Bituminaria bituminosa (L.) C.H. Stirton (Fabaceae) populations: Separation of “true” differences from environmental effects on DNA determination. Environ. Exp. Bot. 2006, 55, 258–265. [Google Scholar] [CrossRef]
- Öncü Öner, T.; Temel, M.; Pamay, S.; Abaci, A.K.; Kaya Akkale, H.B. An Improved Method for Efficient DNA Extraction from Grapevine. Int. J. Life Sci. Biotechnol. 2023, 6, 21–36. [Google Scholar] [CrossRef]
- Sun, L.; Zhu, B.; Zhang, X.; Wang, H.; Yan, A.; Zhang, G.; Wang, X.; Xu, H. The accumulation profiles of terpene metabolites in three Muscat table grape cultivars through HS-SPME-GCMS. Sci. Data 2020, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.A.V.A.; Paiva, A.; Candeias, M.I. Flow cytometry—A simple method for nuclear DNA content evaluation of Vitis vinifera cv. Periquita somatic embryos obtained from anther cultures. Vitis 2003, 42, 99–100. [Google Scholar]
- Faure, O.; Nougarède, A. Nuclear DNA content of somatic and zygotic embryos of Vitis vinifera cv. Grenache noir at the torpedo stage. Protoplasma 1993, 176, 145–150. [Google Scholar] [CrossRef]
- Prado, M.J.; Rodriguez, E.; Rey, L.; González, M.V.; Santos, C.; Rey, M. Detection of somaclonal variants in somatic embryogenesis-regenerated plants of Vitis vinifera by flow cytometry and microsatellite markers. Plant Cell. Tissue Organ Cult. 2010, 103, 49–59. [Google Scholar] [CrossRef]
- Acanda, Y.; Prado, M.J.; González, M.V.; Rey, M. Somatic embryogenesis from stamen filaments in grapevine (Vitis vinifera L. cv. Mencía): Changes in ploidy level and nuclear DNA content. Vitr. Cell. Dev. Biol.-Plant 2013, 49, 276–284. [Google Scholar] [CrossRef]
- González, R.; Vallès, J.; Garnatje, T. Genome Size Variation Assessment in Vitis vinifera L. Landraces in Ibiza and Formentera (Balearic Islands). Plants 2022, 11, 1892. [Google Scholar] [CrossRef]
- Leal, F.; Loureiro, J.; Rodriguez, E.; Pais, M.S.; Santos, C.; Pinto-Carnide, O. Nuclear DNA content of Vitis vinifera cultivars and ploidy level analyses of somatic embryo-derived plants obtained from anther culture. Plant Cell Rep. 2006, 25, 978–985. [Google Scholar] [CrossRef]
- Lodhi, M.A.; Reisch, B.I. Nuclear DNA content of Vitis species, cultivars, and other genera of the Vitaceae. Theor. Appl. Genet. 1995, 90, 11–16. [Google Scholar] [CrossRef]
- Gichuki, D.K.; Ma, L.; Zhu, Z.; Du, C.; Li, Q.; Hu, G.; Zhong, Z.; Li, H.; Wang, Q.; Xin, H. Genome size, chromosome number determination, and analysis of the repetitive elements in Cissus quadrangularis. PeerJ 2019, 7, e8201. [Google Scholar] [CrossRef]
- Hermawaty, D.; Considine, J.A.; Considine, M.J. An Evaluation of Nuclei Preparation of the Dormant Axillary Bud of Grapevine for Cell Cycle Analysis by Flow Cytometry. Front. Plant Sci. 2022, 13, 834977. [Google Scholar] [CrossRef]
- Xie, X.; Agüero, C.B.; Wang, Y.; Walker, M.A. In vitro induction of tetraploids in Vitis × Muscadinia hybrids. Plant Cell. Tissue Organ Cult. 2015, 122, 675–683. [Google Scholar] [CrossRef]
- Grigoriou, A.; Tsaniklidis, G.; Hagidimitriou, M.; Nikoloudakis, N. The cypriot indigenous grapevine germplasm is a multi-clonal varietal mixture. Plants 2020, 9, 1034. [Google Scholar] [CrossRef] [PubMed]
- Zohary, D.; Hopf, M.; Weiss, E. Domestication of Plants in the Old World: The Origin and Spread of Domesticated Plants in Southwest Asia, Europe, and the Mediterranean Basin; Oxford University Press: Oxford, UK, 2012; ISBN 0199549060. [Google Scholar]
- Athinodorou, F.; Foukas, P.; Tsaniklidis, G.; Kotsiras, A.; Chrysargyris, A.; Delis, C.; Kyratzis, A.C.; Tzortzakis, N.; Nikoloudakis, N. Morphological diversity, genetic characterization, and phytochemical assessment of the cypriot tomato germplasm. Plants 2021, 10, 1698. [Google Scholar] [CrossRef] [PubMed]
- Kyriacou, M.C.; Ioannidou, S.; Nikoloudakis, N.; Seraphides, N.; Papayiannis, L.C.; Kyratzis, A.C. Physicochemical characterization and trait stability in a genetically diverse ex situ collection of pomegranate (Punica granatum L.) germplasm from Cyprus. Sci. Hortic. 2020, 263, 109116. [Google Scholar] [CrossRef]
- Anestiadou, K.; Nikoloudakis, N.; Hagidimitriou, M.; Katsiotis, A. Monumental olive trees of Cyprus contributed to the establishment of the contemporary olive germplasm. PLoS ONE 2017, 12, e0187697. [Google Scholar] [CrossRef] [PubMed]
- Nikoloudakis, N.; Aissat, A.; Katsiotis, A. Screening A. ventricosa populations for 2n gametes. Euphytica 2018, 214, 34. [Google Scholar] [CrossRef]
- Constandinou, S.; Nikoloudakis, N.; Kyratzis, A.C.C.; Katsiotis, A. Genetic diversity of Avena ventricosa populations along an ecogeographical transect in Cyprus is correlated to environmental variables. PLoS ONE 2018, 13, e0193885. [Google Scholar] [CrossRef] [PubMed]
- Doležel, J.; Binarová, P.; Lcretti, S. Analysis of Nuclear DNA content in plant cells by Flow cytometry. Biol. Plant. 1989, 31, 113–120. [Google Scholar] [CrossRef]
- Charalambous, I.; Ioannou, N.; Kyratzis, A.C.; Kourtellarides, D.; Hagidimitriou, M.; Nikoloudakis, N. Genome Size Variation across a Cypriot Fabeae Tribe Germplasm Collection. Plants 2023, 12, 1469. [Google Scholar] [CrossRef] [PubMed]
- Doležel, J.; Bartoš, J.; Voglmayr, H.; Greilhuber, J.; Thomas, R.A. Nuclear DNA content and genome size of trout and human (multiple letters). Cytom. Part A 2003, 51, 127–129. [Google Scholar] [CrossRef] [PubMed]
- Vincken, J.-P.; Schols, H.A.; Oomen, R.J.F.J.; McCann, M.C.; Ulvskov, P.; Voragen, A.G.J.; Visser, R.G.F. If Homogalacturonan Were a Side Chain of Rhamnogalacturonan I. Implications for Cell Wall Architecture. Plant Physiol. 2003, 132, 1781–1789. [Google Scholar] [CrossRef]
- Greilhuber, J. “Self-tanning”—A new and important source of stoichiometric error in cytophotometric determination of nuclear DNA content in plants. Plant Syst. Evol. 1988, 158, 87–96. [Google Scholar] [CrossRef]
- Schmitt, M.H.; Ward, D.; Shrader, A.M. Salivary tannin-binding proteins: A foraging advantage for goats? Livest. Sci. 2020, 234, 103974. [Google Scholar] [CrossRef]
- Mole, S.; Butler, L.G.; Iason, G. Defense against dietary tannin in herbivores: A survey for proline rich salivary proteins in mammals. Biochem. Syst. Ecol. 1990, 18, 287–293. [Google Scholar] [CrossRef]
- Shimada, T. Salivary proteins as a defense against dietary tannins. J. Chem. Ecol. 2006, 32, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Hoseini, S.M.; Moghaddam, A.A.; Ghelichpour, M.; Pagheh, E.; Haghpanah, A.; Gharavi, B.; Mansouri, B.; Arghideh, M. Dietary glycine supplementation modulates antioxidant and immune responses of beluga, Huso huso, juveniles. Aquac. Rep. 2022, 23, 101026. [Google Scholar] [CrossRef]
- Barra, G.B.; Santa Rita, T.H.; Vasques, J.d.A.; Chianca, C.F.; Nery, L.F.A.; Costa, S.S.S. EDTA-mediated inhibition of DNases protects circulating cell-free DNA from ex vivo degradation in blood samples. Clin. Biochem. 2015, 48, 976–981. [Google Scholar] [CrossRef] [PubMed]
- Greilhuber, J.; Temsch, E.M.; Loureiro, J.C.M. Nuclear DNA Content Measurement. In Flow Cytometry with Plant Cells: Analysis of Genes, Chromosomes and Genomes; John Wiley & Sons: Hoboken, NJ, USA, 2007; pp. 67–101. [Google Scholar]
- Doležel, J.; Greilhuber, J.; Suda, J. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2007, 2, 2233–2244. [Google Scholar] [CrossRef]
- Lemire, K.A.; Rodriguez, Y.Y.; McIntosh, M.T. Alkaline hydrolysis to remove potentially infectious viral RNA contaminants from DNA. Virol. J. 2016, 13, 88. [Google Scholar] [CrossRef] [PubMed]
- Vrontis, D.; Thrassou, A. The renaissance of Commandaria: A strategic branding prescriptive analysis. J. Glob. Bus. Adv. 2011, 4, 302–316. [Google Scholar] [CrossRef]
- Magris, G.; Jurman, I.; Fornasiero, A.; Paparelli, E.; Schwope, R.; Marroni, F.; Di Gaspero, G.; Morgante, M. The genomes of 204 Vitis vinifera accessions reveal the origin of European wine grapes. Nat. Commun. 2021, 12, 7240. [Google Scholar] [CrossRef]



| Buffer | Chemical Composition |
|---|---|
| Lysis Buffer (LB01); [35] | 15 mM Tris HCl; 2 mM Na2EDTA; 0.5 mM Spermine tetrahydrochloride; 80 mM KCl; 20 mM NaCl; 0.1% (v/v) Triton X-100; 0.1% (v/v) β-mercaptoethanol; 50 μg/mL RNAse; 50 μg/mL Propidium Iodide; pH 7.5 |
| Leal’s buffer; [23] | 100 mM Tri-Sodium Citrate; 50 mM HEPES; 5 mM EDTA; 50 mM Glucose; 15 mM NaCl; 15 mM KCl; 1% (w/v) Polyvinylpyrrolidone (PVP-40); 1% (v/v) Tween 20; 0.1% (v/v) β-mercaptoethanol; 50 μg/mL RNAse; 50 μg/mL Propidium Iodide, pH 7.2 |
| Woody-Plant Buffer (WBP); [3] | 0.2 mM Tris HCl; 2 mM MgCl2; 2.5 mM EDTA; 86 mM NaCl; 10 mM Sodium sulphite; 1% (w/v) Polyvinylpyrrolidone (PVP-40); 1% (v/v) Triton X-100; 0.1% (v/v) β-mercaptoethanol; 50 μg/mL RNAse; 50 μg/mL Propidium Iodide; pH 7.5 |
| Sorbitol-based buffer (SBB); Current-first report | 100 mM Tris-HCl; 0.35 M Sorbitol; 0.05 M glycine; 5 mM EDTA; 90 mM NaCl; 1% (w/v) Polyvinylpyrrolidone (PVP-40); 0.5% (v/v) Tween 20; 0.1% (v/v) β-mercaptoethanol; 50 μg/mL RNAse; 50 μg/mL Propidium Iodide; pH 7.5 |
| Cultivar/Prime Name | Variety Number VIVC | Country/Region of Origin | Colour of Berry Skin | 1C-Value (pg) | 2C-Value (pg) | 2C-Mbp | CV (%) | HSD 1 |
|---|---|---|---|---|---|---|---|---|
| Agiorgitiko | 102 | Greece | Noir | 0.5839 ± 0.0026 | 1.1678 ± 0.0052 | 1142.1700 | 2.900 ± 0.200 | d-f |
| Assyrtiko | 726 | Greece | Blanc | 0.5776 ± 0.0018 | 1.1552 ± 0.0036 | 1129.7910 | 2.507 ± 0.203 | f |
| Cabernet Sauvignon | 1929 | France | Noir | 0.5849 ± 0.0023 | 1.1698 ± 0.0046 | 1144.1110 | 2.723 ± 0.152 | c-f |
| Carignan | 2098 | France | Noir | 0.577 ± 0.005 | 1.154 ± 0.01 | 1128.6550 | 2.453 ± 0.397 | f |
| Chardonnay Blanc | 2455 | France | Blanc | 0.5945 ± 0.0015 | 1.189 ± 0.003 | 1162.7610 | 2.463 ± 0.182 | ab |
| Giannoudi | - | Cyprus | Noir | 0.5887 ± 0.0023 | 1.1774 ± 0.0046 | 1151.4230 | 2.643 ± 0.145 | a-d |
| Grenache | 4461 | Spain | Noir | 0.586 ± 0.0035 | 1.172 ± 0.007 | 1146.3040 | 2.607 ± 0.136 | b-e |
| Kanella | 16,124 | Cyprus | Blanc | 0.5882 ± 0.0028 | 1.1764 ± 0.0056 | 1150.5570 | 2.417 ± 0.144 | a-d |
| Lefkada | - | Cyprus | Noir | 0.5942 ± 0.0018 | 1.1884 ± 0.0036 | 1162.2620 | 2.637 ± 0.543 | ab |
| Maratheftiko | 7374 | Cyprus | Noir | 0.5936 ± 0.0037 | 1.1872 ± 0.0074 | 1161.0500 | 2.497 ± 0.172 | a-c |
| Mavro | 27,628 | Cyprus | Noir | 0.5891 ± 0.0042 | 1.1782 ± 0.0084 | 1152.2250 | 2.667 ± 0.095 | a-d |
| Merlot Noir | 7657 | France | Noir | 0.5869 ± 0.0061 | 1.1738 ± 0.0122 | 1148.0270 | 2.690 ± 0.639 | b-d |
| Morokanella | 16,123 | Cyprus | Noir | 0.5904 ± 0.0009 | 1.1808 ± 0.0018 | 1154.8720 | 2.587 ± 0.341 | a-d |
| Moschofilero | 8068 | Greece | Rose | 0.5948 ± 0.003 | 1.1896 ± 0.006 | 1163.4160 | 2.607 ± 0.090 | ab |
| Ofthalmo | 8782 | Cyprus | Noir | 0.5908 ± 0.0045 | 1.1816 ± 0.009 | 1155.5940 | 2.293 ± 0.107 | ef |
| Pinot Noir | 9279 | France | Noir | 0.5902 ± 0.0017 | 1.1804 ± 0.0034 | 1154.4150 | 3.440 ± 0.104 | a-d |
| Promara | 9737 | Cyprus | Blanc | 0.5898 ± 0.0011 | 1.1796 ± 0.0022 | 1153.6940 | 3.153 ± 1.268 | a-d |
| Riesling | 3264 | France | Blanc | 0.593 ± 0.0027 | 1.186 ± 0.0054 | 1159.9250 | 2.317 ± 0.057 | a-c |
| Sauvignon blanc | 10,790 | France | Blanc | 0.5872 ± 0.0011 | 1.1744 ± 0.0022 | 1148.5260 | 2.413 ± 0.108 | b-d |
| Shiraz | 11,748 | France | Noir | 0.5827 ± 0.0069 | 1.1654 ± 0.0138 | 1139.7350 | 2.893 ± 0.180 | d-f |
| Spourtiko | 16,121 | Cyprus | Blanc | 0.5965 ± 0.0009 | 1.193 ± 0.0018 | 1166.7150 | 3.023 ± 0.785 | a |
| Vasilisa | - | Cyprus | Blanc | 0.5859 ± 0.0013 | 1.1718 ± 0.0026 | 1146.0950 | 2.973 ± 0.873 | b-f |
| Xinomavro | 13,284 | Greece | Noir | 0.5909 ± 0.0021 | 1.1818 ± 0.0042 | 1155.8570 | 2.940 ± 0.583 | a-d |
| Xynisteri | 704 | Cyprus | Blanc | 0.5906 ± 0.0008 | 1.1812 ± 0.0016 | 1155.2420 | 2.660 ± 0.131 | a-d |
| Average | - | - | - | 0.5886 ± 0.0026 | 1.1773 ± 0.0052 | 1151.3926 | 2.688 ± 0.318 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michael, K.; Andreou, C.; Markou, A.; Christoforou, M.; Nikoloudakis, N. A Novel Sorbitol-Based Flow Cytometry Buffer Is Effective for Genome Size Estimation across a Cypriot Grapevine Collection. Plants 2024, 13, 733. https://doi.org/10.3390/plants13050733
Michael K, Andreou C, Markou A, Christoforou M, Nikoloudakis N. A Novel Sorbitol-Based Flow Cytometry Buffer Is Effective for Genome Size Estimation across a Cypriot Grapevine Collection. Plants. 2024; 13(5):733. https://doi.org/10.3390/plants13050733
Chicago/Turabian StyleMichael, Kyriakos, Constantina Andreou, Anastasia Markou, Michalakis Christoforou, and Nikolaos Nikoloudakis. 2024. "A Novel Sorbitol-Based Flow Cytometry Buffer Is Effective for Genome Size Estimation across a Cypriot Grapevine Collection" Plants 13, no. 5: 733. https://doi.org/10.3390/plants13050733
APA StyleMichael, K., Andreou, C., Markou, A., Christoforou, M., & Nikoloudakis, N. (2024). A Novel Sorbitol-Based Flow Cytometry Buffer Is Effective for Genome Size Estimation across a Cypriot Grapevine Collection. Plants, 13(5), 733. https://doi.org/10.3390/plants13050733









