Abstract
The species Prunus mume consists of uniquely aromatic woody perennials with large amounts of free aromatic substances in the flower cells. Uridine diphosphate glycosyltransferase (UGT) modifies these free aromatic substances into water-soluble glycoside-bound volatiles (GBVs) which play an important role in regulating the use of volatiles by plants for information exchange, defense, and stress tolerance. To investigate the changes in the glycosidic state of aromatic substances during the flowering period of P. mume and discern the location and expression of glycoside synthesis genes, we extracted and enzymatically hydrolyzed GBVs of P. mume and then utilized gas chromatography–mass spectrometry (GC–MS) to characterize and analyze the types and contents of GBV glycosides. Further, we identified and classified the members of the UGT gene family of P. mume using the bioinformatic method and analyzed the correlation between the expression of the UGT family genes in P. mume and the changes in glycosidic content. The results showed that the benzenoids were the main aromatic substance that was glycosylated during flowering in P. mume and that glycosidic benzaldehyde was the most prevalent compound in different flower parts and at different flowering stages. The titer of glycoside benzaldehyde gradually increased during the bud stage and reached the highest level at the big bud stage (999.6 μg·g−1). Significantly, titers of glycoside benzaldehyde significantly decreased and stabilized after flowering while the level of free benzaldehyde, in contrast, significantly increased and then reached a plateau after the flowering process was completed. A total of 155 UGT family genes were identified in the P. mume genome, which were divided into 13 subfamilies (A–E, G–N); according to the classification of Arabidopsis thaliana UGT gene subfamilies, the L subfamily contains 17 genes. The transcriptome analysis showed that PmUGTL9 and PmUGTL13 were highly expressed in the bud stage and were strongly correlated with the content of the glycosidic form of benzaldehyde at all stages of flowering. This study provides a theoretical basis to elucidate the function of UGT family genes in P. mume during flower development, to explore the mechanism of the storage and transportation of aromatic compounds in flower tissues, and to exploit industrial applications of aromatic products from P. mume.
1. Introduction
Plants produce terpenoids, phenols, alkaloids, etc., during metabolism [1,2], and these secondary metabolites play important roles in normal plant growth and metabolism and in response to biotic and abiotic stresses [3,4]. The large intracellular accumulation of secondary metabolites disrupts the cell membrane structure and causes toxic effects on the plant cells [5]. An effective way for plants to cope with excessive intracellular secondary metabolites is to modify them into non-toxic and low-active glycoside-bound compounds, a process that requires the transfer of sugar groups into secondary metabolites with the aid of glycosyltransferases [6,7]. Among them, uridine diphosphate glycosyltransferase (UGT) can catalyze the glycosylation of plant secondary metabolites using uridine diphosphate sugar molecules as donors [8]. A total of 14 UGT subfamilies were identified in Arabidopsis thaliana. A benzoate substrate screening of 90 UGT recombinant proteins in Arabidopsis thaliana by Alexandra et al. revealed that members of the L subfamily are highly specific for O-glucoside formation, suggesting that the uridine diphosphate glycosyltransferase L-subfamily (UGTL) recognizes phenylpropane substrates and modifies them to form glycosidic bonds [9].
Small volatile compounds released within plant flower tissues exert complex effects including the exchange of information between organisms, the attraction of beneficial insects, and for biodefenses [10,11]. Aromatic volatiles exist in three forms: the volatile state released into the air, the free state located in cellular endogenous spaces, and the glycoside-bound states (GBVs) that are formed by linking with sugar groups [12]. With the discovery of monoterpene glycoside compounds in roses, several aromatic glycoside compounds have been isolated and identified in grapes and tea, with most GBV glycoside structures being β-D-glucoside and α-L-rhamnoside [6,13,14,15]. Changes in tea, wine, fruit, and floral aromas in production processes are mostly related to the transformation of GBV [16,17,18,19]. Glycosidic phenylpropane volatiles during tomato growth and development were highest at fruit ripening, and tomato fruit flavor quality was enhanced when the relevant β-glucosidase gene was overexpressed, suggesting that aromatic volatiles can be stored in tomatoes in the form of GBV [20].
Prunus mume is one of China’s ten most renowned flowers, with rich cultural connections and widespread popularity [21]. Benzenoid substances are secondary metabolites of phenylalanine generated by several modifications catalyzed by phenylalanine ammonium lyase (PAL) [22], in which benzaldehyde can be reduced to benzyl alcohol, which can also be catalyzed by acetyltransferase to generate benzyl acetate. Several components of this pathway are volatile and collectively constitute the characteristic aroma of P. mume [23].
Benzaldehyde is produced in large quantities at sites of mechanical damage in plants [24]. This compound inhibits the germination of fungal spores [25] and is a major component of new agricultural insecticides [26]. All these studies suggest that benzaldehyde plays an important role in plant defense. Hao [23] identified and quantitatively analyzed the volatile and free aromatic constituents in P. mume and found that the free-state content of the main aromatic constituents of P. mume was significantly greater than the volatile amount. The free-form aromatic components exhibit the highest benzaldehyde content, yet their volatilization efficiency is remarkably low [27], suggesting that, in P. mume, the predominant form of benzaldehyde is in its free state. Mechanisms exist for the interconversion of free and glycosidic states of defense compounds in plant cells [28], for example, when Camellia sinensis is infected with anthracnose, geraniol and benzyl alcohol in the form of glycosides are hydrolyzed and released by endogenous glycosidases to achieve an antimicrobial effect [29]. We speculate that there may also be an interconversion between the glycosidic and free states of benzaldehyde in P. mume as a way to maintain benzaldehyde at a certain concentration to achieve the defense function.
The mechanism of interconversion between free and glycosidic floral components in P. mume is not well understood. In this study, we identified and analyzed the glycosidic aromatic substances in different anthesis stages and parts using GC–MS, which was combined with the content of free aromatic substances to summarize the change rule of glycosidic benzaldehyde in different anthesis stages. UGT family members were identified from P. mume, and a phylogenetic tree was constructed to allow for the analysis of chromosomal localization and collinearity using bioinformatics. This study further utilized transcriptome data to screen the UGTL genes specifically expressed at different times of P. mume, which provided a reference for elucidating the mechanism of UGTL genes involved in the synthesis of aromatic substances’ glycosidic state in P. mume.
2. Results
2.1. Glycosidic Aromatic Substances of P. mume in Different Anthesis Stages
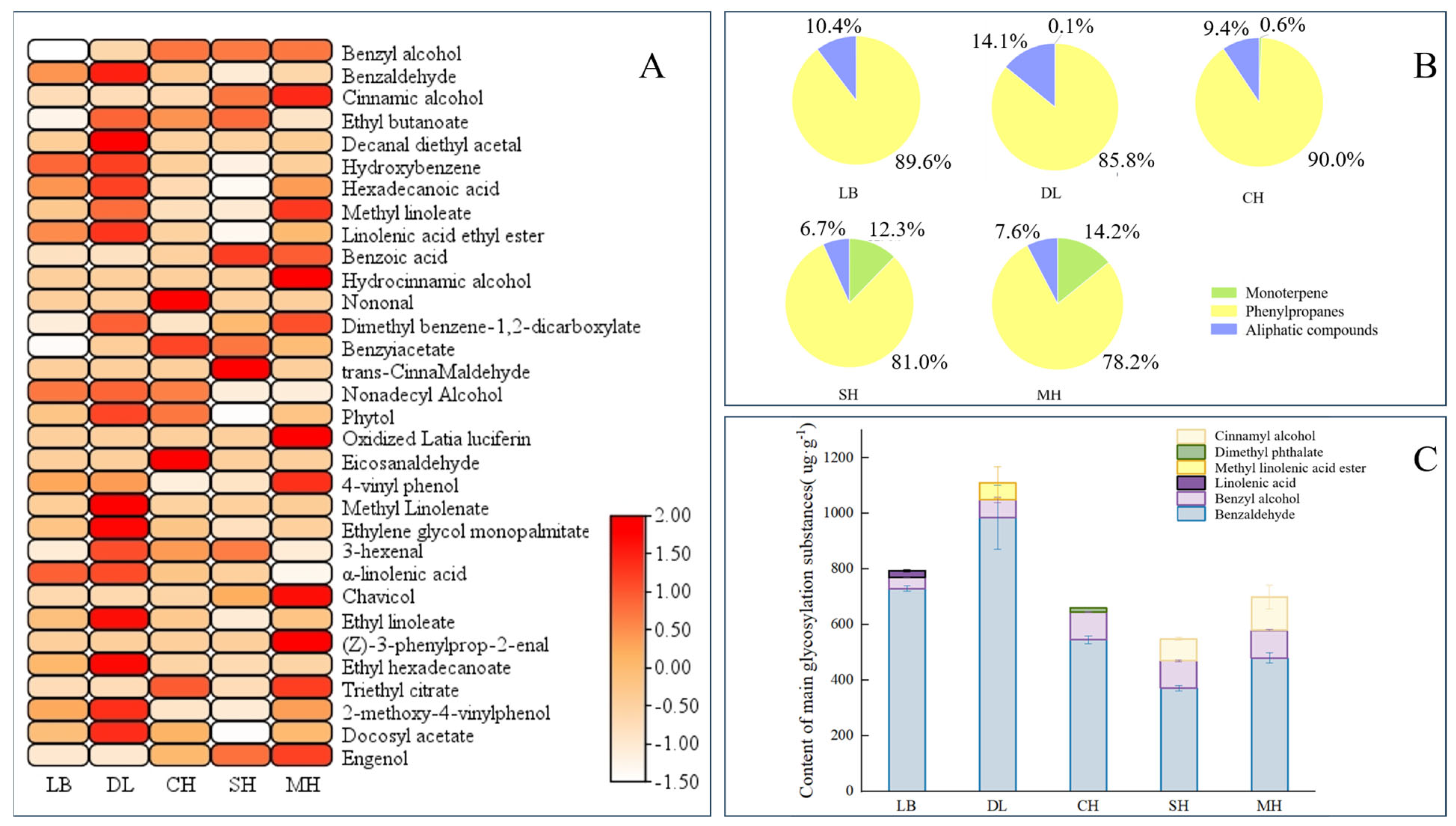
Glycosidic substances were extracted and analyzed from P. mume of different anthesis stages, and a total of 27 glycosidic aromatic substances were detected. According to the glycosidic compound classification method, the glycosidic aromatic substances in P. mume were classified into three categories, namely, monoterpenes, phenylpropanoids, and aliphatic compounds [30]. These included 4 monoterpenes, 10 phenylpropanes, and 13 aliphatic compounds. A total of 20 substances were glycosylated at the big bud stage, and their contents were relatively higher compared to other anthesis stages. The largest number of aromatic substances was glycosylated at the primordial stage, with 25 substances glycosylated, including 3 monoterpenes, 9 phenylpropanes, and 13 aliphatic compounds (Figure 1A).
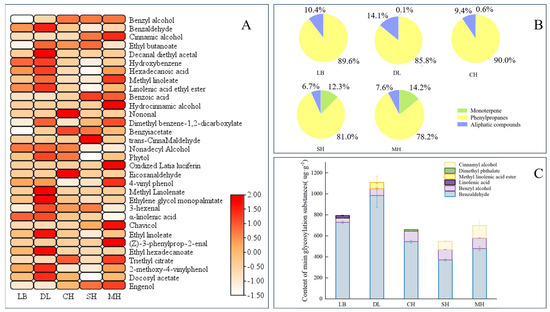
Figure 1.
Analysis of aroma glycosides at different blooming stages of P. mume. (A): Heatmap analysis of aroma glycosides at different blooming stages of P. mume. Each column represents a flowering period. The emission values were normalized using the log10 transformation. The color of the heatmap ranges from white (value, −1.5) to red (value, 2) in the natural logarithmic scale. Data are presented with means of three biological replicates. (B): Relative content of aromatic glycosidic properties at different blooming stages of P. mume. (C): Glucoside content of main aromatic substances at different blooming stages of P. mume. The top three glycosidic substances at each flowering stage are listed in the figure. LB, dew petal stage; DL, large bud stage; CH, first bloom stage; SH, full bloom stage; MH, last bloom stage.
The relative contents of glycosylated phenylpropanes were the highest at different anthesis stages of P. mume, and all of them were greater than 78%, with the highest relative content of 89.6% in the early flowering period, which is when the main glycosylated aromatic substances were found in P. mume. Glycosidic monoterpenes started appearing at the large bud stage and gradually increased with the flowering process, with the highest content of 14.2% compared to other flowering stages at the last bloom stage (Figure 1B). Benzaldehyde and benzyl alcohol were the main glycosidic aromatic substances in P. mume, and the content of glycosidic benzaldehyde was the highest in different anthesis stages, with the highest content in the big bud stage showing a trend of gradual decrease with the continuation of the anthesis stage (Figure 1C). By comparing the types and absolute and relative contents of glycosidic states in different anthesis stages, the results are consistent with the fact that benzene ring-like substances are the predominant volatile aroma components in developing flower tissues of P. mume, indicating that P. mume could store benzyl alcohol and benzaldehyde in the form of glycosidic states in the flora.
2.2. Quantitative Analysis of Endogenous Benzaldehyde in Different Floral Parts of P. mume
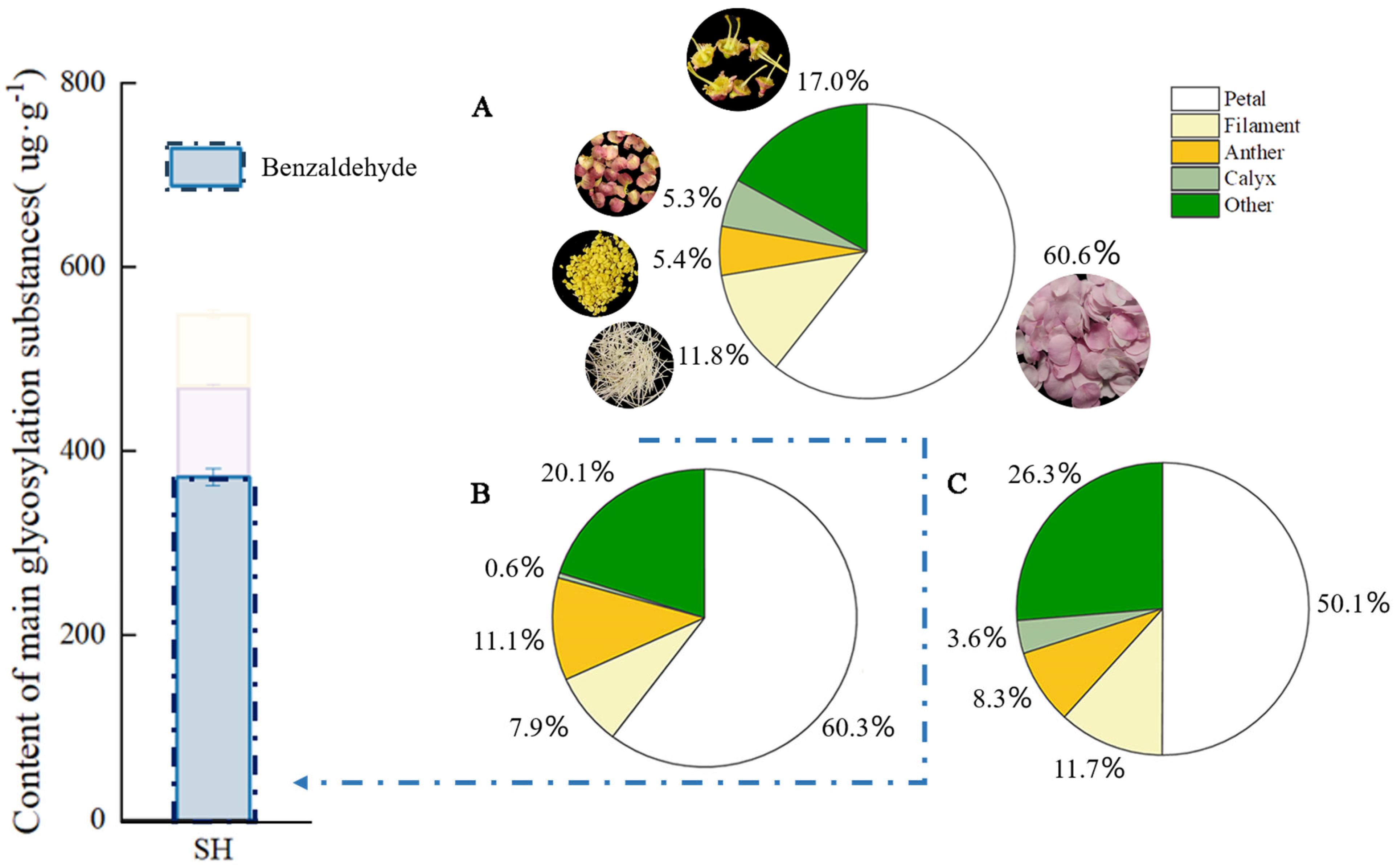
Quantitative analyses were conducted on the mass, glycosidic, and free benzaldehyde contents in the five organs of P. mume at full bloom. As can be seen from Figure 2, petals had the largest mass proportion of 60.6% among the parts of P. mume at full bloom, while their glycosidic and free benzaldehyde contents were the highest, at 60.3% and 50.1%, respectively, followed by benzaldehyde endogenous contents in other flower parts (disk + pistil). The mass ratio of the anther part was 5.4%, but its content of glycosidic and free benzaldehyde reached 11.1% and 8.3%, respectively; the mass ratio of the calyx was similar to that of the anther, but its content of glycosidic and free benzaldehyde was only 0.6% and 3.6%, respectively; 11.3% of free benzaldehyde and 7.9% of glycosidic benzaldehyde were detected in the filament.
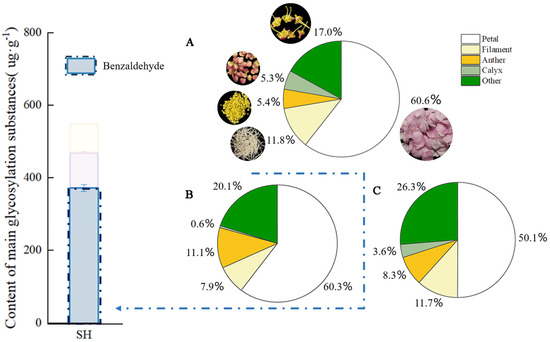
Figure 2.
Free and glycoside benzaldehyde quantitative analysis in different parts of P. mume. The blue bars in the histogram represent the glycosidic state benzaldehyde content in the full bloom stage of P. mume (A): Proportion of weight of different parts of flowers. The circular picture around the pie chart shows the materials used. (B): The content of glycoside benzaldehyde in different parts of flowers. (C): The content of free benzaldehyde in different parts of flowers. SH, full bloom stage.
2.3. Quantitative Analysis of Glycosidic State of Key Components of P. mume in Different Anthesis Stages
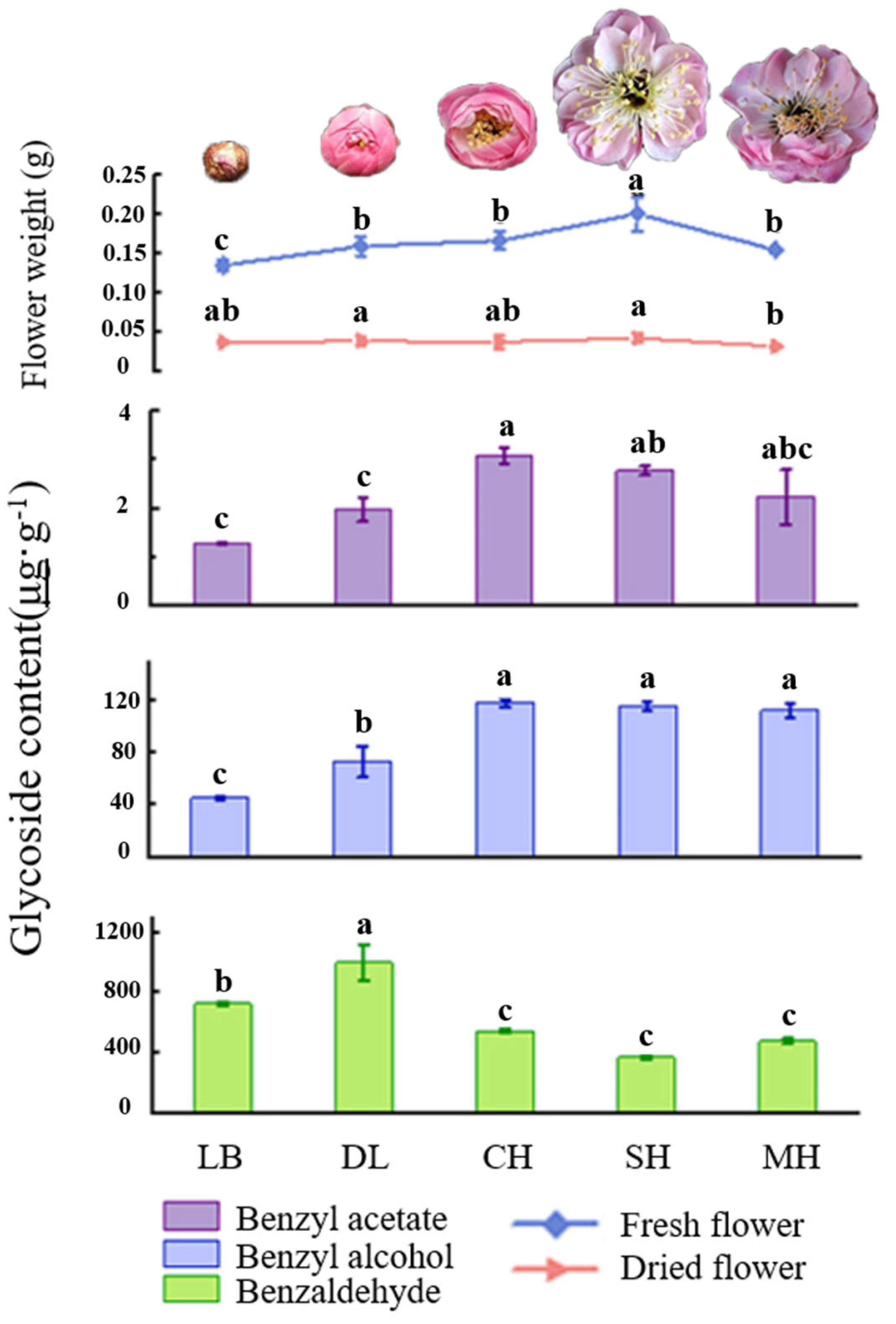
Phenylpropanes were the main aromatic substances in floral tissues of P. mume, as was found after analyzing the glycosidic state substances of the characteristic aroma after hydrolysis by β-glucosidase. The content of glycosidic benzaldehyde was significantly higher than that of benzyl alcohol and benzyl acetate in flower tissues at different developmental stages of P. mume, in which the content of glycosidic benzaldehyde showed a general trend of increasing and then decreasing, and the content of benzaldehyde in the dew petal stage and bud stage was significantly higher than that in other stages and reached the maximum value of 999.6 μg·g−1 in the big bud period. The glycosidic contents of benzyl alcohol and benzyl acetate increased gradually over the period, and they were significantly higher than those in the first-flowering stage and the last-flowering stage, reaching a maximum of 118.37 μg·g−1 and 3.1 μg·g−1, respectively, in the first-flowering stage. The differences between the first-flowering stage and the last-flowering stage were not significant. The fresh weight of the flowers gradually increased with the flowering stage and reached their maximum at the full bloom stage. This was consistent with the trend of the contents of benzyl alcohol and benzyl acetate in the glycosidic states, and the fresh and dry weights of the flowers at the final flowering stage were both significantly lower than those at the full bloom stage (Figure 3).
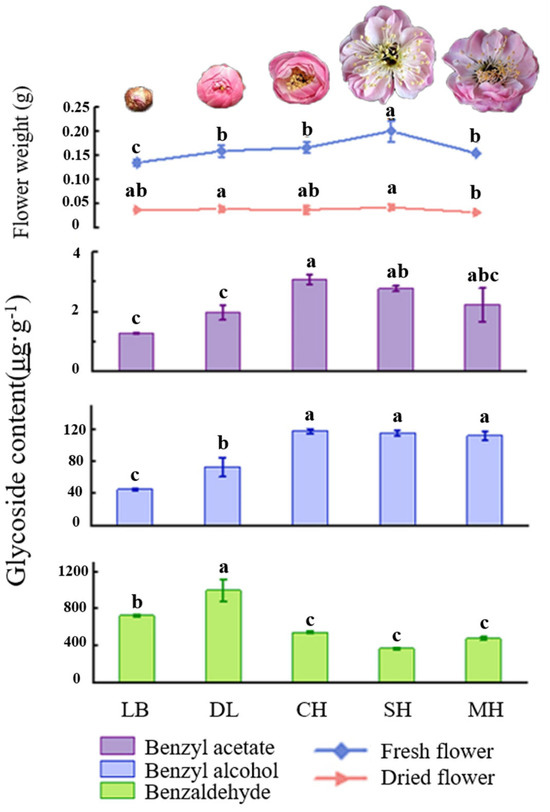
Figure 3.
Quality and key glycosylated aromatic components’ quantitative analysis at different blooming stages of P. mume. Data are mean ± standard deviation (SD) calculated from three biological replicates. Vertical lines represent standard deviation. The different lowercase letters indicate significant difference at 0.05 level. LB, dew petal stage; DL, large bud stage; CH, first bloom stage; SH, full bloom stage; MH, last bloom stage. The P. mume in the picture is a display of material from each stage of flowering.
2.4. Quantitative Analysis of Endogenous Benzaldehyde at Different Anthesis Stages of P. mume
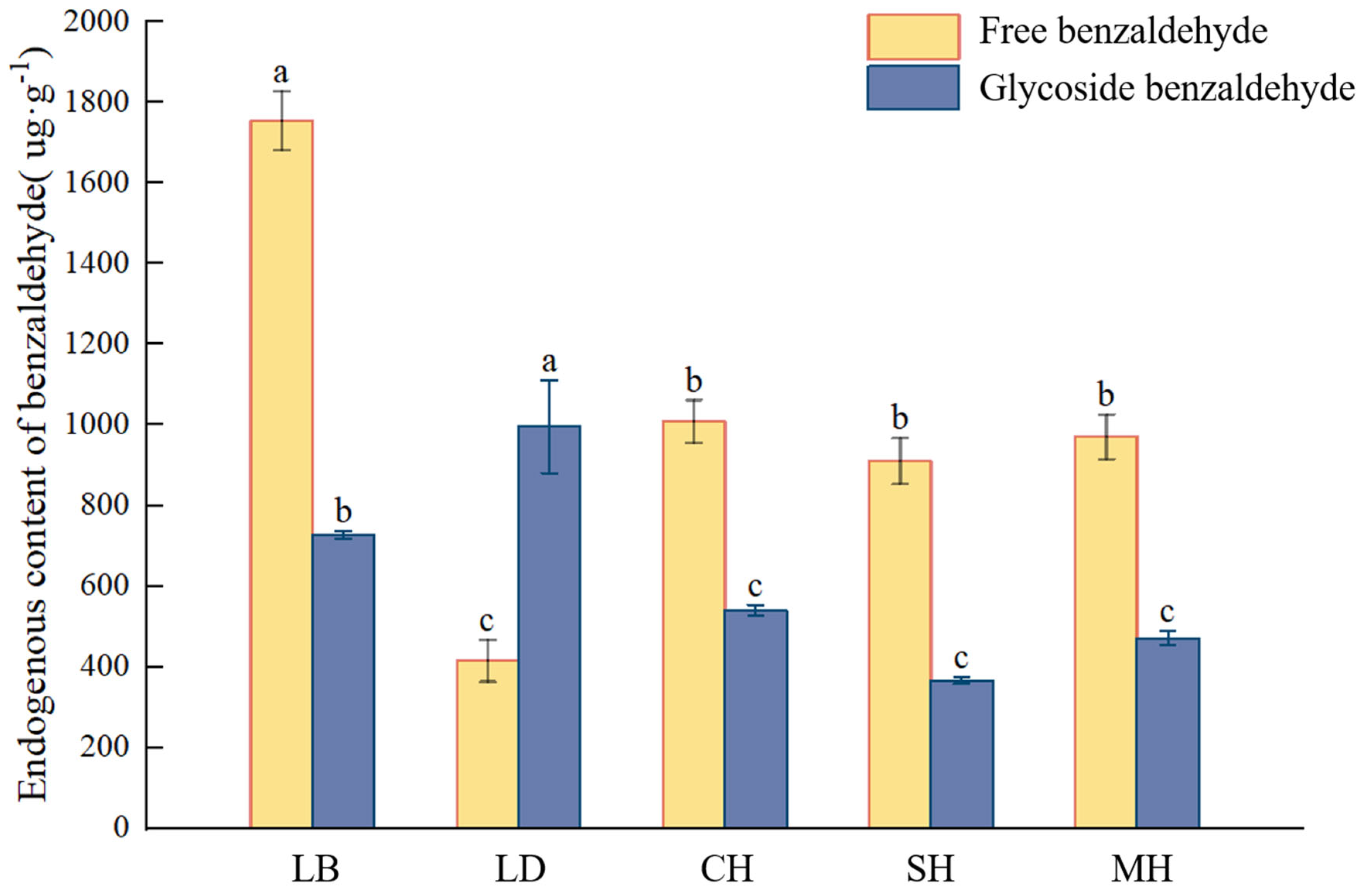
This study quantified the content of glycosidic benzaldehyde at five flowering anthesis stages of P. mume and analyzed it in combination with the free state and volatile amount of benzaldehyde in each period. As can be seen from Figure 4, the free benzaldehyde content was significantly higher in the dew petal period than in other anthesis stages, reaching 1754.16 μg·g−1. As the flower buds expanded to large buds, the free-state content decreased abruptly, and the content of glycosidic benzaldehyde in the same period was the highest compared to that in the other anthesis stages, reaching 995.48 μg·g−1. The free benzaldehyde content stabilized after the blooming period, while the glycosidic benzaldehyde content continued to show a decreasing trend as the flowering period progressed.
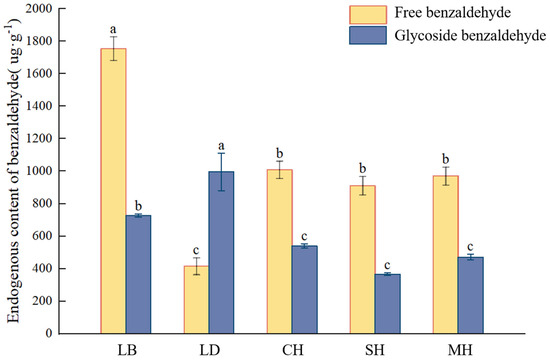
Figure 4.
Volatile, free, and glycosylated benzaldehyde quantitative analyses at different blooming stages of P. mume. Data are mean ± standard deviation (SD) calculated from three biological replicates. Vertical lines represent standard deviation. The different lowercase letters indicate significant difference at 0.05 level. LB, dew petal stage; DL, large bud stage; CH, first bloom stage; SH, full bloom stage; MH, last bloom stage.
2.5. Identification and Subfamily Classification of PmUGT Family Members
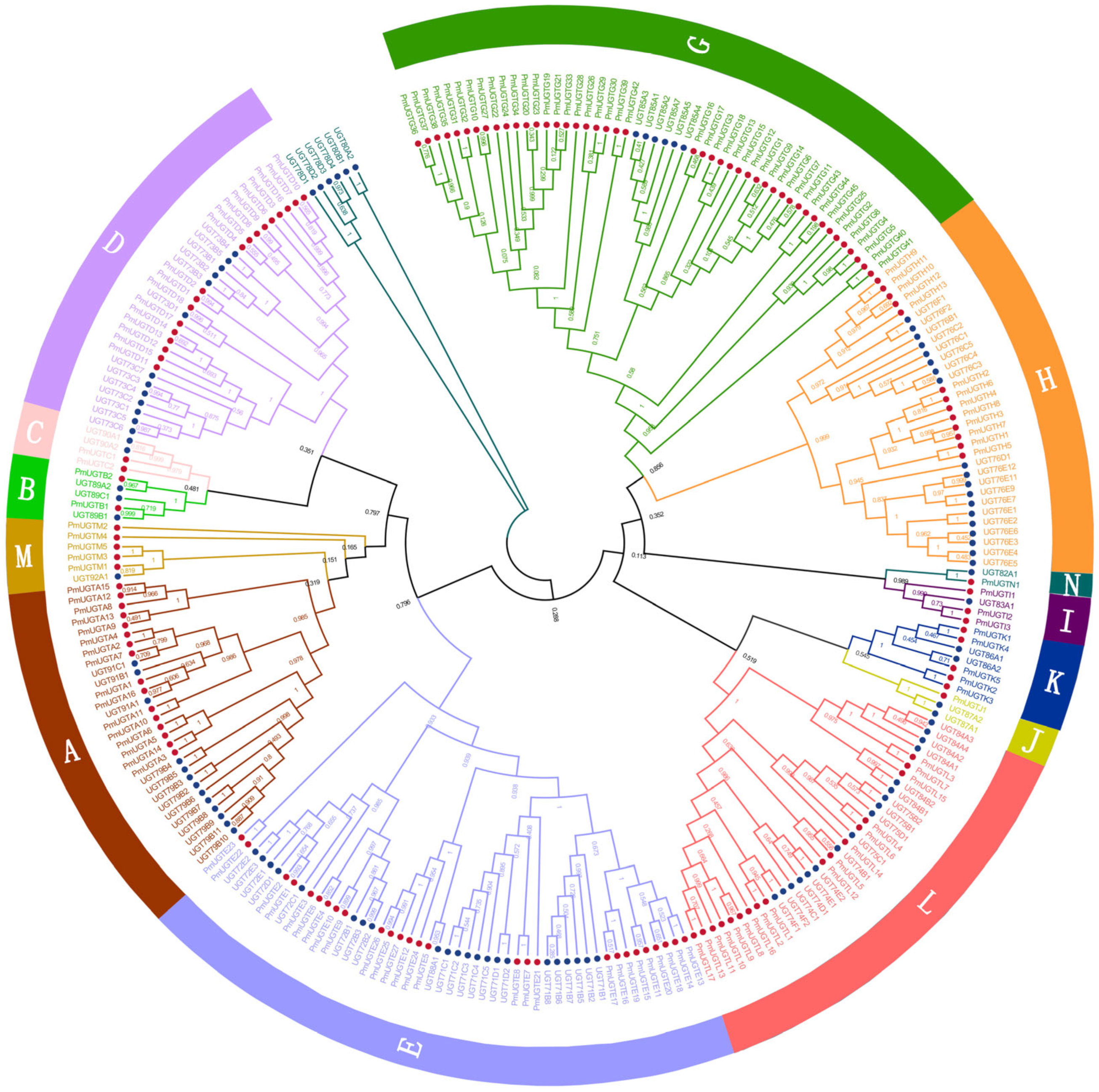
In order to comprehensively characterize the UGT gene family in P. mume, the Pfam structural model of UGT proteins and 114 UGT protein sequences in Arabidopsis thaliana were used as references for comparison and screening, and 155 members of the PmUGTs family were finally obtained. A phylogenetic tree of the UGT family in P. mume and Arabidopsis was constructed using the NJ method of MEGA11, and the results show that the UGT family genes were classified into 13 subfamilies (Figure 5), including UGTA-E and UGTG-N. The PmUGTG subfamily had the largest number of members, with 45 members, which accounted for 29.03% of the whole UGT family, and the PmUGTJ subfamily had the smallest number of members, with only 1 member. The PmUGTE and PmUGTL subfamilies had 27 and 17 members, respectively. No member of subfamily F was identified in P. mume.
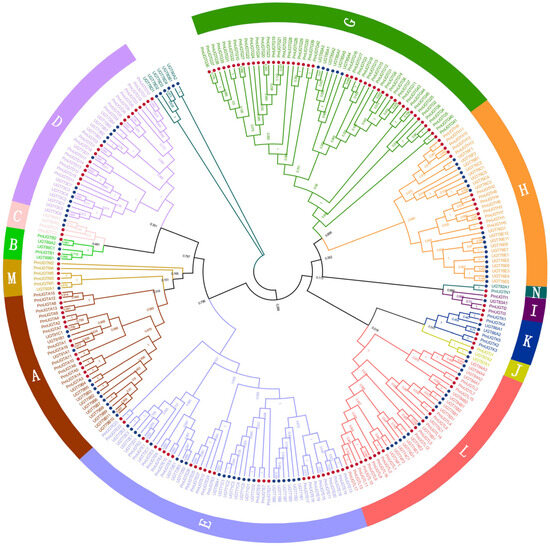
Figure 5.
Phylogenetic tree of PmUGTs and AtUGTs. Phylogenetic trees were constructed using the neighbor-joining (NJ) method and repeated 1000 times. All PmUGTs were classified into 13 subfamilies (A–E, G–N), and each subfamily is indicated by a different color.
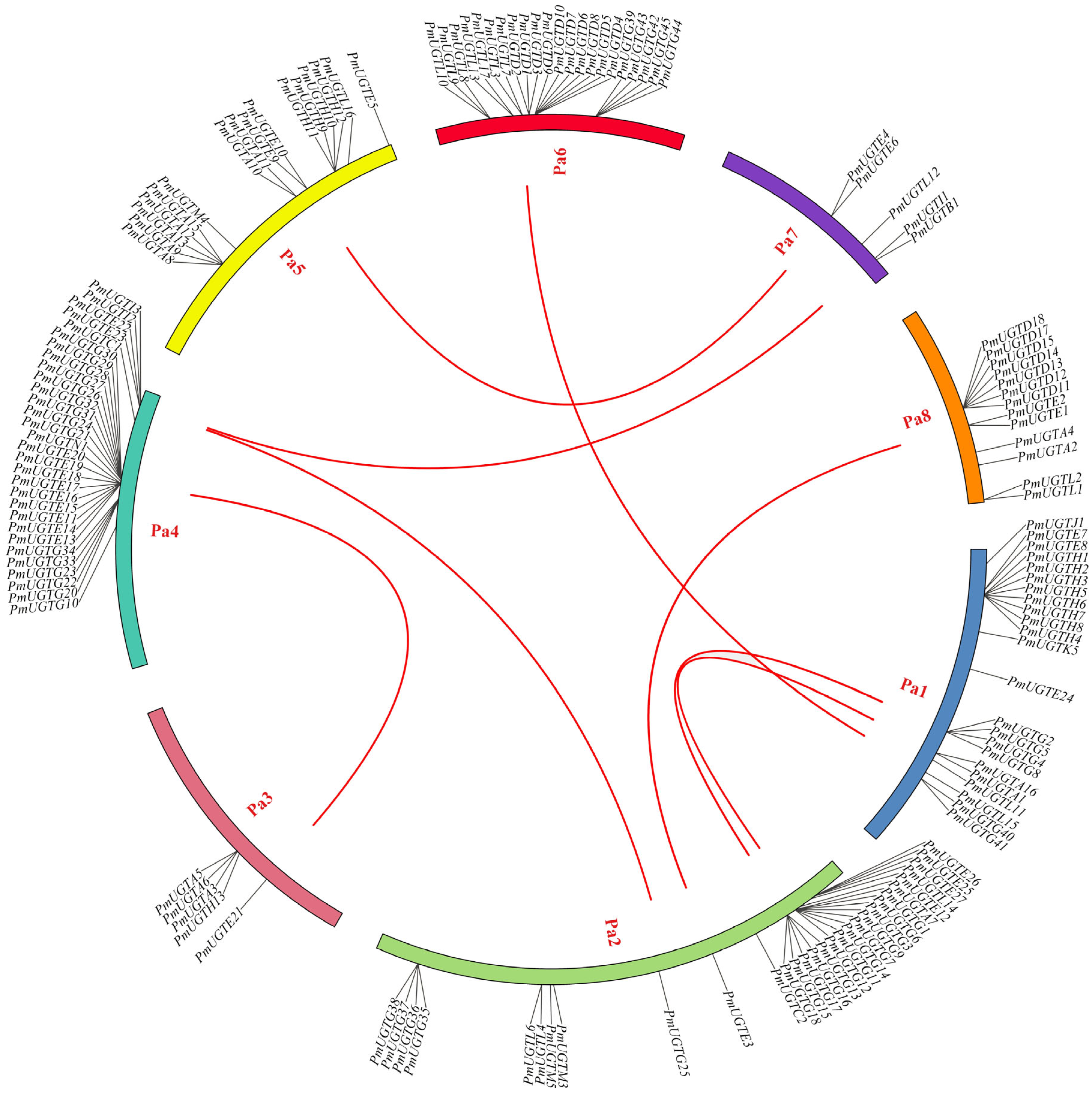
2.6. Chromosome Localization and Colinearity Analysis of the PmUGT Family
According to the chromosome localization analysis of PmUGTs (Figure 6), 144 of the 155 PmUGTs genes were distributed on eight chromosomes, and the number of genes localized on each chromosome was uneven, with the largest number of PmUGTs members on chromosomes 4 and 2, both with 30 members, and only 5 PmUGTs members on chromosomes 3 and 7. The distribution of different subfamily members on the chromosomes was irregular, with the PmUGTL members distributed on all six chromosomes, but with a maximum of seven PmUGTs members on chromosome 6. Some family members existed as gene clusters, such as PmUGTR13-20 on chromosome 4 with a total of eight E family members in one cluster and PmUGTH1-8 on chromosome 1 in one cluster, while PmUGTG′ family members existed in gene clusters on chromosomes 2, 4, and 6.
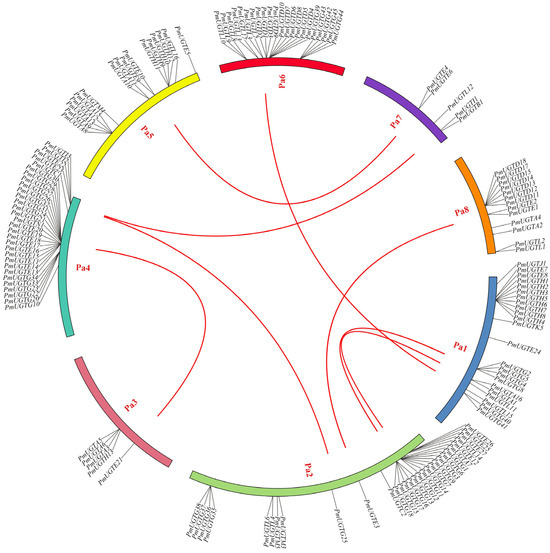
Figure 6.
The chromosome distribution and collinearity analysis of PmUGT family genes. Individual chromosomes are represented by different colors, and the red lines in the figure represent segmental repeats.
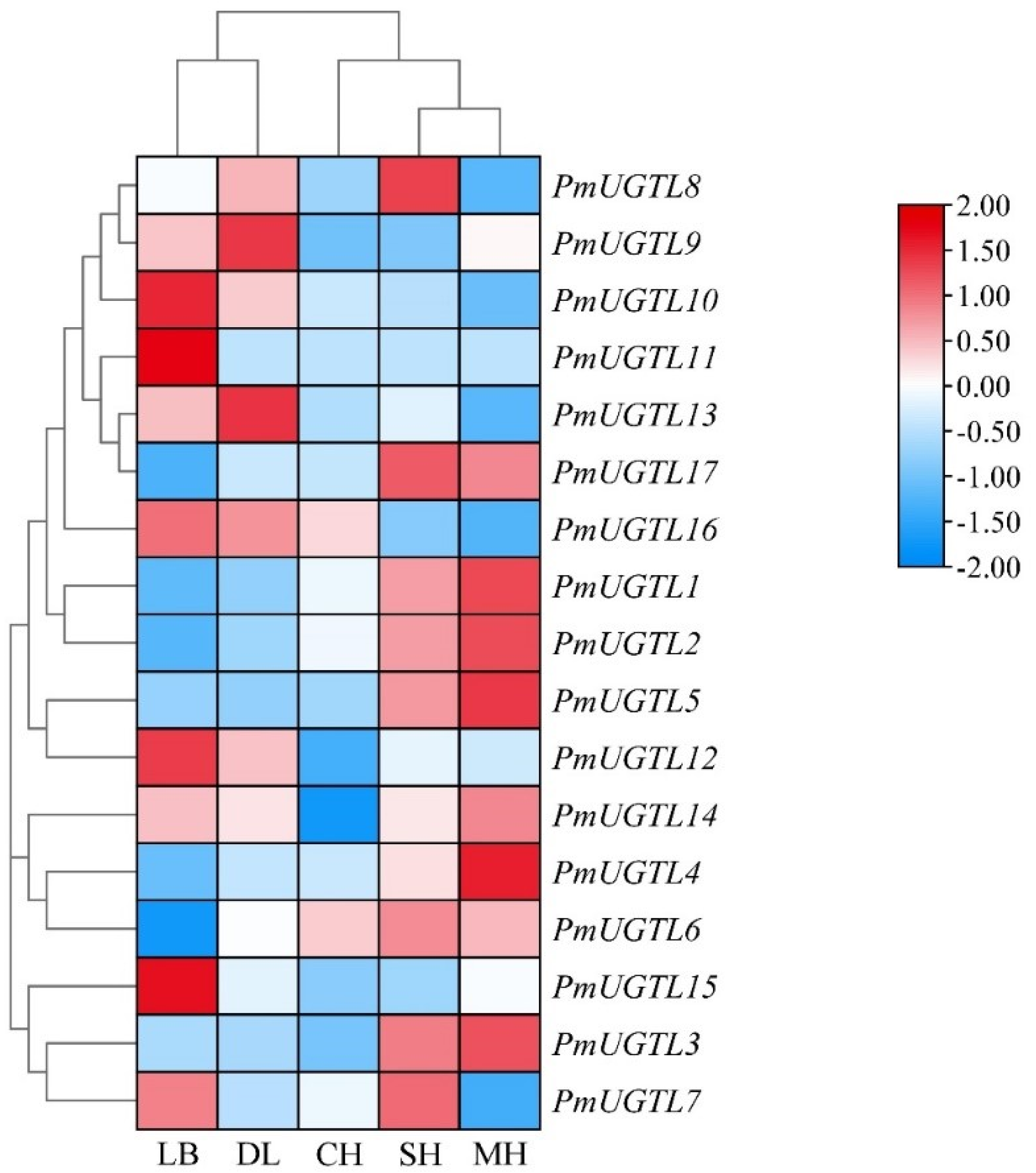
2.7. Expression Analysis of PmUGTL Genes at Different Anthesis Stages of P. mume
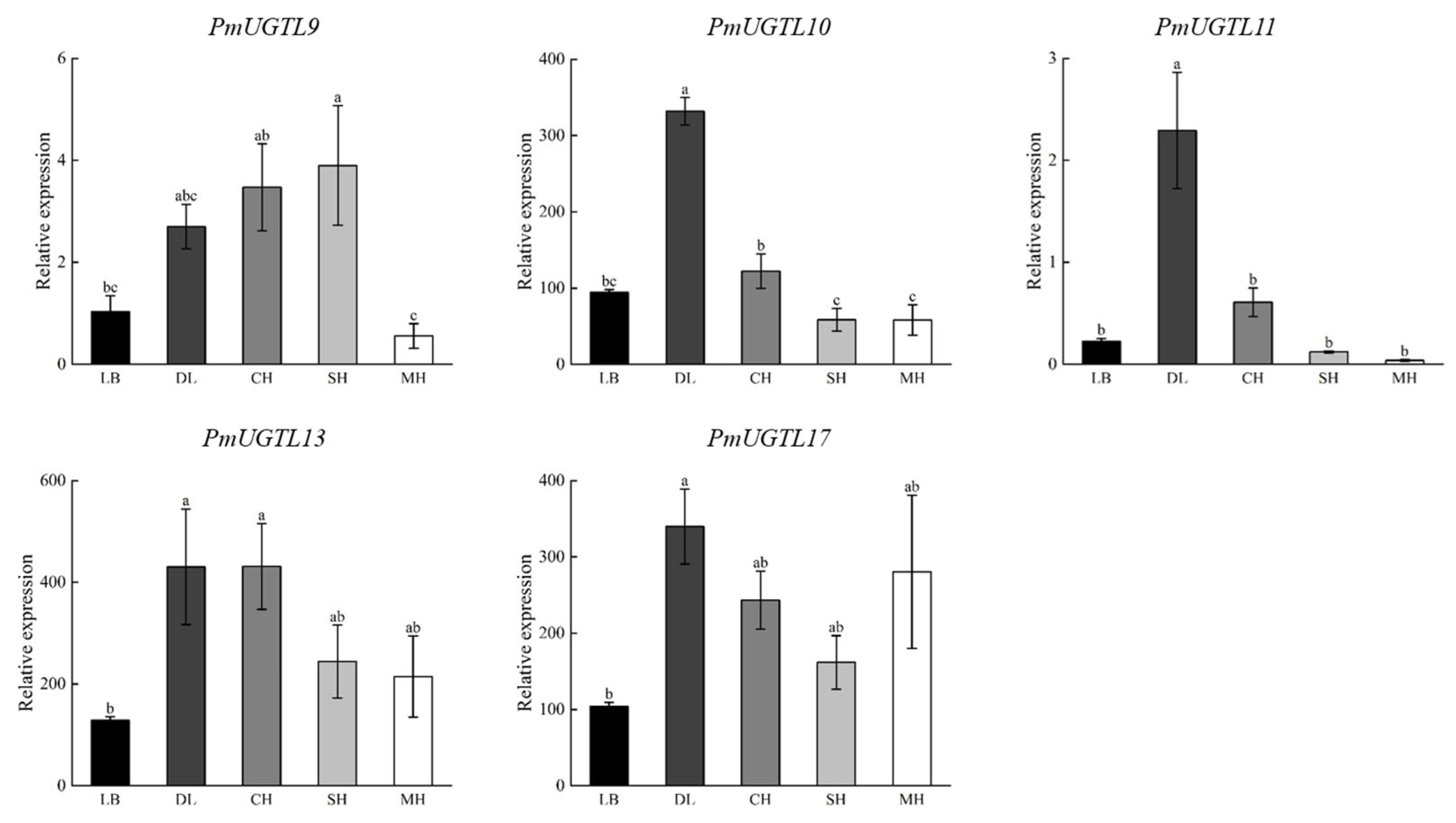
In order to further study the UGTL genes’ expression patterns at different anthesis stages of P. mume, 17 UGTL genes at five anthesis stages were visualized based on transcriptome sequencing data (Figure 7). The results show that the expressions of PmUGTL9 and PmUGTL13 were higher at the large bud stage than at other anthesis stages; that the expressions of PmUGTL10, 11, 12, and 15 were higher at the dewlap stage; and that the expression of PmUGTL16 was the highest at the primordial stage compared with other genes. PmUGTL1, 2, 3, 4, and 5 were all expressed at higher levels at the last bloom stage.
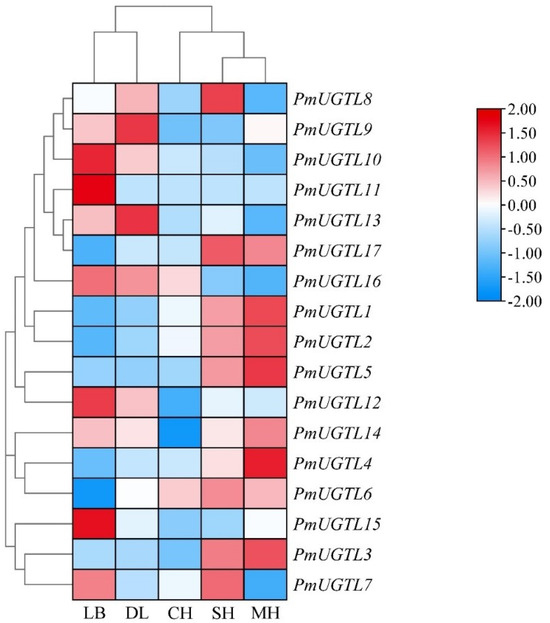
Figure 7.
Expression calorimetry of PmUGTLs gene of P. mume at different stages. The emission values were normalized through log10 transformation. The color of the heatmap ranges from blue (value, −2) to red (value, 2) in the natural logarithmic scale. Data are presented as means of three biological replicates. LB, dew petal stage; DL, large bud stage; CH, first bloom stage; SH, full bloom stage; MH, last bloom stage.
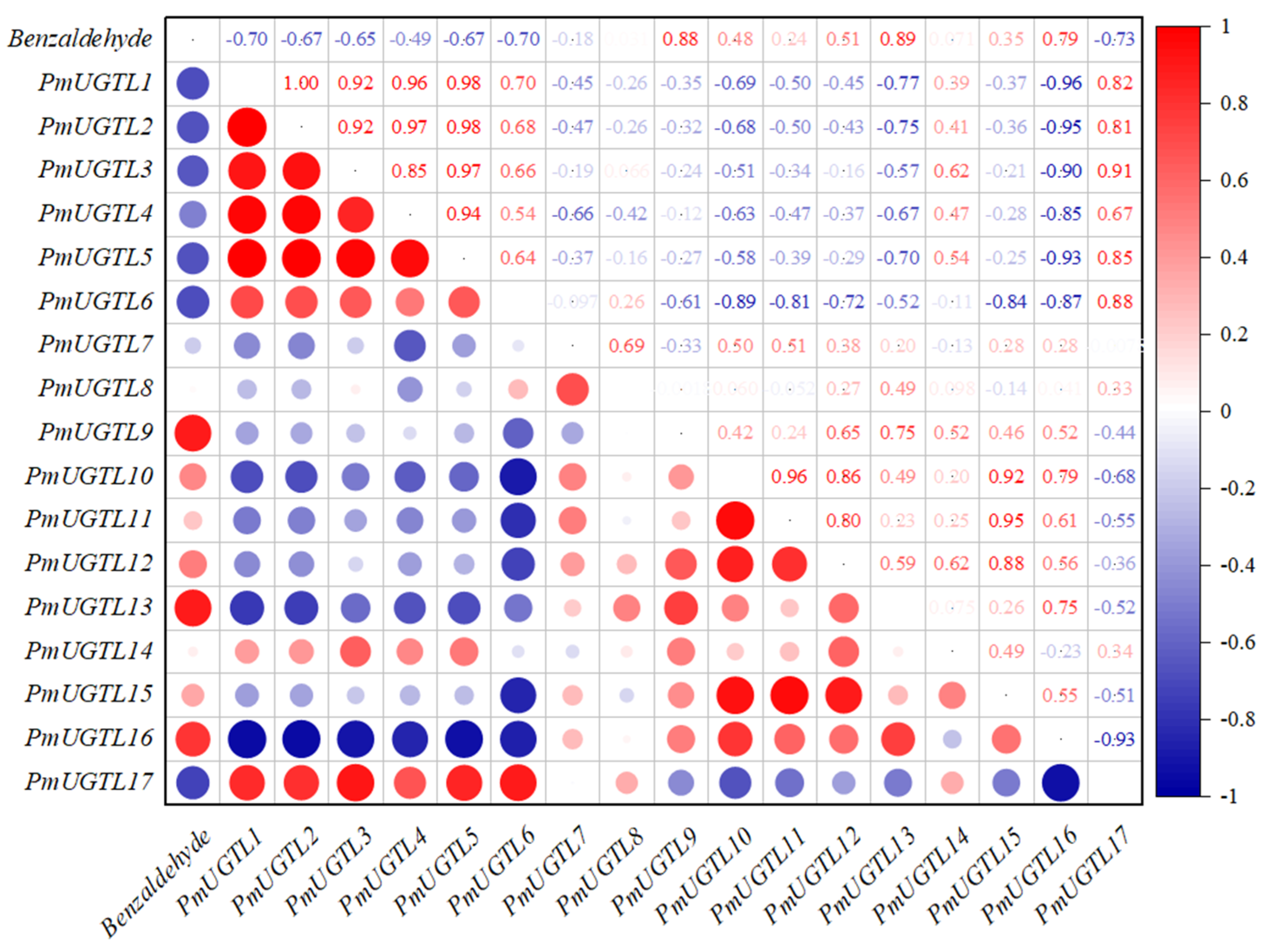
2.8. Correlation Analysis between Benzaldehyde Glycosidation and PmUGTL Gene Expression in Prunus mume
A correlation analysis between the glycosidic benzaldehyde syntheses at each period of P. mume and PmUGTL genes’ expression was conducted (Figure 8). The glycosidic benzaldehyde synthesis was strongly positively correlated with PmUGTL9 and 13 positively correlated with PmUGTL16. Notably, PmUGTL1, 2, 3, 5, and 6 were all strongly negatively correlated with glycosidic benzaldehyde synthesis, and the expression levels of PmUGTL1, 2, and 5 were all relatively high at the last flowering stage.
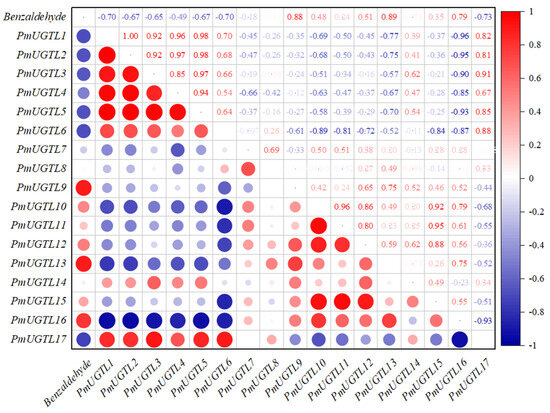
Figure 8.
Correlation analysis between synthesis efficiency of key glycosides and PmUGTL gene expression in P. mume. The emission values were normalized through log10 transformation. The color of the heatmap ranges from dark blue (value, −1) to red (value, 1) in the natural logarithmic scale. Data are presented as means of three biological replicates.
2.9. Validation of qRT-RCR of Key Genes of PmUGTLs
Combining the results of the correlation analysis, gene expression calorimetry, and changes in the period of benzaldehyde glycosylation, PmUGTL9 and 13 were identified as candidate genes, and qRT-PCR was performed for the candidate genes and their homologous genes (Figure 9). It was found that the expression levels of PmUGTL10, 13, and 17 were significantly higher than that of other genes, as PmUGTL10 and 13 showed the same trend with benzaldehyde glycosidation, and PmUGTL17 was highly expressed in the end-flowering period compared to other genes.
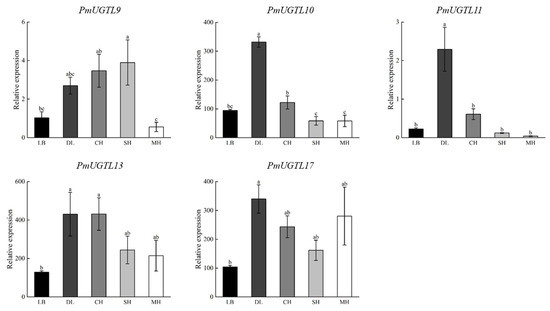
Figure 9.
qRT-RCR of key P. mume gene. Data are mean ± standard deviation (SD) calculated from three biological replicates. Vertical lines represent standard deviation. The different lowercase letters indicate significant diference at 0.05 level. LB, dew petal stage; DL, large bud stage; CH, first bloom stage; SH, full bloom stage; MH, last bloom stage.
3. Discussion
Plants emit an overabundance of volatile compounds that play different roles in plant protection when the free aromatics accumulate in the cells at a certain concentration [31,32]. Plants can also convert excess volatiles, which are toxic to cells, into GBVs as a form of protection when plants suffer abiotic or biotic stress [33]. A previous study found that benzaldehyde is prevalent in plants of the Rosaceae family and that most of them defend themselves by accumulating benzaldehyde [34]. Large amounts of amygdalin are present in peach kernels and almonds. Once the cell structure is broken down, the amygdalin in the vesicles is hydrolyzed by glycoside-degrading enzymes in the cytoplasm, producing bitter-tasting benzaldehyde and toxic prussic acid [28]. The results of this study indicate that benzaldehyde is the main glycosidic substance present in P. mume. The glycosidic and free benzaldehyde contents constituted 60.3% and 50.1% of the total amount of benzaldehyde in full-bloom petals, respectively. In comparison, the concentrations of both glycosidic and free benzaldehyde were significantly higher in the anthers than in other parts of P. mume.
The site where the synthesis of GBV for partial defense takes place is not the same as that for storage. Defensive cyanogenic glycosides can be transported to the roots via the phloem in cassava leaves [35]. Brown found that newborn leaves and ungerminated seeds of Arabidopsis had the highest concentrations of mustard oleoresin, whereas germinated seeds and senescent leaves had significantly reduced levels of mustard oil glycosides [36]. Doyle suggests that plants move defenses to the most protective tissues at different times [37]. The glycosylation of defense substances coordinates in vivo homeostasis and in vitro defense functions in plants, and defense through the hydrolysis of toxic substances is a convenient strategy for plants to cope with stress [38]. In P. mume, the anthers constitute a very small percentage by weight, but the anthers contain higher concentrations of benzaldehyde in the free and glycosidic forms. A previous study found that benzaldehyde was the component with the highest relative volatilization in the anthers of P. mume, with a volatilization of 65.20%, which was significantly higher than the remaining four parts of P. mume [27]. Therefore, we hypothesize that P. mume attracts insects by releasing aromatic substances and stores high concentrations of free and glycosidic benzaldehyde in the anthers to protect the anthers, which is effective in preventing insects from eating the anthers while pollinating them through insects.
Based on our findings, we hypothesize that P. mume stores large amounts of glycosidic and free benzaldehyde in the anthers during flowering to prevent insects from nibbling the pollen. In the present study, glycosidic and free benzaldehyde in P. mume were found to show different patterns of change over time. The free benzaldehyde content continued to decrease, while the glycosidic benzaldehyde content continued to increase and was significantly higher than at other times, starting in bud development and continuing to the large-bud stage. This suggests that there is significant benzaldehyde glycosylation during the budding phase, which is consistent with the period when GBV production is high in the rose [13]. The content of glycosidic benzaldehyde significantly decreased and showed a stable level after flowering, while the content of free benzaldehyde significantly increased and showed a stable level after flowering, which is consistent with the pattern of change of benzene GBV obtained by John et al. in aromatic tobacco [39]. During the flowering period, P. mume is able to volatilize a large number of aromatic compounds [40]. Benzaldehyde is the upstream substrate of benzyl alcohol and phenylmethyl acetate, which are the main aromatic components in the synthesis of P. mume [23]. Based on the above, we hypothesize that a large amount of glycosidic benzaldehyde can act as a reserve of aromatic precursors during the bud stage and that the glycosidic state is gradually hydrolyzed during flowering to replenish the free benzaldehyde for phenylmethyl acetate synthesis.
Widespread in plants, UGT is involved in regulating metabolites such as flavonoids, terpenoids, phenylpropanoids, and phytohormones with diverse biological functions [41]. The UGT family has been identified in several species. A total of 99 were identified in Arabidopsis thaliana [42], and 168 in peach, which is more closely related to P. mume. [43]. In our study, however, 155 UGT family genes were identified in P. mume. The P. mume UGTs were classified into subfamilies with reference to Arabidopsis thaliana. In Arabidopsis, the L subfamily recognizes phenylpropane as a substrate. A large amount of free benzaldehyde, which is structurally similar to phenylpropane compounds, was found in P. mume, we chose to target the UGTL subfamily in P. mume for further analysis. Correlating the gene expression of PmUGTLs with the content of glycosidic benzaldehyde in this study, we obtained PmUGTL9 and PmUGTL13, which had a very strong positive correlation between gene expression and synthesis efficiency, and PmUGTL1, 2, 3, 5, and 6, which all had strong negative correlations with the amount of glycosidic benzaldehyde synthesized, and all five genes were highly expressed at the time of the end of flowering, which is not consistent with the time of the massive glycosidation of benzaldehyde. UGT85A19 (Accession: B2XBQ5) in Prunus dulcis is homologous to candidate gene PmUGTL13, which is involved in the synthesis of the (R)-bitter amygdalin precursor and associated with the strong bitter flavor of almonds [44]. Benzaldehyde is a substrate for the synthesis of bittersweet amygdalin and has a characteristic bitter amygdaloid flavor. Therefore, the gene PmUGTL13 is more likely to be associated with the synthesis of the glycosidic state of benzaldehyde in P. mume.
4. Materials and Methods
4.1. Plant Materials
Multiple healthy and vigorous ten-year-old P. mume on the campus of Shanxi Agricultural University were used. During the flowering period from February to March 2022, the flowers were collected from 7 to 9 a.m. on a sunny morning at the dew petal stage (LB), large bud stage (DL), first bloom stage (CH), full bloom stage (SH), and last bloom stage (MH) (Figure 3). The full-bloom-stage flowers that were of uniform sizes were selected and rapidly divided into petals, filaments, anthers, calyx, and other parts on ice (Figure 2), and they were quickly frozen in liquid nitrogen and stored in an ultra-low temperature refrigerator at −80 °C.
4.2. Instruments and Reagents
β-D-glucosidase was purchased from Shanghai Yuanye Biotechnology Co., Ltd., Shanghai, China; methanol (chromatographic pure), ethyl acetate (chromatographic pure), citric acid, disodium hydrogen phosphate, and anhydrous sodium carbonate were obtained from Tianjin Damao Chemical Reagent Factory, Tianjin, China; concentrator plus vacuum concentrator, ultra-low temperature refrigerator, electronic balance, ultrasonic cleaner, pipette gun were obtained from Eppendorf, Hamburg, Germany; thermostatic bath was obtained from PolyScience, Niles, IL, USA; acid meter, Trace1300 gas chromatograph, Trace ISQ mass spectrometer (GC-MS), and NanoDrop 2000 nucleic acid analyzer were obtained from Thermo, Waltham, MA, USA; high-efficiency plant total RNA extraction kit, UnionScript First-strand cDNA synthesis mix for qPCR kit, and real-time fluorescence quantitative SYBR GreenⅠMaster kit were purchased from Beijing Jinsha Biological Co., Ltd., Beijing, China; LightCycler 480 quantitative PCR instrument was obtained from Roche, Basel, Switzerland.
4.3. Extraction of Glycosidic Aromatic Substances
The cinnamon glycoside extraction method was referred to and optimized, we inactivated the endogenous enzymes of P. mume to eliminate the effect of endogenous enzymes on the results [17]. The material was weighed as 0.5 g samples from the different anthesis stages, with 0.2 g samples of the different parts of P. mume, which were fully ground together with liquid nitrogen into a glass test tube. To this, 5 mL of 80% methanol solution was added, and heated with an alcohol lamp for 2 min, all of which was transferred to a 10 mL centrifuge tube for ultrasonic shaking for 15 min to assist the extraction, followed by a low-temperature centrifugation of 10,000× g rpm at 4 °C for 15 min. Then, the supernatant was partitioned into 2 mL centrifuge tubes and placed in a vacuum concentrator and then concentrated in a vacuum at 30 °C until it was completely dry to remove the methanol.
4.4. Hydrolysis of Glycosidic Aromatic Substances
The crude extracted glycosides were dissolved with 2 mL of 0.2 mol·L−1 citric acid and 0.2 mol·L−1 disodium hydrogen phosphate buffer, to which 5 U β-glucosidase was added. This was covered with 3 mL of ethyl acetate and fully extracted by using ethyl acetate in a 37 °C water bath for 24 h. The extracts were dried with anhydrous sodium carbonate, filtered with an organic system filtration membrane, and transferred to the liquid feed vials for GC–MS analysis.
4.5. GC-MS Analysis
The analysis of the endogenous components was performed using GC–MS, and the GC conditions were as follows: the carrier gas was helium, and the VF-5MS column (30 m × 0.25 mm × 0.25 μm) was used as the chromatographic column. The heating-up program was set as follows: the starting temperature was 40 °C for 2 min, the temperature was raised to 180 °C at the rate of 4 °C·min−1 and maintained for 3 min, and then the temperature was raised to 220 °C at a rate of 10 °C min−1. The MS conditions were as follows: the ionization mode was EI, 45–500 amu for the scan range for the mass-to-charge ratio (m/z), 70 eV for the electronic energy, and the GC–MS interface temperature and the ion source temperature were both 250 °C. The endogenous components in P. mume were identified using the X caliber software system(v2.0.7); the components were determined by searching through the NIST08 standard spectral library and with manual spectral analysis. The aromatic components were quantified using the external standard method.
4.6. Statistics and Analysis of Data
Three replications were performed for each sample, and the experimental data were integrated and calculated using Microsoft Excel 2019 and analyzed for significance, while Duncan’s multiple comparisons (p < 0.05) were used using SPSS 22.0 software and plotted using Origin Pro 2021.
4.7. Transcriptome Measurement of P. mume at Different Nthesis Stages
Total RNA was extracted from five flowering stages of P. mume, and the samples were set up in three sets of biological replicates for transcriptome sequencing using the Illumina HiSeqTM2500 platform (Bemac Biotech Co., Ltd., Imabari, Japan). The sequencing results were analyzed using BMK Cloud (www.biocloud.net (accessed on 1 November 2022)). The sequencing results were uploaded to the SRA database of NCBI, GenBank: PRJNA905928.
4.8. Identification of UGT Gene Family Members
P. mume whole genome data were obtained from the P. mume genome database (http://Prunusmumegenome.bjfu.edu.cn (accessed on 4 March 2023)), and UGT protein structural domain files (PF00201,UDPGT.HMM) were obtained from the Pfam protein structure database (http://pfam.xfam.org/ (accessed on 4 March 2023)) [45]. HMMER 3.0 was used to construct the UGT Hidden Markov Model and was used in the P. mume Genome Protein Database for comparative searches with an E-value of 10−5. Arabidopsis UGT transporter protein sequences were obtained from the Arabidopsis thaliana database (https://www.arabidopsis.org/ (accessed on 5 March 2023)), and the P. mume Genome Protein Database was searched with BLASTP. The resultant files were submitted to the NCBI Batch CD-search (https://www.ncbi.nlm.nih.gov/ (accessed on 8 March 2023)) database, where candidate sequences were screened for conserved structural domains, and sequences with incomplete PSPG box structures were removed.
4.9. Multiple Sequence Alignment and Subfamily Classification of the UGT Gene Family
The identified PmUGT gene candidate sequences were compared with the protein sequences of AtUGT family members using ClustalW, and the comparison results were used to construct a phylogenetic tree of the PmUGT gene family using the neighbor-joining method (NJ) in the MEGA11 software, with the bootstrap value set to 1000 times. The Poisson model was selected for the alternative model, partial deletion was used, and the default settings were used for the rest of the parameters. Referring to the distribution of Arabidopsis subfamily UGT genes on the phylogenetic tree, PmUGTs were categorized into 13 subfamilies (PmUGTA-E, PmUGTG-N).
4.10. Chromosomal Localization and Colinearity Analysis of the UGT Gene
All P. mume UGT family genes were localized in chromosomes using Circos based on P. mume genome annotation information. Sequence pairs were made within the P. mume genome using the TBtools sequence comparison tool Blast Compare 2 Seq (E-value < 10−5) to obtain homologous gene pairs, which were analyzed using the covariance analysis tool MCScanX to obtain covariance regions within the genome, and the intraspecies relationships of the UGT family genes were visualized using TBtools(v2.003).
4.11. Analysis of the Expression of UGTLs Genes in P. mume at Different Anthesis Stages with Time
Gene expression analysis was carried out using transcriptome data from five anthesis stages of P. mume. Based on the data of differentially expressed genes, the RPKM values of gene expression of PmUGTLs were extracted, and the heatmap of gene expression was drawn. A correlation analysis of the transcriptome data of P. mume at different anthesis stages with the benzaldehyde glycosidation content at different anthesis stages was performed and plotted with Origin Pro 2021.
4.12. Quantitative Expression Analysis of Candidate Genes for UGTLs
The software Primer Premier 5 was used to design the qRT-PCR primers for the key genes, and the specificity of the primers was verified using the BLAST online tool on the NCBI website. The high-efficiency plant total RNA extraction kit was selected to extract the total RNA of Prunus mume at each flowering stage, the quality of the resulting RNA was examined using NanoDrop 2000 Nucleic Acid Meter, repeated three times for each group. The unionscript first-strand cDNA synthesis mix for the qPCR kit was used for the first-strand reverse transcription, and the qRT-PCR of the key genes was performed with the real-time fluorescence quantitative SYBR GreenⅠMaster kit. The quantification was carried out by the Roche Light Cycler 480 instrument, and the UBC gene was used as an internal reference gene [46]. The reaction procedures were as follows: pre-denaturation at 95 °C for 3 min; denaturation at 94 °C for 20 s; 60 °C annealing and extension for 34 s; a total of 40 cycles. The gene relative expression level was calculated using the 2−ΔΔCt method with the P. mume dew petal stage as the control [47].
5. Conclusions
P. mume is a unique woody aromatic plant, and phenylpropanoids/benzenoids are the main components of the GBVs. The content of glycoside benzaldehyde was found to be the highest at different stages and parts of the plant, and the titer of glycoside benzaldehyde gradually increased during the bud stage and reached the highest level at the big bud stage. We identified 155 UGT family members in P. mume and divided these family members into 13 subfamilies. Furthermore, combined with the correlation analysis between the synthesis efficiency of glycosides and the expression of PmUGTL genes in the different blooming stages, our results indicate that PmUGTL9 and 13 were the key candidate genes. The expression profile of PmUGTL13 was extremely consistent with the synthesis efficiency of benzaldehyde glycosides, so we posit that PmUGTL13 is largely involved in the synthesis of benzaldehyde glycosides. This research further elucidates the important role played by benzaldehyde glycosidation in maintaining stable intracellular benzaldehyde concentrations, floral defense, and the stable volatilization of aromatic components. In addition, it provides a theoretical basis to elucidate the function of UGT family genes in P. mume during flower development, to explore the mechanism of the storage and transportation of aromatic compounds in flower tissues, and to exploit industrial applications of aromatic products from P. mume.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants13050703/s1, Table S1: Gene primer sequence, Table S2: Absolute content of glycosidic aromatic substances at different anthesis stages of P. mume, Table S3: Absolute content of glycosidic aromatic substances in different parts of P. mume, Table S4: Physicochemical properties and prediction of subcellular location in P. mume.
Author Contributions
Conceptualization, R.H. and H.J.; methodology, H.J.; software, L.F., X.L. and J.W.; validation, L.F. and X.L.; data curation, X.G.; writing—original draft preparation, H.J. and X.G.; writing—review and editing, H.J. and X.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 31870696.
Data Availability Statement
Data reported are available in Section 2 and Supplementary Materials.
Acknowledgments
The authors appreciate those contributors who make the genome data accessible in public databases.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, L.; Fang, X.; Yang, C.; Li, J.; Chen, X. Biosynthesis and regulation of secondary terpenoid metabolism in plants. Sci. Sin. 2013, 43, 1030–1046. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Shi, L.; Liu, Y. Application of functional genomic and metabolomic techniques to the studies on biosynthesis and regulation of plant secondary metabolites. J. Beijing For. Univ. 2007, 29, 153–159. [Google Scholar] [CrossRef]
- Lai, C.; Zhang, J.; Shentu, X.; Hao, P.; Pang, K.; Yu, X. Review of how secondary metabolites defend plants against herbivorous insects. Chin. J. Appl. Entomol. 2022, 59, 969–978. [Google Scholar] [CrossRef]
- Lu, X.; Jiang, W.; Tang, K. Research progress of jasmonates’ regulation on the plant secondary metabolis. J. Shanghai Jiaotong Univ. (Agric. Sci.) 2011, 29, 87–91. [Google Scholar] [CrossRef]
- Davide, S.; Stéphanie, M.M.; Tristan, R.; Pierre, W.T.; Jonathan, B.; Jean-Pierre, M.; Jean-Michel, M.; de Gilles, R. Centrifugal partition chromatography applied to the isolation of oak wood aroma precursors. Food Chem. 2013, 141, 2238–2245. [Google Scholar] [CrossRef]
- Shoji, O.; Eiichiro, O.; Manabu, H.; Jun, M.; Koujirou, T.; Hiromi, T.; Yukie, O.; Hideo, D.; Tatsuo, A.; Kenji, M.; et al. Volatile glycosylation in tea plants: Sequential glycosylations for the biosynthesis of aroma β-Primeverosides are catalyzed by two camellia sinensis glycosyltransferases. Plant Physiol. 2015, 168, 464–477. [Google Scholar] [CrossRef]
- Qin, J.; Sun, C.; Zhang, M.; Wang, Y. Classification, function and evolution of plant UDP-glycosyltransferase. Genom. Appl. Biol. 2018, 37, 440–450. [Google Scholar] [CrossRef]
- Keiko, Y.S.; Kousuke, H. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011, 66, 182–193. [Google Scholar] [CrossRef]
- Alexandra, L.; Denise, H.; Rosamond, G.J.; Gilu, L.G.; Luisa, E.; Eng-Kiat, L.; Fabián, E.V.; Dianna, J.B. The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J. 2006, 48, 286–295. [Google Scholar] [CrossRef]
- Florian, P.S. The evolution of floral scent and insect chemical communication. Ecol. Lett. 2010, 13, 643–656. [Google Scholar] [CrossRef]
- Tang, F.; Chen, Y.; Li, D.; Huang, N. Allelopathic effects of volatiles from Eucalyptus grandis × E. urophylla’s leaves on seed germination and seedling growth of three kinds of plants. Med. Plant. 2014, 5, 33. [Google Scholar]
- Cai, J.; Liu, B.; Ling, P.; Su, Q. Analysis of free and bound volatiles by gas chromatography and gas chromatography–mass spectrometry in uncased and cased tobaccos. J. Chromatogr. A 2002, 947, 267–275. [Google Scholar] [CrossRef]
- Francis, M.; Allcock, C. Geraniol β-D-glucoside; occurrence and synthesis in rose flowers. Phytochemistry 1969, 8, 1339–1347. [Google Scholar] [CrossRef]
- Williams, P.J.; Strauss, C.R.; Wilson, B.; Massy-Westropp, R.A. Novel monoterpene disaccharide glycosides of vitis vinifera grapes and wines. Phytochemistry 1982, 21, 2013–2020. [Google Scholar] [CrossRef]
- Guo, W.; Sakata, K.; Watanabe, N.; Nakajima, R.; Yagi, A.; Ina, K.; Luo, S. Geranyl 6-O-β-d-xylopyranosyl-β-d-glucopyranoside isolated as an aroma precursor from tea leaves for oolong tea. Phytochemistry 1993, 33, 1373–1375. [Google Scholar] [CrossRef] [PubMed]
- Gueguen, Y.; Chemardin, P.; Janbon, G.; Arnaud, A.; Galzy, P. A very efficient β-glucosidase catalyst for the hydrolysis of flavor precursors of wines and fruit juices. J. Agric. Food Chem. 1996, 44, 2336–2340. [Google Scholar] [CrossRef]
- Yuan, J.; Luo, J.; Zhu, L.; Xi, W.; Zeng, X.; Xiong, K.; Wang, C.; Zheng, R. Free and glycosylated aroma components in petals of three Osmanthus fragrans cultivars. Sci. Silvae Sin. 2021, 57, 33–42. [Google Scholar] [CrossRef]
- Zhou, Y.; Zeng, L.; Gui, J.; Liao, Y.; Li, J.; Tang, J.; Meng, Q.; Dong, F.; Yang, Z. Functional characterizations of β-glucosidases involved in aroma compound formation in tea (Camellia sinensis). Food Res. Int. 2017, 96, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Jiang, Q.; Lin, Q.; Ma, Q.; Wang, L.; Weng, S.; Huang, G.; Li, L.; Chen, F. Enzymatic hydrolysis and auto-isomerization during β-glucosidase treatment improve the aroma of instant white tea infusion. Food Chem. 2021, 342, 128565. [Google Scholar] [CrossRef] [PubMed]
- Tikunov, Y.M.; Molthoff, J.; de Vos, R.C.; Beekwilder, J.; van Houwelingen, A.; van der Hooft, J.J.; Nijenhuis-de Vries, M.; Labrie, C.W.; Verkerke, W.; van de Geest, H. Non-smoky glycosyltransferase1 prevents the release of smoky aroma from tomato fruit. Plant Cell 2013, 25, 3067–3078. [Google Scholar] [CrossRef]
- Zhang, T.; Huo, T.; Ding, A.; Hao, R.; Wang, J.; Cheng, T.; Bao, F.; Zhang, Q. Genome-wide identification, characterization, expression and enzyme activity analysis of coniferyl alcohol acetyltransferase genes involved in eugenol biosynthesis in Prunus mume. PLoS ONE 2019, 10, e0223974. [Google Scholar] [CrossRef]
- Zhao, Y.; Pan, H.; Zhang, Q.; Sun, M.; Pan, C. Studies on the volatile constituents from cultivars of Prunus mume. J. Trop. Subtrop. Bot. 2010, 18, 310–315. [Google Scholar] [CrossRef]
- Hao, R.; Zhang, Q.; Yang, W.; Wang, J.; Pan, H.; Cheng, T. Study on the difference in characteristic scent between Prunus mume and its interspecific hybrids. J. Nucl. Agric. Sci. 2014, 28, 808–816. [Google Scholar] [CrossRef]
- Hu, Z.; Shen, Y.; Luo, Y.; Shen, F.; Gao, H.; Gao, R. Aldehyde volatiles emitted in succession from mechanically damaged leaves of poplar cuttings. J. Plant Biol. 2008, 51, 269–275. [Google Scholar] [CrossRef]
- Liu, T.; Zou, L.; Tian, D.; Can, Q.; Zhu, M.; Mo, M.; Zhang, K. Proteomic changes in Arthrobotrys oligospora conidia in response to benzaldehyde-induced fungistatic stress. J. Proteom. 2019, 192, 358–365. [Google Scholar] [CrossRef]
- Ullah, I.; Khan, A.L.; Ali, L.; Khan, A.R.; Waqas, M.; Hussain, J.; Lee, I.J.; Shin, J.H. Benzaldehyde as an insecticidal, antimicrobial, and antioxidant compound produced by Photorhabdus temperata M1021. J. Microbiol. 2015, 53, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Hao, R.; Yang, S.; Zhang, Z.; Zhang, Y.; Chang, J.; Qiu, C. Identification and specific expression patterns in flower organs of ABCG genes related to floral scent from Prunus mume. Sci. Hortic. 2021, 288, 110218. [Google Scholar] [CrossRef]
- János, V. Plant cyanogenic glycosides. Toxicon 2000, 38, 11–36. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Y.; Qi, L.; Wan, X. Antifungal activities of major tea leaf volatile constituents toward Colletorichum camelliae Massea. J. Agric. Food Chem. 2006, 54, 3936–3940. [Google Scholar] [CrossRef]
- Liang, Z.; Fang, Z.; Pai, A.; Luo, J.; Gan, R.; Gao, Y.; Lu, J.; Zhang, P. Glycosidically bound aroma precursors in fruits: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2022, 62, 215–243. [Google Scholar] [CrossRef]
- Xie, Y.; Tian, L.; Han, X.; Yang, Y. Research advances in allelopathy of volatile organic compounds (VOCs) of plants. Horticulturae 2021, 7, 278. [Google Scholar] [CrossRef]
- Nicholas, J.B.; Steven, J.R. C6-volatiles derived from the lipoxygenase pathway induce a subset of defense-related genes. Plant J. 1998, 16, 561–569. [Google Scholar] [CrossRef]
- Axel, M.; Wilhelm, B. Plant defense against herbivores: Chemical aspects. Annu. Rev. Plant Biol. 2012, 63, 431–450. [Google Scholar] [CrossRef]
- Ilker, D.; Murat, K. Prulaurasin content of leaves, kernels and pulps of Prunus lauracerasus L. (Cherry Laurel) during ripening. J. Res. Pharm. 2019, 23, 69–75. [Google Scholar] [CrossRef]
- Morten, E.J.; Hussam, H.N.; Barbara, A.H. Transport of defense compounds from source to sink: Lessons learned from glucosinolates. Trends Plant Sci. 2015, 20, 508–514. [Google Scholar] [CrossRef]
- Paul, D.B.; Jim, G.T.; Michael, R.; Jonathan, G. Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 2003, 62, 471–481. [Google Scholar] [CrossRef]
- Doyle, M. Adaptive patterns in alkaloid physiology. Am. Nat. 1974, 108, 305–320. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, S.; Yang, J.; Kang, C.; Huang, L.; Guo, L. Glycosylation of plant secondary metabolites: Regulating from chaos to harmony. Environ. Exp. Bot. 2022, 194, 104703. [Google Scholar] [CrossRef]
- John, H.L.; Thomas, R.H.; Harold, R.B.; Roger, A.A.; David, F.H. Glycosidically bound volatile components of Nicotiana sylvestris and N. Suaveolens flowers. Phytochemistry 1992, 31, 1537–1540. [Google Scholar] [CrossRef]
- Hao, R.; Chang, J.; Qiu, C.; Yang, S. An identification and expression analysis of the ABCG genes related to benzaldehyde transportation among three Prunus species. Horticulturae 2022, 8, 475. [Google Scholar] [CrossRef]
- Eng-Kiat, L.; Dianna, J.B. A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 2004, 23, 2915–2922. [Google Scholar] [CrossRef]
- Yi, L.; Sandie, B.; Eng-Kiat, L.; Dianna, J.B. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana* 210. J. Biol. Chem. 2001, 276, 4338–4343. [Google Scholar] [CrossRef]
- Wu, B.; Gao, L.; Gao, J.; Xu, Y.; Liu, H.; Cao, X.; Zhang, B.; Chen, K. Genome-wide identification, expression patterns, and functional analysis of UDP glycosyltransferase family in peach (Prunus persica L. Batsch). Front. Plant Sci. 2017, 8, 389. [Google Scholar] [CrossRef] [PubMed]
- Tricia, K.F.; Abbas, Y.; Michelle, G.W.; Jennifer, R.G.; Brent, N.K.; Margaret, S.; Christopher, M.F. A seed coat cyanohydrin glucosyltransferase is associated with bitterness in almond (Prunus dulcis) kernels. Funct. Plant Biol. 2008, 35, 236–246. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R. Pfam: A domain-centric method for analyzing proteins and proteomes. Comp. Genom. 2007, 396, 43–58. [Google Scholar] [CrossRef]
- Wang, T.; Lu, J.; Xu, Z.; Yang, W.; Wang, J.; Cheng, T.; Zhang, Q. Selection of suitable reference genes for miRNA expression normalization by qRT-PCR during flower development and different genotypes of Prunus mume. Sci. Hortic. 2014, 169, 130–137. [Google Scholar] [CrossRef]
- Mohd, A.; Glyn, M.; Sibte, H. Analysis of rpoS and bolA gene expression under various stress-induced environments in planktonic and biofilm phase using 2−ΔΔCT method. Mol. Cell. Biochem. 2011, 357, 275–282. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).