Molecular Characterization Analysis and Adaptive Responses of Spodoptera frugiperda (Lepidoptera: Noctuidae) to Nutritional and Enzymatic Variabilities in Various Maize Cultivars
Abstract
1. Introduction
2. Results
2.1. Molecular Characterization Analysis of Host Strain of S. frugiperda
2.2. Effects of Various Maize Varieties on Nutritional Indices of 3rd Instar Larvae of S. frugiperda
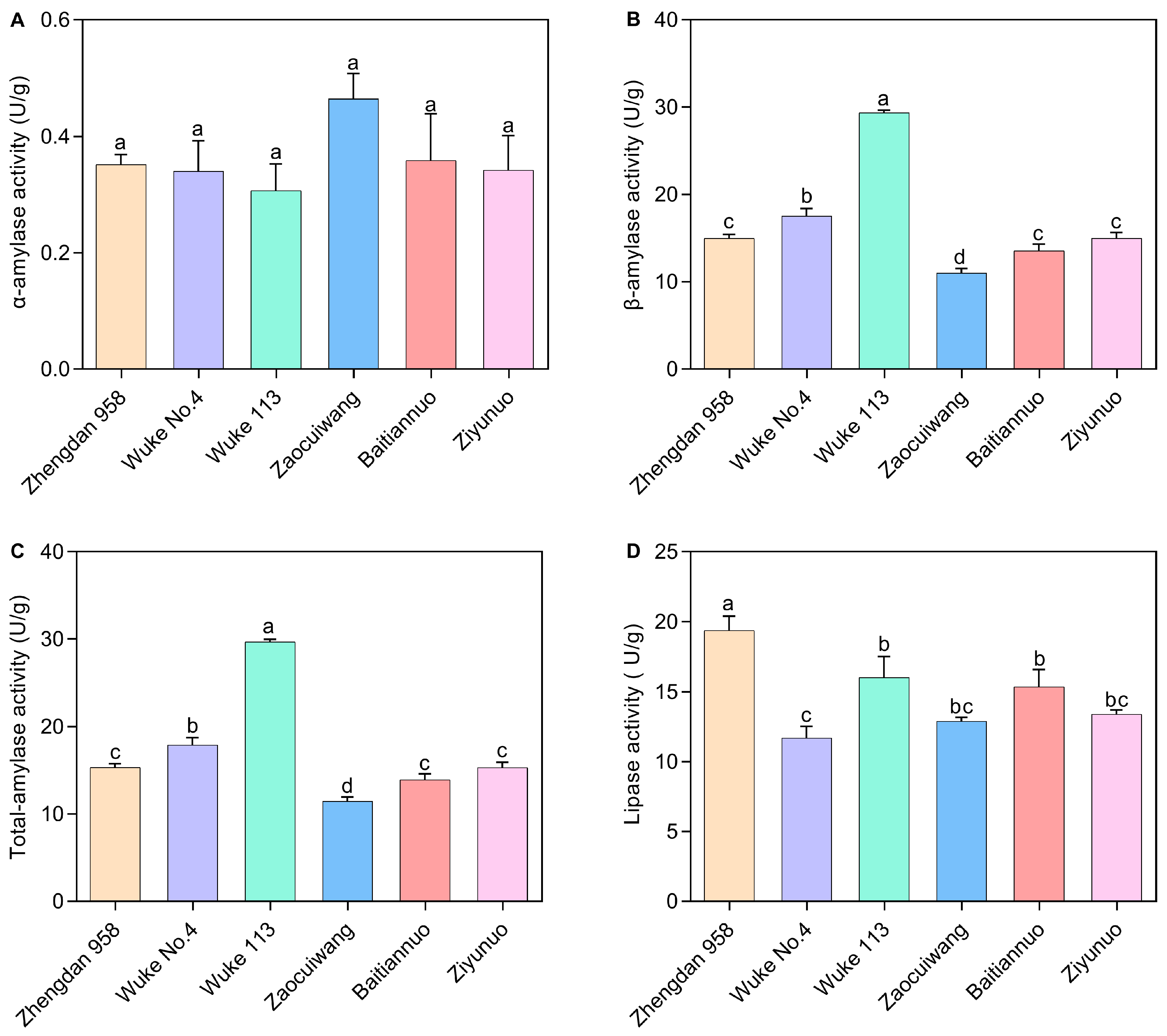
2.3. Effects of Six Maize Varieties on the Digestive Enzyme Activity of S. frugiperda
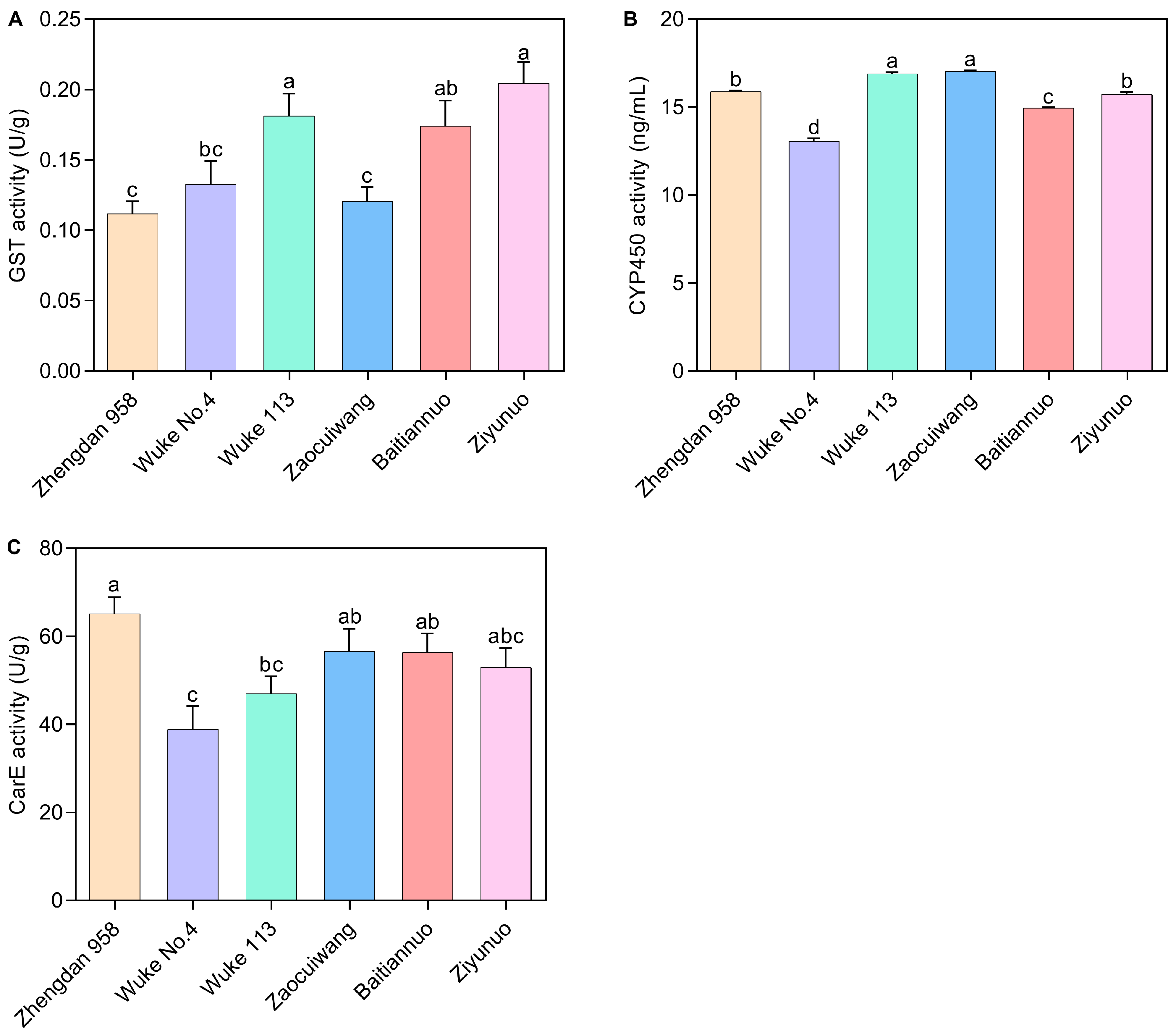
2.4. Effects of Six Maize Varieties on Detoxification Enzyme Activity of S. frugiperda
2.5. Correlation among Chemical Substances in Maize Leaves, the Nutritional Indices, Digestive, and Detoxifying Enzymes of S. frugiperda
3. Discussion
4. Materials and Methods
4.1. Insect Rearing
4.2. Host Plants and Growth Condition
4.3. Host Strain Identification and Sequence Alignment
4.4. Assessment of Nutritional Indices of 3rd Instar Larvae of S. frugiperda on Various Maize Varieties
4.5. Measurement of Digestive Enzyme Activity
4.5.1. Midgut Dissection and Enzyme Extraction
4.5.2. Amylase Activity Assay
4.5.3. Lipase Activity Assay
4.6. Determination of Detoxification Enzyme Activity
4.6.1. Enzyme Solution Preparation
4.6.2. Glutathione-S-Transferase (GST) Activity Assay
4.6.3. CarE Activity Determination
4.6.4. CYP450 Activity Determination
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goergen, G.; Kumar, P.L.; Sankung, S.B.; Togola, A.; Tamò, M. First report of outbreaks of the fall armyworm Spodoptera frugiperda (JE Smith) (Lepidoptera, Noctuidae), a new alien invasive pest in West and Central Africa. PLoS ONE 2016, 11, e165632. [Google Scholar] [CrossRef]
- Otuka, A. Prediction of the Overseas Migration of the Fall Armyworm, Spodoptera frugiperda, to Japan. Insects 2023, 14, 804. [Google Scholar] [CrossRef]
- Yadava, P.; Abhishek, A.; Singh, R.; Singh, I.; Kaul, T.; Pattanayak, A.; Agrawal, P.K. Advances in maize transformation technologies and development of transgenic maize. Front. Plant Sci. 2017, 7, 1949. [Google Scholar] [CrossRef]
- Scott, P.; Pratt, R.C.; Hoffman, N.; Montgomery, R. Specialty corns. In Corn; Elsevier: Amsterdam, The Netherlands, 2019; pp. 289–303. [Google Scholar]
- Montezano, D.G.; Sosa-Gómez, D.R.; Specht, A.; Roque-Specht, V.F.; Sousa-Silva, J.C.; Paula-Moraes, S.D.; Peterson, J.A.; Hunt, T.E. Host plants of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas. Afr. Entomol. 2018, 26, 286–300. [Google Scholar] [CrossRef]
- Prasanna, B.M.; Bruce, A.; Beyene, Y.; Makumbi, D.; Gowda, M.; Asim, M.; Martinelli, S.; Head, G.P.; Parimi, S. Host plant resistance for fall armyworm management in maize: Relevance, status and prospects in Africa and Asia. Theor. Appl. Genet. 2022, 135, 3897–3916. [Google Scholar] [CrossRef]
- Guo, J.; Zhao, J.; He, K.; Zhang, F.; Wang, Z. Potential invasion of the crop-devastating insect pest fall armyworm Spodoptera frugiperda to China. Plant Prot. 2018, 44, 1–10. [Google Scholar]
- Cui, L.; Rui, C.H.; Li, Y.P.; Wang, Q.Q.; Yang, D.B.; Yan, X.J.; Guo, Y.W.; Yuan, H.Z. Research and application of chemical control technology against Spodoptera frugiperda (Lepidoptera: Noctuidae) in foreign countries. Plant Prot. 2019, 45, 7–13. [Google Scholar]
- Wu, P.; Wu, F.; Fan, J.; Zhang, R. Potential economic impact of invasive fall armyworm on mainly affected crops in China. J. Pest. Sci. 2021, 94, 1065–1073. [Google Scholar] [CrossRef]
- Nagoshi, R.N.; Meagher, R.L. Behavior and distribution of the two fall armyworm host strains in Florida. Fla. Entomol. 2004, 87, 440–449. [Google Scholar] [CrossRef]
- Nagoshi, R.N. The fall armyworm triose phosphate isomerase (Tpi) gene as a marker of strain identity and inter strain mating. Ann. Entomol. Soc. Am. 2010, 103, 283–292. [Google Scholar] [CrossRef]
- Lu, Y.J.; Kochert, G.D.; Isenhour, D.J.; Adang, M.J. Molecular characterization of a strain-specific repeated DNA sequence in the fall armyworm Spodoptera frugiperda (Lepidoptera: Noctuidae). Insect. Mol. Biol. 1994, 3, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Dumas, P.; Legeai, F.; Lemaitre, C.; Scaon, E.; Orsucci, M.; Labadie, K.; Gimenez, S.; Clamens, A.; Henri, H.; Vavre, F. Spodoptera frugiperda (Lepidoptera: Noctuidae) host-plant variants: Two host strains or two distinct species? Genetica 2015, 143, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Nagoshi, R.N.; Murúa, M.G.; Hay-Roe, M.; Juárez, M.L.; Willink, E.; Meagher, R.L. Genetic characterization of fall armyworm (Lepidoptera: Noctuidae) host strains in Argentina. J. Econ. Entomol. 2012, 105, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jin, M.H.; Zhang, D.D.; Jiang, Y.Y.; Liu, J.; Wu, K.M.; Xiao, Y.T. Molecular identification of invasive fall armyworm Spodoptera frugiperda in Yunnan Province. Plant Prot. 2019, 45, 19–24. [Google Scholar]
- Li, X.; Garvey, M.; Kaplan, I.; Li, B.; Carrillo, J. Domestication of tomato has reduced the attraction of herbivore natural enemies to pest-damaged plants. Agr. Forest. Entomol. 2018, 20, 390–401. [Google Scholar] [CrossRef]
- Zhang, Y.; Lou, Y. Research progress in chemical interactions between plants and phytophagous insects. J. Appl. Ecol. 2020, 31, 2151–2160. [Google Scholar]
- Carrasco, D.; Larsson, M.C.; Anderson, P. Insect host plant selection in complex environments. Curr. Opin. Insect. Sci. 2015, 8, 1–7. [Google Scholar] [CrossRef]
- Hwang, S.Y.; Liu, C.H.; Shen, T.C. Effects of plant nutrient availability and host plant species on the performance of two Pieris butterflies (Lepidoptera: Pieridae). Biochem. Syst. Ecol. 2008, 36, 505–513. [Google Scholar] [CrossRef]
- Karlsson Green, K. The effects of host plant species and larval density on immune function in the polyphagous moth Spodoptera littoralis. Ecol. Evol. 2021, 11, 10090–10097. [Google Scholar] [CrossRef]
- Gong, B.; Zhang, G. Interactions between plants and herbivores: A review of plant defense. Acta Phytoecol. Sin. 2014, 34, 325–336. [Google Scholar] [CrossRef]
- Wang, W.; He, P.; Liu, T.; Jing, X.; Zhang, S. Comparative studies of ovipositional preference, larval feeding selectivity, and nutritional indices of Spodoptera frugiperda (Lepidoptera: Noctuidae) on 6 crops. J. Econ. Entomol. 2023, 116, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.M.; Jin, R.H.; Guo, Z.P.; Cai, T.W.; Zhang, Y.H.; Gao, J.Y.; Huang, G.Y.; Wan, H.; He, S.; Xie, Y.L. Insecticide susceptibility and mechanism of Spodoptera frugiperda on different host plants. J. Agr. Food. Chem. 2022, 70, 11367–11376. [Google Scholar] [CrossRef]
- Xie, W.; Zhi, J.R.; Ye, J.Q.; Zhou, Y.M.; Li, C.; Liang, Y.J.; Yue, W.B.; Li, D.Y.; Zeng, G.; Hu, C.X. Age-stage, two-sex life table analysis of Spodoptera frugiperda (JE Smith) (Lepidoptera: Noctuidae) reared on maize and kidney bean. Chem. Biol. Technol. Agr. 2021, 8, 44. [Google Scholar] [CrossRef]
- Scriber, J.M.; Slansky Jr, F. The nutritional ecology of immature insects. Annu. Rev. Entomol. 1981, 26, 183–211. [Google Scholar] [CrossRef]
- Lu, Z.H.; He, S.Q.; Guo, J.; Chen, Y.P.; Hu, X.Q.; Li, H.; Xie, Q.; Gui, F.R. Effects of host plants on activities of three groups of enzymes in Spodoptera frugiperda (Smith)larvae. J. South. Agric. 2020, 51, 2461–2469. [Google Scholar]
- Heidel-Fischer, H.M.; Vogel, H. Molecular mechanisms of insect adaptation to plant secondary compounds. Curr. Opin. Insect. Sci. 2015, 8, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Li, X.C.; Schuler, M.A.; Berenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253. [Google Scholar] [CrossRef]
- Moreau, J.; Benrey, B.; Thiéry, D. Grape variety affects larval performance and also female reproductive performance of the European grapevine moth Lobesia botrana (Lepidoptera: Tortricidae). Bull. Entomol. Res. 2006, 96, 205–212. [Google Scholar] [CrossRef]
- Awmack, C.S.; Leather, S.R. Host plant quality and fecundity in herbivorous insects. Annu. Rev. Entomol. 2002, 47, 817–844. [Google Scholar] [CrossRef]
- Altaf, N.; Idrees, A.; Ullah, M.I.; Arshad, M.; Afzal, A.; Afzal, M.; Rizwan, M.; Li, J. Biotic potential induced by different host plants in the fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae). Insects 2022, 13, 921. [Google Scholar] [CrossRef]
- Chiriboga Morales, X.; Tamiru, A.; Sobhy, I.S.; Bruce, T.J.; Midega, C.A.; Khan, Z. Evaluation of African maize cultivars for resistance to fall armyworm Spodoptera frugiperda (JE Smith) (Lepidoptera: Noctuidae) larvae. Plants 2021, 10, 392. [Google Scholar] [CrossRef] [PubMed]
- He, L.M.; Wang, T.L.; Chen, Y.C.; Ge, S.S.; Wyckhuys, K.A.G.; Wu, K.M. Larval diet affects development and reproduction of East Asian strain of the fall armyworm, Spodoptera frugiperda. J. Integr. Agr. 2021, 20, 736–744. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, X.G.; Lv, G.Q.; Hao, X.Z.; Li, S.H.; Li, G.P.; Feng, H.Q. Comparison of population fitness of Spodoptera frugiperda (Lepidoptera: Noctuidae) feeding on wheat and different varieties of maize. Plant Prot. 2020, 46, 126–131. [Google Scholar]
- De La Rosa-Cancino, W.; Rojas, J.C.; Cruz-Lopez, L.; Castillo, A.; Malo, E.A. Attraction, feeding preference, and performance of Spodoptera frugiperda larvae (Lepidoptera: Noctuidae) reared on two varieties of maize. Environ. Entomol. 2016, 45, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Omoto, C.; Bernardi, O.; Salmeron, E.; Sorgatto, R.J.; Dourado, P.M.; Crivellari, A.; Carvalho, R.A.; Willse, A.; Martinelli, S.; Head, G.P. Field-evolved resistance to Cry1Ab maize by Spodoptera frugiperda in Brazil. Pest Manag. Sci. 2016, 72, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.F.; Fang, M.; Yao, L.; Qiu, K.; Zheng, Z.Y.; Jin, T.; Li, G.T. Effects of feeding different corn organizations on growth, development and nutritional indexes of Spodoptera frugiperda. Plant Prot. 2020, 46, 24–27. [Google Scholar]
- Zhang, Q.; Zhang, Y.; Quandahor, P.; Gou, Y.; Li, C.; Zhang, K.; Liu, C. Oviposition Preference and Age-Stage, Two-Sex Life Table Analysis of Spodoptera frugiperda (Lepidoptera: Noctuidae) on Different Maize Varieties. Insects 2023, 14, 413. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Zhang, K.X.; Ma, Y.; Zhang, Q.Y.; Liu, W.H.; Jiang, H.X.; Yang, L.K.; Liu, C.Z. Feeding selectivity of Spodoptera frugiperda on different maize cultivars and its relationship with chemical substances in maize leaves. Chi Plant Prot. 2021, 41, 10–16. [Google Scholar]
- Yang, P.Y.; Zhu, X.M.; Guo, J.F.; Wang, Z.Y. Strategy and advice for managing the fall armyworm in China. Plant Prot. 2019, 45, 1–6. [Google Scholar]
- Guan, F.; Zhang, J.; Shen, H.; Wang, X.; Padovan, A.; Walsh, T.K.; Tay, W.T.; Gordon, K.H.J.; James, W.; Czepak, C. Whole-genome sequencing to detect mutations associated with resistance to insecticides and Bt proteins in Spodoptera frugiperda. Insect Sci. 2021, 28, 627–638. [Google Scholar] [CrossRef]
- Jiang, Y.Y.; Liu, J.; Xie, M.M.; Li, Y.H.; Yang, J.J.; Zhang, M.L.; Qiu, K. Observation on law of diffusion damage of Spodoptera frugiperda in China in 2019. Plant Prot. 2019, 45, 10–19. [Google Scholar]
- Li, X.J.; Wu, M.F.; Ma, J.; Gao, B.Y.; Wu, Q.L.; Chen, A.D.; Liu, J.; Jiang, Y.Y.; Zhai, B.P.; Early, R. Prediction of migratory routes of the invasive fall armyworm in eastern China using a trajectory analytical approach. Pest Manag. Sci. 2020, 76, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Razmjou, J.; Naseri, B.; Hemati, S.A. Comparative performance of the cotton bollworm, Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae) on various host plants. J. Pest. Sci. 2014, 87, 29–37. [Google Scholar] [CrossRef]
- Hoo, C.F.S.; Fraenkel, G. The consumption, digestion, and utilization of food plants by a polyphagous insect, Prodenia eridania (Cramer). J. Insect. Physiol. 1966, 12, 711–730. [Google Scholar] [CrossRef]
- Behmer, S.T. Insect herbivore nutrient regulation. Annu. Rev. Entomol. 2009, 54, 165–187. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.M.D.; Bueno, A.D.F.; Andrade, K.; Stecca, C.D.S.; Neves, P.M.O.J.; Oliveira, M.C.N.D. Biology and nutrition of Spodoptera frugiperda (Lepidoptera: Noctuidae) fed on different food sources. Sci. Agr. 2017, 74, 18–31. [Google Scholar] [CrossRef]
- Hemmati, S.A.; Shishehbor, P.; Stelinski, L.L. Life table parameters and digestive enzyme activity of Spodoptera littoralis (Boisd) (Lepidoptera: Noctuidae) on selected legume cultivars. Insects 2022, 13, 661. [Google Scholar] [CrossRef]
- Kang, L. The chemical defenses of plants to phytophagous insects. Chin. Bull. Bot. 1995, 12, 22. [Google Scholar]
- Borzoui, E.; Naseri, B.; Nouri-Ganbalani, G. Effects of food quality on biology and physiological traits of Sitotroga cerealella (Lepidoptera: Gelechiidae). J. Econ. Entomol. 2017, 110, 266–273. [Google Scholar] [PubMed]
- Truzi, C.C.; Holzhausen, H.G.; Álvaro, J.C.; De Laurentis, V.L.; Vieira, N.F.; Vacari, A.M.; De Bortoli, S.A. Food consumption utilization, and life history parameters of Helicoverpa armigera (Lepidoptera: Noctuidae) reared on diets of varying protein level. J. Insect. Sci. 2019, 19, 12. [Google Scholar] [CrossRef]
- Wu, L.; Li, L.S.; Wang, L.Y.; Yuan, Y.F.; Chen, M. Effects of gallic acid on the nutritional efficiency and detoxification enzymes in Hyphantria cunea larvae. J. Environ. Entomol. 2020, 42, 471–479. [Google Scholar]
- Xue, M.; Pang, Y.H.; Wang, H.T.; Li, Q.L.; Liu, T.X. Effects of four host plants on biology and food utilization of the cutworm, Spodoptera litura. J. Insect. Sci. 2010, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Sarate, P.J.; Tamhane, V.A.; Kotkar, H.M.; Ratnakaran, N.; Susan, N.; Gupta, V.S.; Giri, A.P. Developmental and digestive flexibilities in the midgut of a polyphagous pest, the cotton bollworm, Helicoverpa armigera. J. Insect. Sci. 2012, 12, 42. [Google Scholar] [CrossRef] [PubMed]
- Shahbaz, M.; Nouri Ganbalani, G.; Naseri, B. Comparative damage and digestive enzyme activity of Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae) on 12 tomato cultivars. Entomol. Res. 2019, 49, 401–408. [Google Scholar] [CrossRef]
- Zhao, X.D.; Sun, Y.H.; Chen, C.Y.; Tian, S.; Tao, R.; Hao, D.J. Cloning and spatio-temporal expression of serine protease gene HcSP1 and its expression in response to feeding on different host plants in Hyphantria cunea (Lepidoptera: Arctiidae). Acta Entomol. Sin. 2019, 62, 160–169. [Google Scholar]
- Lu, Z.; Chen, Y.; Zhou, A.; He, S.; Li, H.; Bao, Y.; Gui, F. Effects of multi-generation feeding with different host plants on activities of enzyme in Spodoptera frugiperda larvae. J. Environ. Entomol. 2020, 42, 1361–1368. [Google Scholar]
- Kotkar, H.M.; Sarate, P.J.; Tamhane, V.A.; Gupta, V.S.; Giri, A.P. Responses of midgut amylases of Helicoverpa armigera to feeding on various host plants. J. Insect. Physiol. 2009, 55, 663–670. [Google Scholar] [CrossRef]
- Alfonso, J.; Ortego, F.; Sanchez-Monge, R.; Garcia-Casado, G.; Pujol, M.; Castañera, P.; Salcedo, G. Wheat and barley inhibitors active towards α-amylase and trypsin-like activities from Spodoptera frugiperda. J. Chem. Ecol. 1997, 23, 1729–1741. [Google Scholar] [CrossRef]
- Namin, F.R.; Naseri, B.; Razmjou, J.; Cohen, A. Nutritional performance and activity of some digestive enzymes of the cotton bollworm, Helicoverpa armigera, in response to seven tested bean cultivars. J. Insect. Sci. 2014, 14, 93. [Google Scholar] [CrossRef]
- Lazarevic, J.; Peric-Mataruga, V.; Vlahovic, M.; Mrdakovic, M.; Cvetanovic, D. Effects of rearing density on larval growth and activity of digestive enzymes in Lymantria dispar L. (Lepidoptera: Lymantriidae). Folia. Biol. 2004, 52, 105–112. [Google Scholar]
- Broadway, R.M. Dietary regulation of serine proteinases that are resistant to serine proteinase inhibitors. J. Insect. Physiol. 1997, 43, 855–874. [Google Scholar] [CrossRef]
- Jongsma, M.A.; Bolter, C. The adaptation of insects to plant protease inhibitors. J. Insect. Physiol. 1997, 43, 885–895. [Google Scholar] [CrossRef]
- Howe, G.A.; Jander, G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 2008, 59, 41–66. [Google Scholar] [CrossRef]
- Huang, Y.F.; Xu, Z.B.; Lin, X.Y.; Feng, Q.L.; Zheng, S.C. Structure and expression of glutathione S-transferase genes from the midgut of the Common cutworm, Spodoptera litura (Noctuidae) and their response to xenobiotic compounds and bacteria. J. Insect. Physiol. 2011, 57, 1033–1044. [Google Scholar] [CrossRef]
- Sun, X.Q.; Zhang, M.X.; Yu, J.Y.; Jin, Y.; Ling, B.; Du, J.P.; Li, G.H.; Qin, Q.M.; Cai, Q.N. Glutathione S-transferase of brown planthoppers (Nilaparvata lugens) is essential for their adaptation to gramine-containing host plants. PLoS ONE 2013, 8, e64026. [Google Scholar] [CrossRef]
- Tao, X.Y.; Xue, X.Y.; Huang, Y.P.; Chen, X.Y.; Mao, Y.B. Gossypol-enhanced P450 gene pool contributes to cotton bollworm tolerance to a pyrethroid insecticide. Mol. Ecol. 2012, 21, 4371–4385. [Google Scholar] [CrossRef]
- Zou, X.P.; Xu, Z.B.; Zou, H.W.; Liu, J.S.; Chen, S.N.; Feng, Q.L.; Zheng, S.C. Glutathione S-transferase SlGSTE1 in Spodoptera litura may be associated with feeding adaptation of host plants. Insect Biochem. Mol. Biol. 2016, 70, 32–43. [Google Scholar] [CrossRef]
- Snyder, M.J.; Glendinning, J.I. Causal connection between detoxification enzyme activity and consumption of a toxic plant compound. J. Comp. Physiol. 1996, 179, 255–261. [Google Scholar] [CrossRef]
- Xiang, Y.Y.; Niu, H.H.; Jin, B.L.; Zhang, Y.C.; Yin, P.F. Effects of Honeysuckle Varieties on Protective and Detoxifying Enzyme Activities in Heterolocha Jinyinhuaphaga Chu (Lepidoptera: Geometridae) Larvae. J. Chem. Ecol. 2023, 49, 205–213. [Google Scholar] [CrossRef]
- Wang, S.Y.; Zhu, Q.Z.; Tan, Y.T.; Ma, Q.L.; Wang, R.F.; Zhang, M.F.; Xu, H.H.; Zhang, Z.X. Artificial diets and rearing technique of Spodoptera frugiperda (JE Smith) in laboratory. J. Environ. Entomol. 2019, 41, 742–747. [Google Scholar]
- Nagoshi, R.N.; Koffi, D.; Agboka, K.; Tounou, K.A.; Banerjee, R.; Jurat-Fuentes, J.L.; Meagher, R.L. Comparative molecular analyses of invasive fall armyworm in Togo reveal strong similarities to populations from the eastern United States and the Greater Antilles. PLoS ONE 2017, 12, e181982. [Google Scholar] [CrossRef]
- Fahmi, F.; Kusumah, R.Y.M.; Buchori, D. Genetic variation of pest fall armyworm Spodoptera frugiperda (JE Smith) (Lepidoptera: Noctuidae) in different landscapes in Bogor. J. Entomol. Indones. 2023, 20, 1. [Google Scholar] [CrossRef]
- Jiang, J.; Huang, B.; Wu, Q.; Li, S.; Gu, J.; Huang, L. Identification of Spodoptera frugiperda (Lepidoptera: Noctuidae) and its two host strains in China by PCR-RFLP. J. Econ. Entomol. 2023, 116, 983–992. [Google Scholar] [CrossRef]
- Waldbauer, G.P. The consumption and utilization of food by insects. Adv. Insect Physiol. 1968, 5, 229–288. [Google Scholar]
- Wang, C.Z.; Qin, J.D. Identification of major protease activities in the midgut of Helicoverpa armigera larvae. Acta Entomol. Sin. 1996, 39, 7–11. [Google Scholar]
- Bernfeld, P. Amylases, α and β. Methods Enzymol. 1955, 1, 149–158. [Google Scholar]
- Bai, Y.; Wang, W.X. Methods for determination of protease, amylase, lipase and cellulase activities in the gut of Apostichopus japonicas. Feed Ind. 2012, 33, 28–32. [Google Scholar]
- Han, Y.; Yu, W.; Zhang, W.; Yang, Y.; Walsh, T.; Oakeshott, J.G.; Wu, Y. Variation in P450-mediated fenvalerate resistance levels is not correlated with CYP337B3 genotype in Chinese populations of Helicoverpa armigera. Pest. Biochem. Physiol. 2015, 121, 129–135. [Google Scholar] [CrossRef]
- Van Asperen, K. A study of housefly esterases by means of a sensitive colorimetric method. J. Insect Physiol. 1962, 8, 401–416. [Google Scholar] [CrossRef]
- Aitio, A. A simple and sensitive assay of 7-ethoxycoumarin deethylation. Anal. Biochem. 1978, 85, 488–491. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Zhang, Y.; Zhang, K.; Liu, H.; Gou, Y.; Li, C.; Haq, I.U.; Quandahor, P.; Liu, C. Molecular Characterization Analysis and Adaptive Responses of Spodoptera frugiperda (Lepidoptera: Noctuidae) to Nutritional and Enzymatic Variabilities in Various Maize Cultivars. Plants 2024, 13, 597. https://doi.org/10.3390/plants13050597
Zhang Q, Zhang Y, Zhang K, Liu H, Gou Y, Li C, Haq IU, Quandahor P, Liu C. Molecular Characterization Analysis and Adaptive Responses of Spodoptera frugiperda (Lepidoptera: Noctuidae) to Nutritional and Enzymatic Variabilities in Various Maize Cultivars. Plants. 2024; 13(5):597. https://doi.org/10.3390/plants13050597
Chicago/Turabian StyleZhang, Qiangyan, Yanlei Zhang, Kexin Zhang, Huiping Liu, Yuping Gou, Chunchun Li, Inzamam Ul Haq, Peter Quandahor, and Changzhong Liu. 2024. "Molecular Characterization Analysis and Adaptive Responses of Spodoptera frugiperda (Lepidoptera: Noctuidae) to Nutritional and Enzymatic Variabilities in Various Maize Cultivars" Plants 13, no. 5: 597. https://doi.org/10.3390/plants13050597
APA StyleZhang, Q., Zhang, Y., Zhang, K., Liu, H., Gou, Y., Li, C., Haq, I. U., Quandahor, P., & Liu, C. (2024). Molecular Characterization Analysis and Adaptive Responses of Spodoptera frugiperda (Lepidoptera: Noctuidae) to Nutritional and Enzymatic Variabilities in Various Maize Cultivars. Plants, 13(5), 597. https://doi.org/10.3390/plants13050597







