PvARL1 Increases Biomass Yield and Enhances Alkaline Tolerance in Switchgrass (Panicum virgatum L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Plasmid Construction and Plant Transformation in Switchgrass
2.3. Abiotic Tolerance Assay in Switchgrass
2.4. RNA Extraction and RT-qPCR
2.5. Na+ and K+ Content Measurements
2.6. Chlorophyll Content and Fv/Fm Measurements
2.7. Relative Water Content and Electrolyte Leakage Measurements
2.8. MDA and H2O2 Accumulation Analysis
2.9. Subcellular Localization
2.10. Statistical Analysis
3. Results
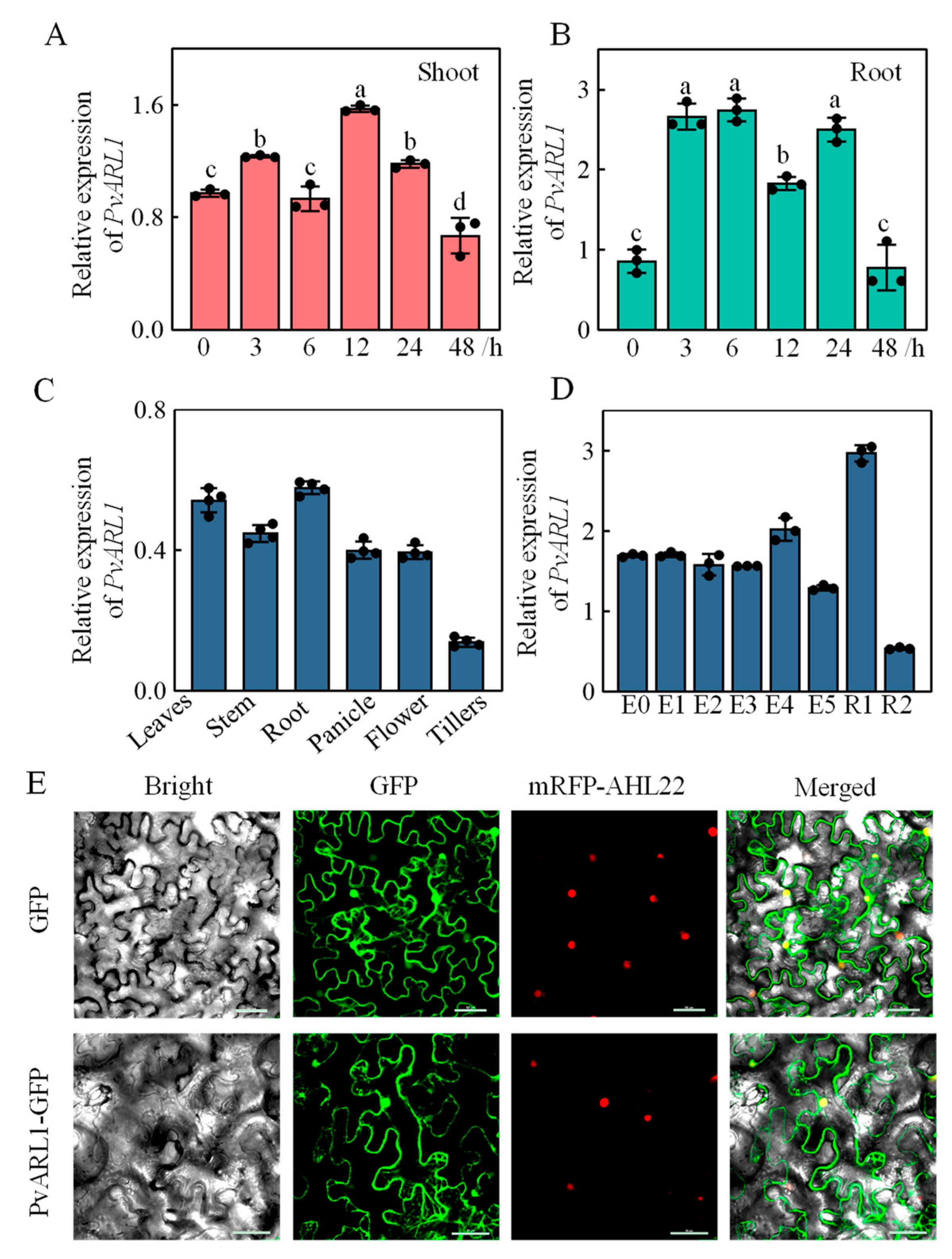
3.1. PvARL1 Responses to Alkaline Stress
3.2. Expression Pattern and Subcellular Location of PvARL1
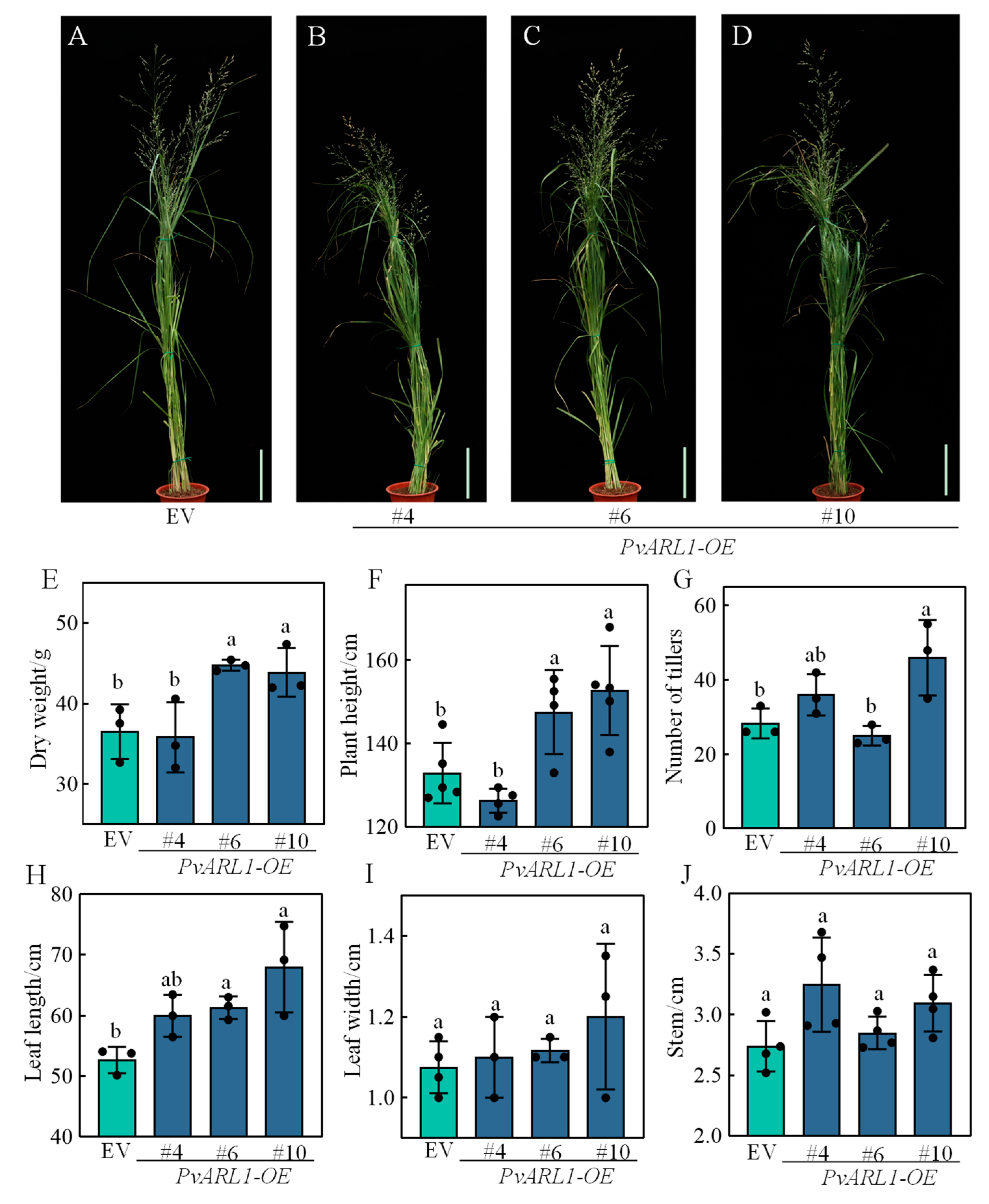
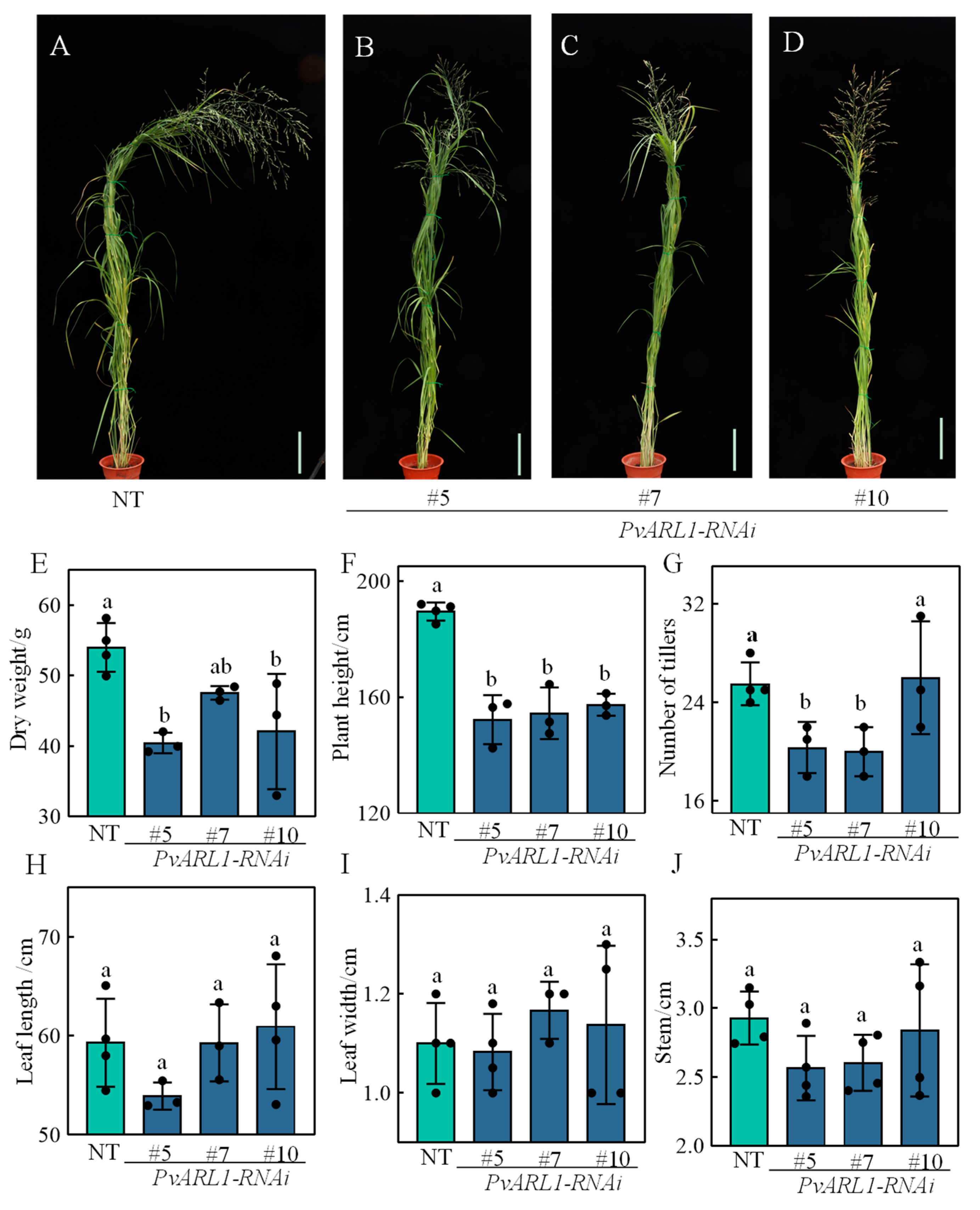
3.3. PvARL1 Positively Regulates Plant Growth
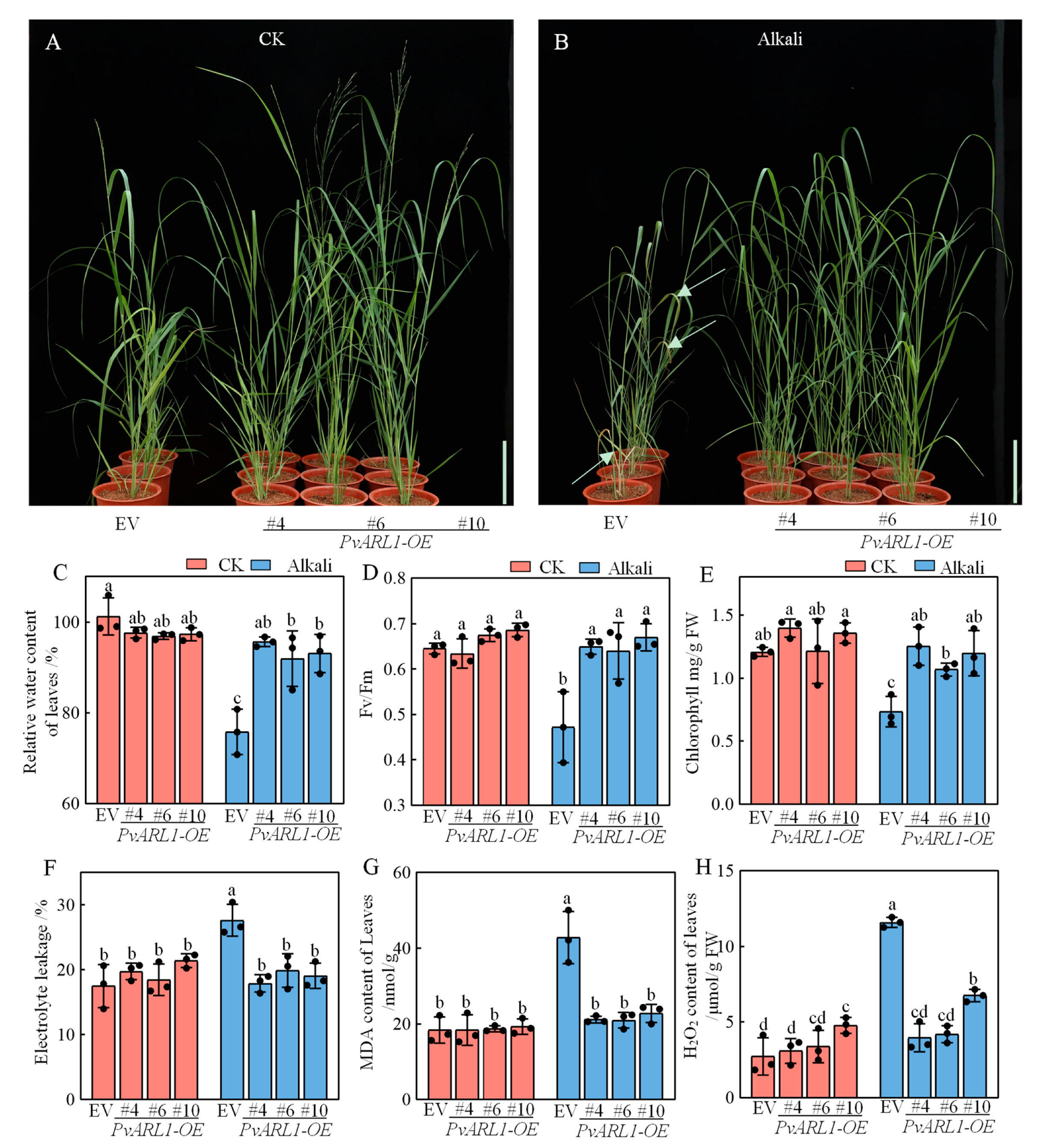
3.4. PvARL1 Alleviates Leaf Damage under Alkali Conditions
3.5. PvARL1 Overexpression Decreases Oxidative Damage after Alkali Treatment
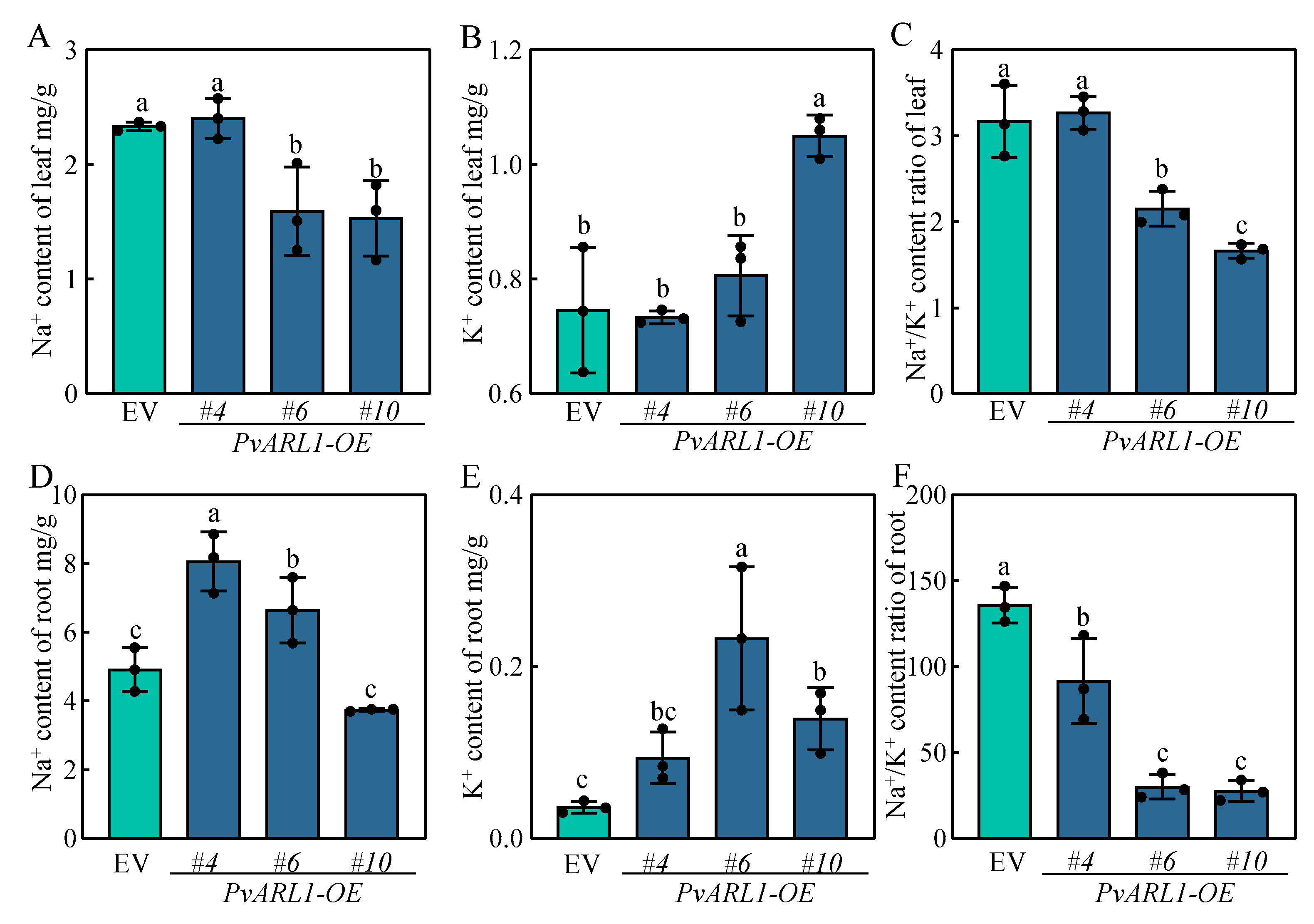
3.6. PvARL1 Maintained Ion Homeostasis in Switchgrass
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, H.; Yu, F.; Xie, P.; Sun, S.; Qiao, X.; Tang, S.; Chen, C.; Yang, S.; Mei, C.; Yang, D.; et al. A Gγ protein regulates alkaline sensitivity in crops. Science 2023, 379, eade8416. [Google Scholar] [CrossRef] [PubMed]
- Caon, L.; Forlano, N.; Cori Keene, M.S.; Sorokin, A.; Verbeke, I.; Ward, C. Status of the World’s Soil Resources; Food and Agriculture Organization of the United Nations: Rome, Italy, 2015; pp. 1–650. [Google Scholar]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Lu, X.; Ma, L.; Zhang, C.; Yan, H.; Bao, J.; Gong, M.; Wang, W.; Li, S.; Ma, S.; Chen, B. Grapevine (Vitis vinifera) responses to salt stress and alkali stress: Transcriptional and metabolic profiling. BMC Plant Biol. 2022, 22, 528. [Google Scholar] [CrossRef]
- Zhao, X.; Xu, Y.; He, G.; He, K.; Xiao, L.; Hu, R.; Li, S. Genome-wide characterization and expression profiling of the GRAS gene family in salt and alkali stresses in Miscanthus sinensis. Int. J. Mol. Sci. 2022, 23, 14521. [Google Scholar] [CrossRef]
- Lu, P.; Dai, S.-Y.; Yong, L.-T.; Zhou, B.-H.; Wang, N.; Dong, Y.-Y.; Liu, W.-C.; Wang, F.-W.; Yang, H.-Y.; Li, X.-W. A Soybean Sucrose Non-Fermenting Protein Kinase 1 Gene, GmSNF1, positively regulates plant response to salt and salt–alkali stress in transgenic plants. Int. J. Mol. Sci. 2023, 24, 12482. [Google Scholar] [CrossRef]
- An, Y.; Yang, X.X.; Zhang, L.; Zhang, J.; Du, B.; Yao, L.; Li, X.T.; Guo, C. Alfalfa MsCBL4 enhances calcium metabolism but not sodium transport in transgenic tobacco under salt and saline-alkali stress. Plant Cell Rep. 2020, 39, 997–1011. [Google Scholar] [CrossRef]
- Ketehouli, T.; Zhou, Y.G.; Dai, S.Y.; Carther, K.F.I.; Sun, D.Q.; Li, Y.; Nguyen, Q.V.H.; Xu, H.; Wang, F.W.; Liu, W.C.; et al. A soybean calcineurin B-like protein-interacting protein kinase, GmPKS4, regulates plant responses to salt and alkali stresses. J. Plant Physiol. 2021, 256, 153331. [Google Scholar] [CrossRef]
- Gao, P.; Bai, X.; Yang, L.; Lv, D.; Pan, X.; Li, Y.; Cai, H.; Ji, W.; Chen, Q.; Zhu, Y. Osa-MIR393: A salinity- and alkaline stress-related microRNA gene. Mol. Biol. Rep. 2010, 38, 237–242. [Google Scholar] [CrossRef]
- Guo, R.; Yang, Z.; Li, F.; Yan, C.; Zhong, X.; Liu, Q.; Xia, X.; Li, H.; Zhao, L. Comparative metabolic responses and adaptive strategies of wheat (Triticum aestivum) to salt and alkali stress. BMC Plant Biol. 2015, 15, 170. [Google Scholar] [CrossRef]
- Kahn, R.A.; Gilman, A.G. The protein cofactor necessary for ADP-ribosylation of Gs by cholera toxin is itself a GTP binding protein. J. Biol. Chem. 1986, 261, 7906–7911. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Ueda, T.; Yahara, N.; Nakano, A. Arf1 GTPase plays roles in the protein traffic between the endoplasmic reticulum and the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 2002, 31, 499–515. [Google Scholar] [CrossRef] [PubMed]
- D’Souza-Schorey, C.; Chavrier, P. ARF proteins: Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006, 7, 347–358. [Google Scholar] [CrossRef]
- Singh, M.K.; Jürgens, G. Specificity of plant membrane trafficking—ARFs, regulators and coat proteins. Semin. Cell Dev. Biol. 2018, 80, 85–93. [Google Scholar] [CrossRef]
- Bourne, H.R.; Sanders, D.A.; McCormick, F. The GTPase superfamily: Conserved structure and molecular mechanism. Nature 1991, 349, 117–127. [Google Scholar] [CrossRef]
- Li, Y.; Kelly, W.G.; Logsdon, J.M., Jr.; Schurko, A.M.; Harfe, B.D.; Hill-Harfe, K.L.; Kahn, R.A. Functional genomic analysis of the ADP-ribosylation factor family of GTPases: Phylogeny among diverse eukaryotes and function in C. elegans. FASEB J. 2004, 18, 1834–1850. [Google Scholar] [CrossRef]
- Shen, X.; Chen, R.; Chen, X.; Munir, N.; Zhang, S.; Xu, X.; Lin, Z.; Zhang, J.; Li, X.; Lin, Y.; et al. Molecular evolution and expression analysis of ADP-ribosylation factors (ARFs) from longan embryogenic callus. Gene 2021, 777, 145461. [Google Scholar] [CrossRef]
- Wang, T.-Z.; Xia, X.-Z.; Zhao, M.-G.; Tian, Q.-Y.; Zhang, W.-H. Expression of a Medicago falcata small GTPase gene, MfARL1 enhanced tolerance to salt stress in Arabidopsis thaliana. Plant Physiol. Biochem. 2013, 63, 227–235. [Google Scholar] [CrossRef]
- Guan, C.; Li, X.; Tian, D.-Y.; Liu, H.-Y.; Cen, H.-F.; Tadege, M.; Zhang, Y.-W. ADP-ribosylation factors improve biomass yield and salinity tolerance in transgenic switchgrass (Panicum virgatum L.). Plant Cell Rep. 2020, 39, 1623–1638. [Google Scholar] [CrossRef]
- Liu, Y.R.; Cen, H.F.; Yan, J.P.; Zhang, Y.W.; Zhang, W.J. Inside out: High-efficiency plant regeneration and Agrobacterium-mediated transformation of upland and lowland switchgrass cultivars. Plant Cell Rep. 2015, 34, 1099–1108. [Google Scholar] [CrossRef]
- Hardin, C.F.; Fu, C.; Hisano, H.; Xiao, X.; Shen, H.; Stewart, C.N., Jr.; Parrott, W.; Dixon, R.A.; Wang, Z.-Y. Standardization of Switchgrass Sample Collection for Cell Wall and Biomass Trait Analysis. Bioenergy Res. 2013, 6, 755–762. [Google Scholar] [CrossRef]
- Liu, Y.; Li, D.; Yan, J.; Wang, K.; Luo, H.; Zhang, W. MiR319-mediated ethylene biosynthesis, signalling and salt stress response in switchgrass. Plant Biotechnol. J. 2019, 17, 2370–2383. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Wallace, P.A.; Teakle, N.L.; Colmer, T.D. Measuring soluble ion concentrations (Na+, K+, Cl−) in salt-treated plants. Plant Stress Toler. Methods Protoc. 2010, 639, 371–382. [Google Scholar]
- Li, Z.; Baldwin, C.M.; Hu, Q.; Liu, H.; Luo, H. Heterologous expression of Arabidopsis H+ plus-pyrophosphatase enhances salt tolerance in transgenic creeping bentgrass (Agrostis stolonifera L.). Plant Cell Environ. 2010, 33, 272–289. [Google Scholar] [CrossRef] [PubMed]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Reiner, D.J.; Lundquist, E.A. Small GTPases. WormBook 2018, 2018, 1–65. [Google Scholar] [CrossRef]
- Ganotra, J.; Sharma, B.; Biswal, B.; Bhardwaj, D.; Tuteja, N. Emerging role of small GTPases and their interactome in plants to combat abiotic and biotic stress. Protoplasma 2022, 260, 1007–1029. [Google Scholar] [CrossRef]
- Wu, K.; Xu, P.; Zang, A.; Chen, H.; Cai, W. The small G protein AtRAN1 regulates vegetative growth and stress tolerance in Arabidopsis thaliana. PLoS ONE 2016, 11, e0154787. [Google Scholar]
- Xu, P.; Ma, W.; Liu, J.; Hu, J.; Cai, W. Overexpression of a small GTP-binding protein Ran1 in Arabidopsis leads to promoted elongation growth and enhanced disease resistance against P. syringae DC3000. Plant J. 2021, 108, 977–991. [Google Scholar] [CrossRef]
- Wang, Q.; Xue, X.; Li, Y.; Dong, Y.; Zhang, L.; Zhou, Q.; Deng, F.; Ma, Z.; Qiao, D.; Hu, C.; et al. A maize ADP-ribosylation factor ZmArf2 increases organ and seed size by promoting cell expansion in Arabidopsis. Physiol. Plant. 2016, 156, 97–107. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Mangu, V.R.; Zandkarimi, H.; Prasad, M.; Baisakh, N. Structure, organization and evolution of ADP-ribosylation factors in rice and foxtail millet, and their expression in rice. Sci. Rep. 2016, 6, 24008. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, E.; Francini, A.; Lorenzini, G.; Nali, C. PSII photochemistry and carboxylation efficiency in Liriodendron tulipifera under ozone exposure. Environ. Exp. Bot. 2011, 70, 217–226. [Google Scholar] [CrossRef]
- Yan, H.; Liu, B.; Cui, Y.; Wang, Y.; Sun, S.; Wang, J.; Tan, M.; Wang, Y.; Zhang, Y. LpNAC6 reversely regulates the alkali tolerance and drought tolerance of Lilium pumilum. J. Plant Physiol. 2022, 270, 153635. [Google Scholar] [CrossRef]
- Gong, B.; Li, X.; VandenLangenberg, K.M.; Wen, D.; Sun, S.; Wei, M.; Li, Y.; Yang, F.; Shi, Q.; Wang, X. Overexpression of S-adenosyl-L-methionine synthetase increased tomato tolerance to alkali stress through polyamine metabolism. Plant Biotechnol. J. 2014, 12, 694–708. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhang, S.; Ma, X.; Wang, Y.; Kong, F.; Meng, Q. A stress-associated NAC transcription factor (SlNAC35) from tomato plays a positive role in biotic and abiotic stresses. Physiol. Plant. 2016, 158, 45–64. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Oxidant and antioxidant signalling in plants: A re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ. 2005, 28, 1056–1071. [Google Scholar] [CrossRef]
- Diaz-Vivancos, P.; Barba-Espin, G.; Antonio Hernandez, J. Elucidating hormonal/ROS networks during seed germination: Insights and perspectives. Plant Cell Rep. 2013, 32, 1491–1502. [Google Scholar] [CrossRef]
- Xu, J.; Yang, J.; Duan, X.; Jiang, Y.; Zhang, P. Increased expression of native cytosolic Cu/Zn superoxide dismutase and ascorbate peroxidase improves tolerance to oxidative and chilling stresses in cassava (Manihot esculenta Crantz). BMC Plant Biol. 2014, 14, 208. [Google Scholar] [CrossRef]
- Brauer, E.K.; Ahsan, N.; Dale, R.; Kato, N.; Coluccio, A.E.; Pineros, M.A.; Kochian, L.V.; Thelen, J.J.; Popescu, S.C. The Raf-like Kinase ILK1 and the High Affinity K+ Transporter HAK5 are required for innate immunity and abiotic stress response. Plant Physiol. 2016, 171, 1470–1484. [Google Scholar] [CrossRef]
- Bajji, M.; Kinet, J.M.; Lutts, S. Osmotic and ionic effects of NaCl on germination, early seedling growth and ion content of Atriplex halimus (Chenopodiaceae). Can. J. Bot. 2002, 80, 297–304. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Guan, C.; Liu, H.; Wang, T.; Lin, M.; Zhou, D.; Zhang, Y.; Bi, X. PvARL1 Increases Biomass Yield and Enhances Alkaline Tolerance in Switchgrass (Panicum virgatum L.). Plants 2024, 13, 566. https://doi.org/10.3390/plants13050566
Li X, Guan C, Liu H, Wang T, Lin M, Zhou D, Zhang Y, Bi X. PvARL1 Increases Biomass Yield and Enhances Alkaline Tolerance in Switchgrass (Panicum virgatum L.). Plants. 2024; 13(5):566. https://doi.org/10.3390/plants13050566
Chicago/Turabian StyleLi, Xue, Cong Guan, Huayue Liu, Tingting Wang, Mengzhuo Lin, Die Zhou, Yunwei Zhang, and Xiaojing Bi. 2024. "PvARL1 Increases Biomass Yield and Enhances Alkaline Tolerance in Switchgrass (Panicum virgatum L.)" Plants 13, no. 5: 566. https://doi.org/10.3390/plants13050566
APA StyleLi, X., Guan, C., Liu, H., Wang, T., Lin, M., Zhou, D., Zhang, Y., & Bi, X. (2024). PvARL1 Increases Biomass Yield and Enhances Alkaline Tolerance in Switchgrass (Panicum virgatum L.). Plants, 13(5), 566. https://doi.org/10.3390/plants13050566






