Bioinformatic Analysis of Codon Usage Bias of HSP20 Genes in Four Cruciferous Species
Abstract
1. Introduction
2. Results
2.1. Identification and Nucleotide Composition of HSP20 Genes
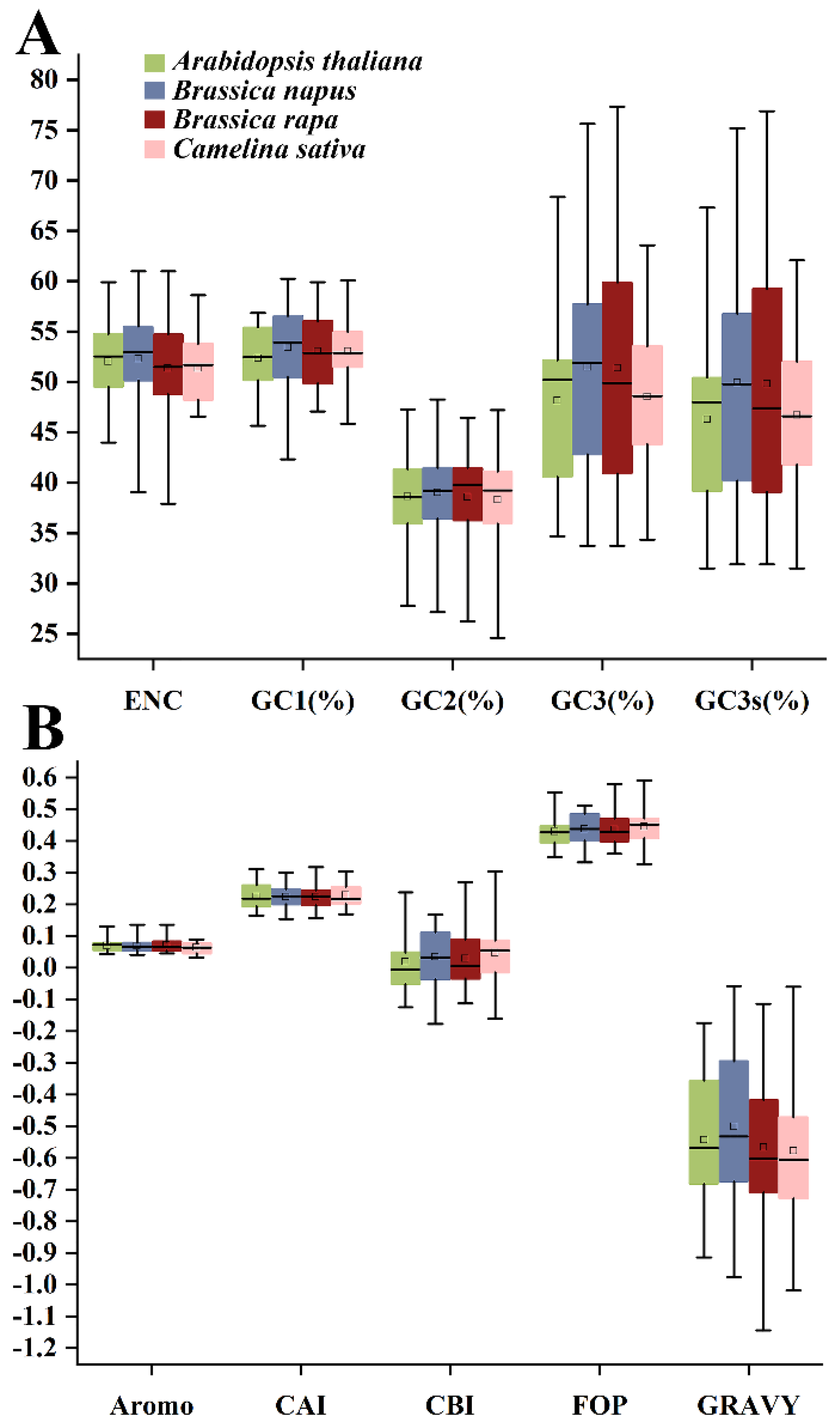
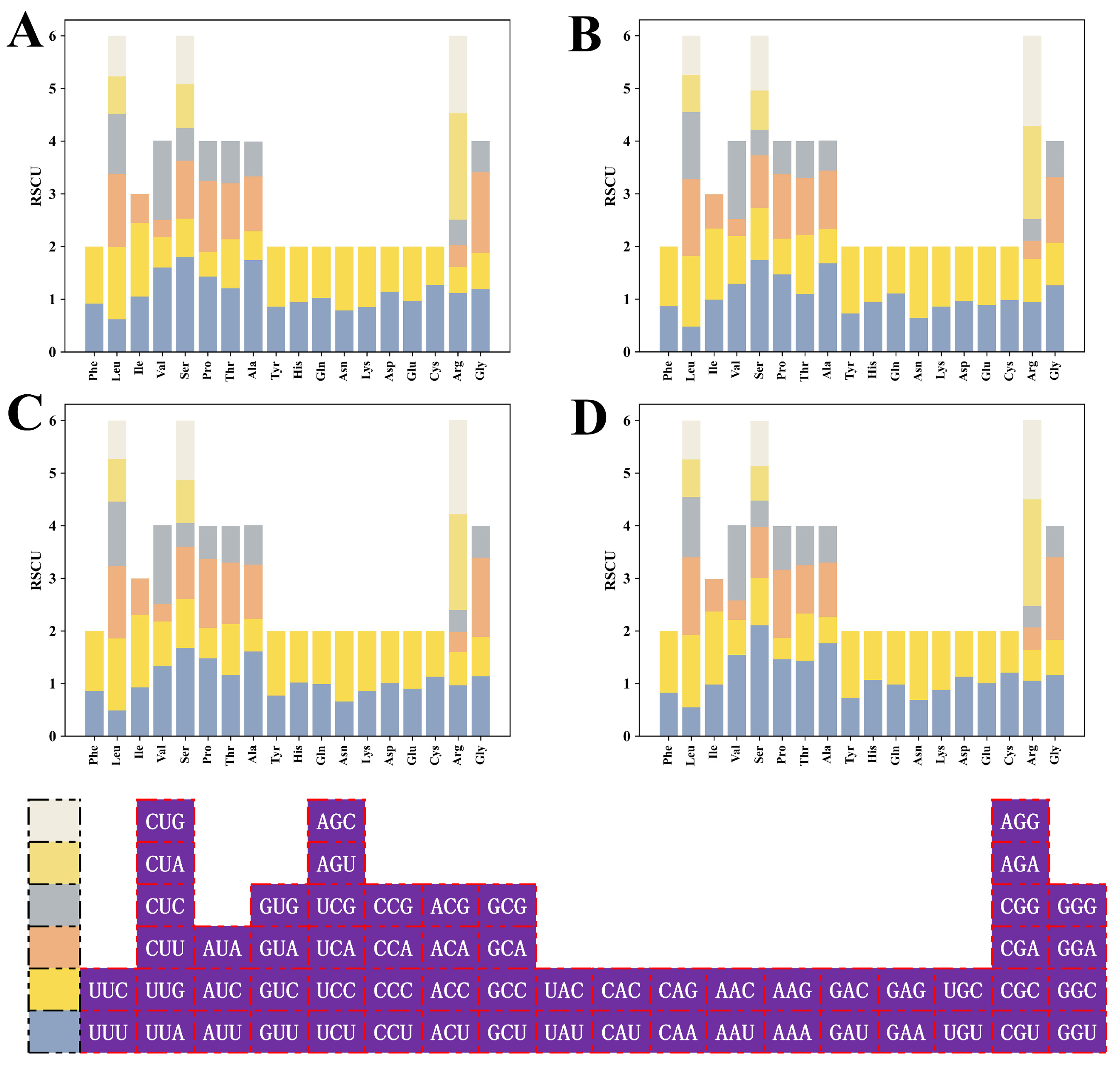
2.2. Codon Usage Indices of HSP20 Genes
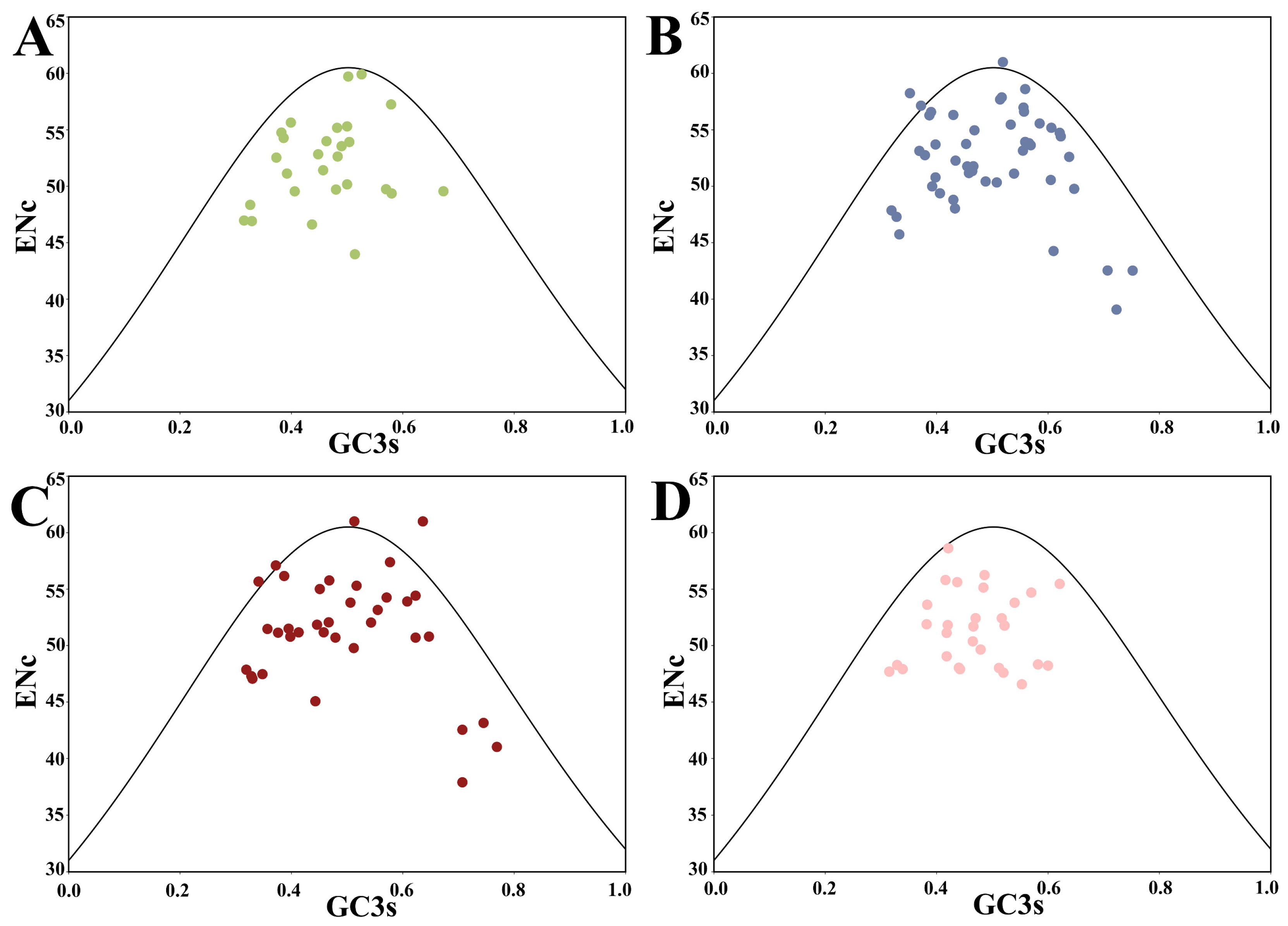
2.3. ENc-GC3s Plot Analysis
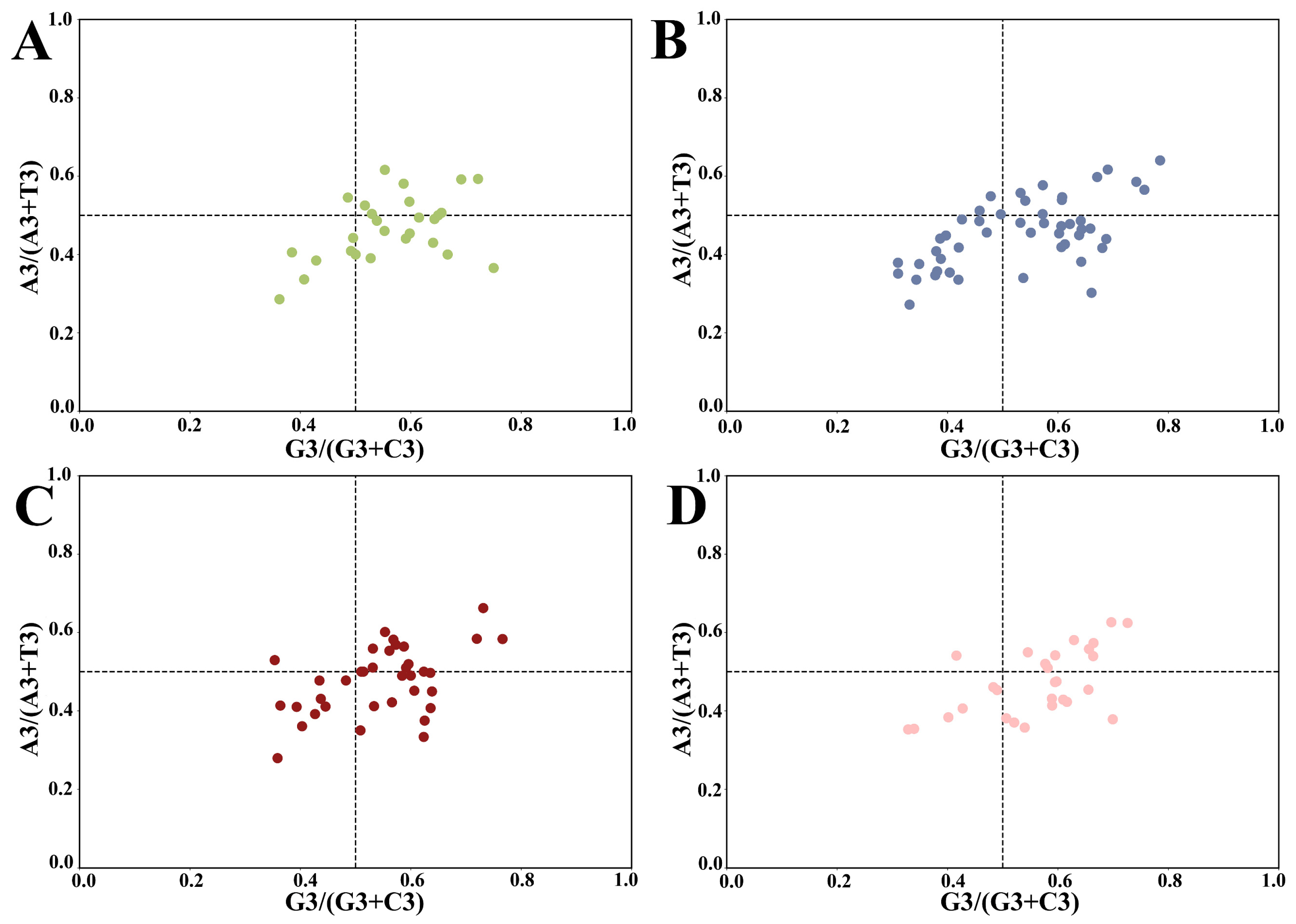
2.4. PR2-Plot Analysis
2.5. Neutrality Plot
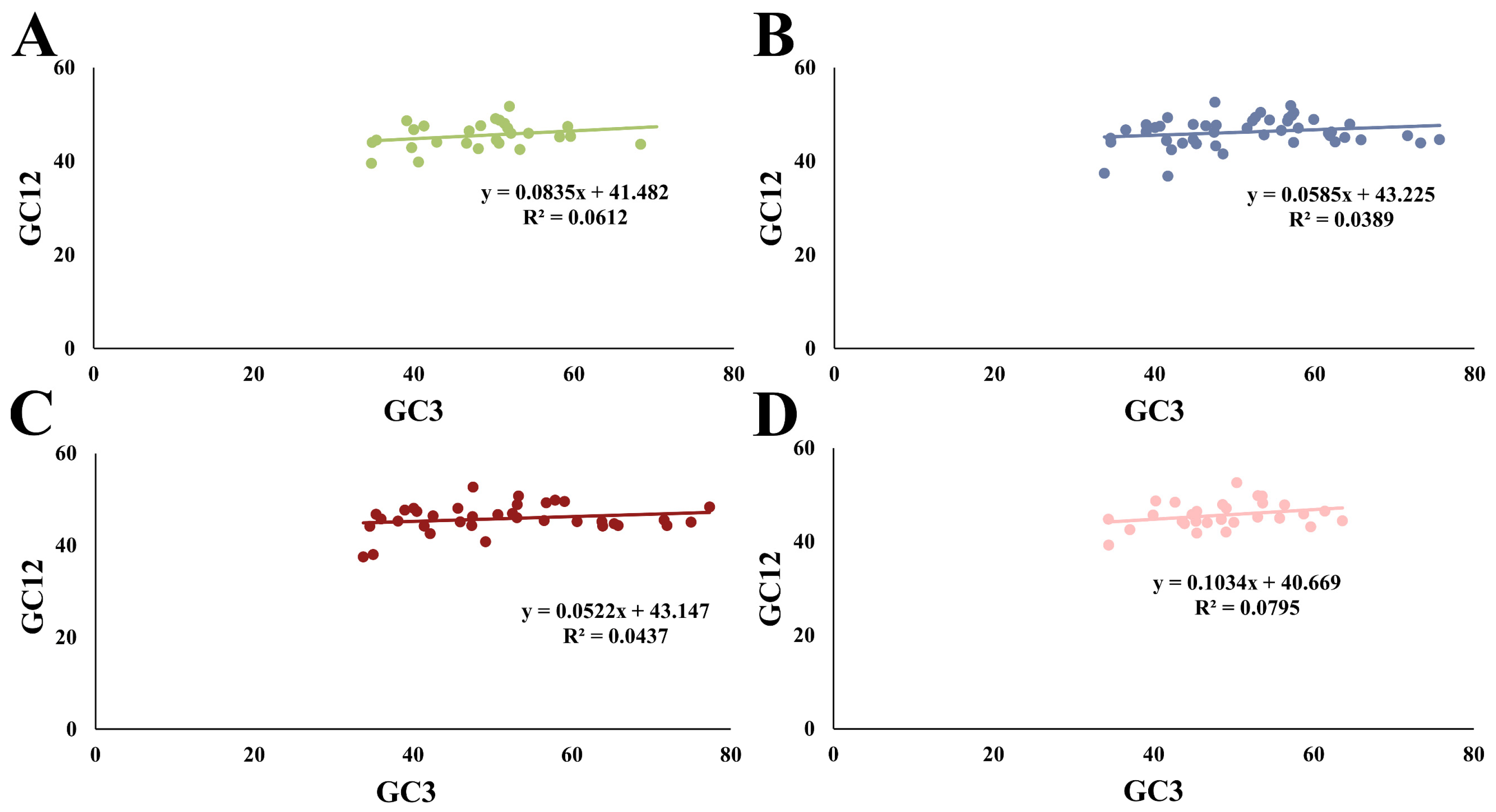
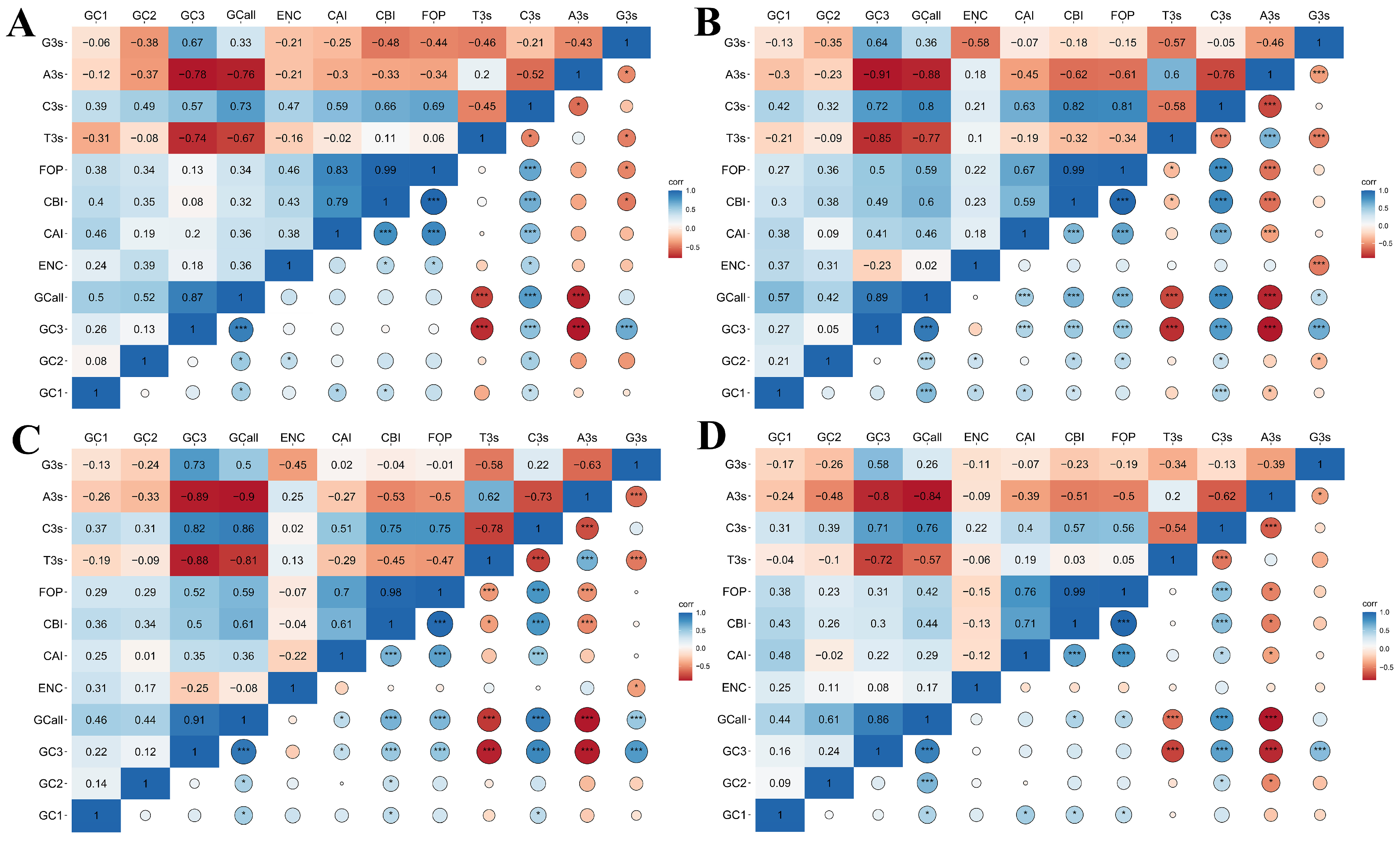
2.6. Correlation Coefficient Analysis of Codon Usage Indices
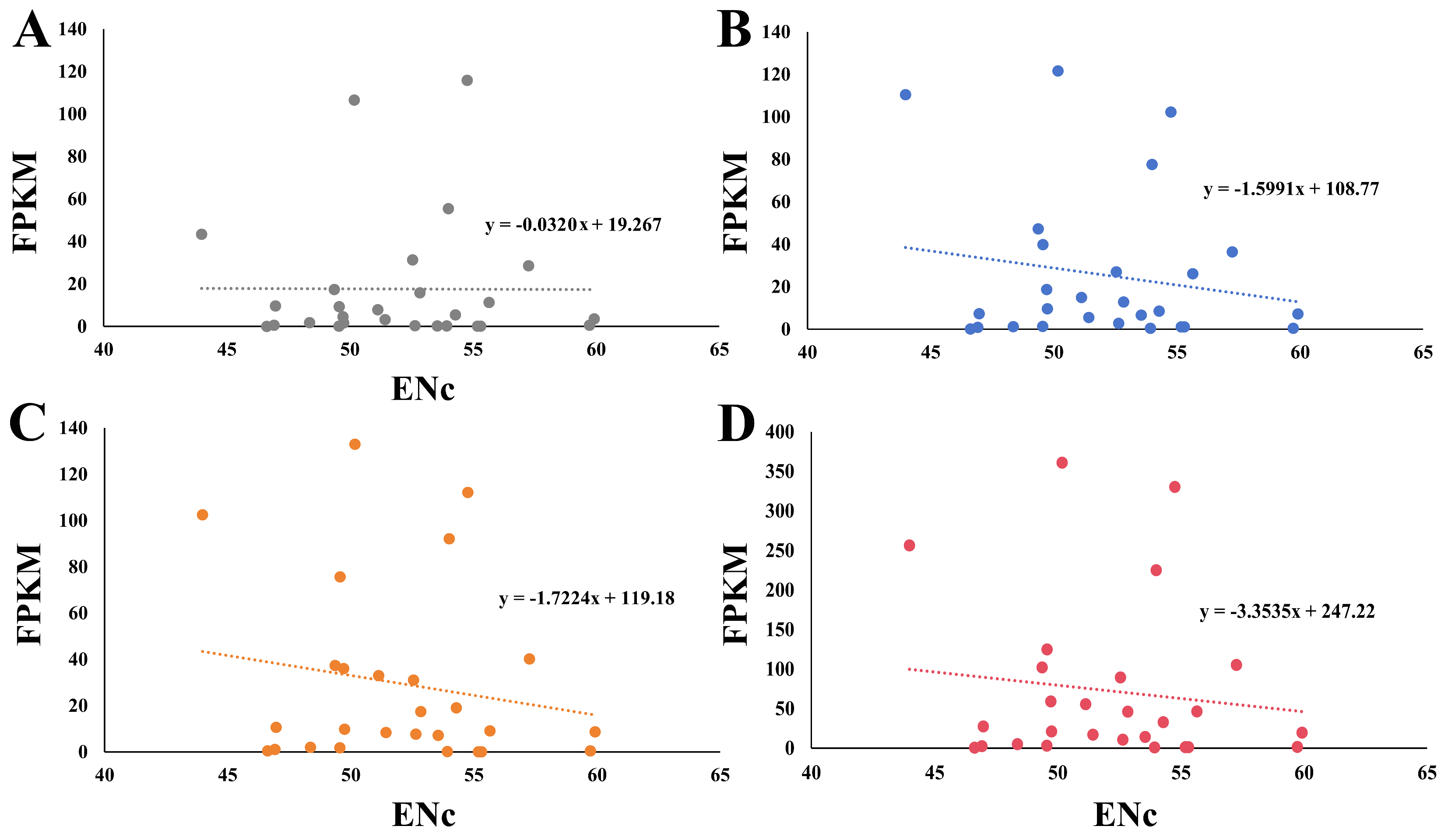
2.7. Relationship between ENc and Gene Expression Level
2.8. Identification of Optimal Codons
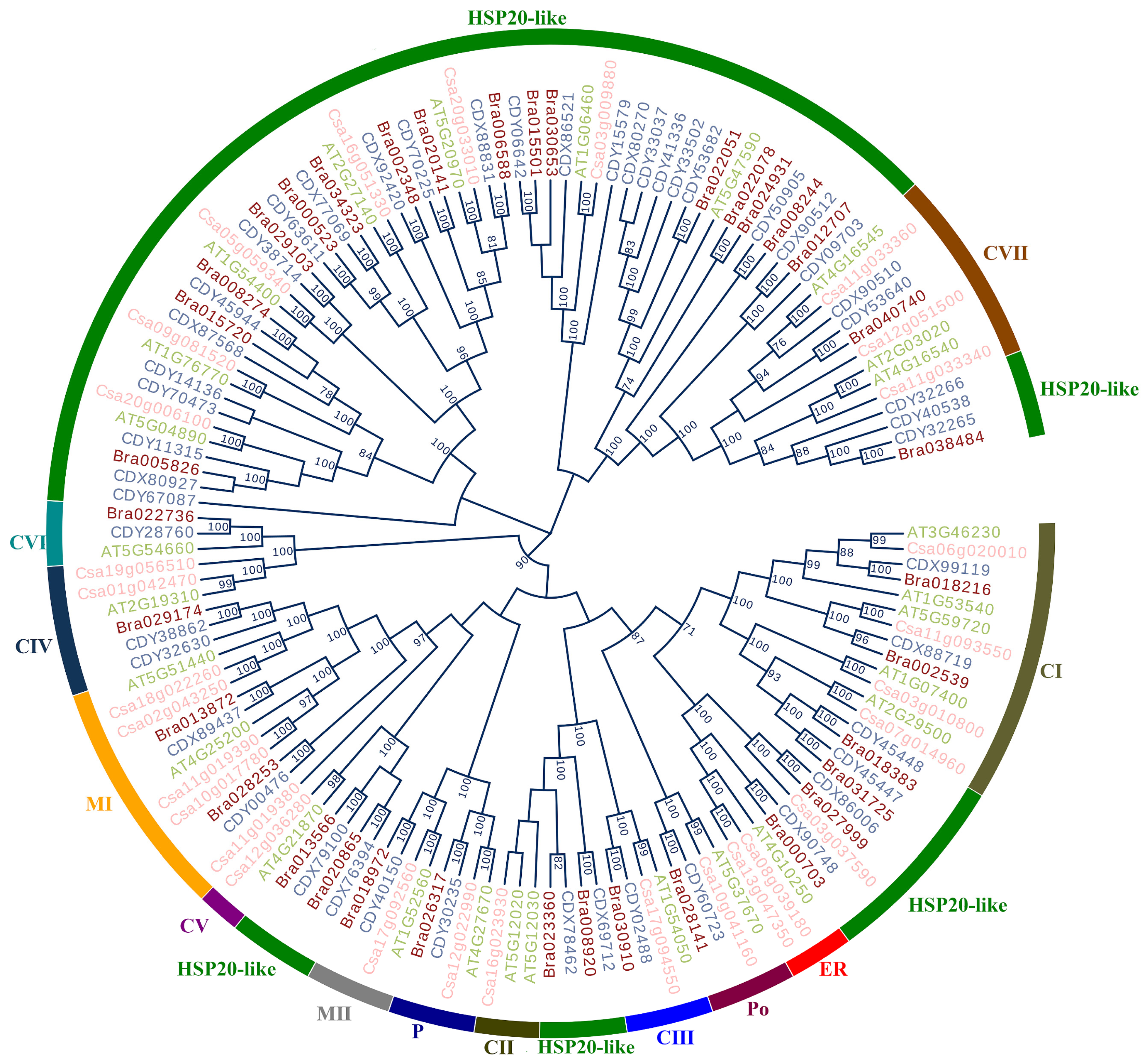
2.9. Comparison of Evolutionary Relationship and Cluster Analysis
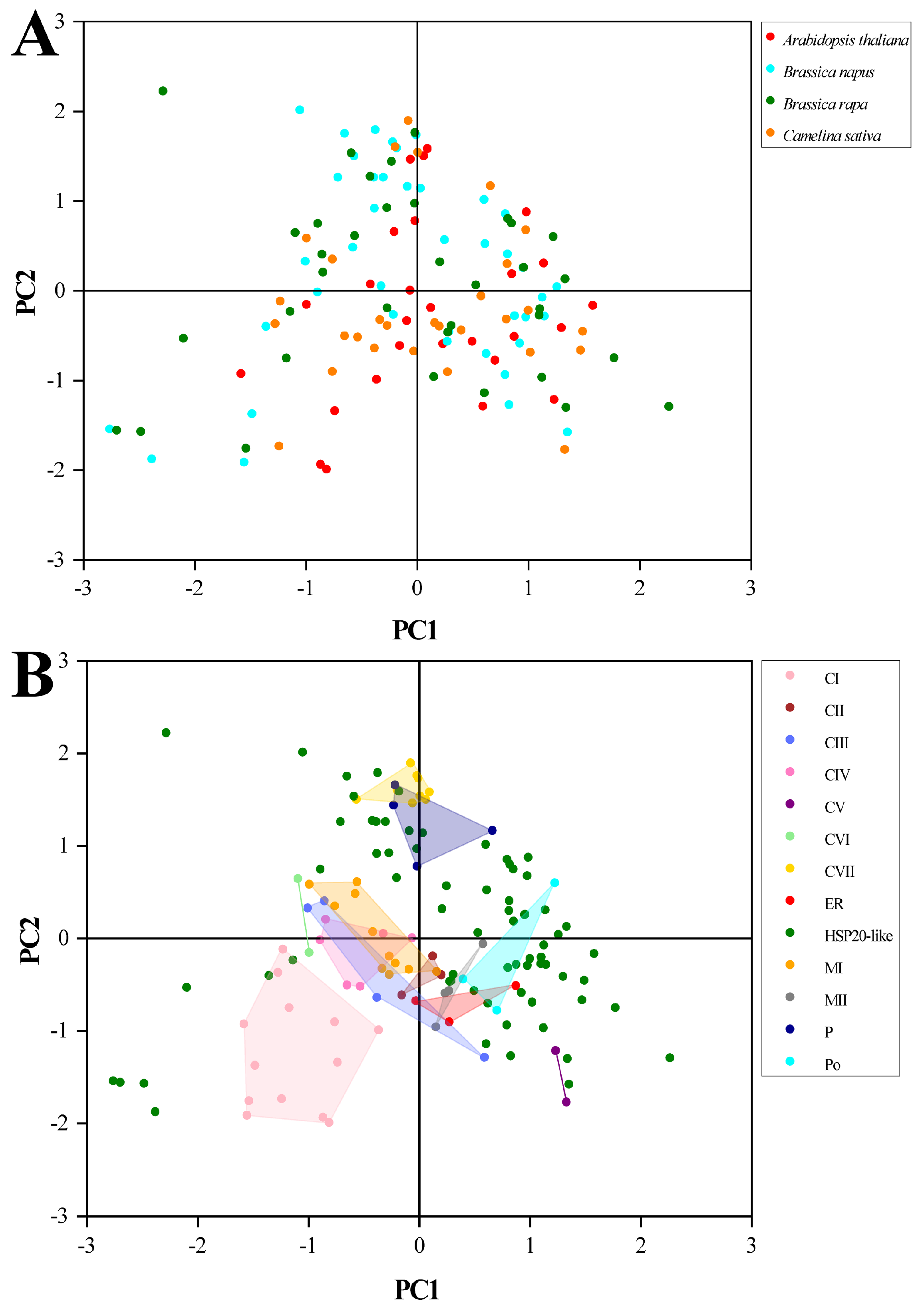
2.10. PCA of RSCU Values with Species and Subfamilies
3. Discussion
4. Materials and Methods
4.1. Identification of HSP20 Genes
4.2. Codon Usage Bias Analysis
4.3. ENc-GC3s Plot
4.4. PR2-Plot Analysis
4.5. Neutrality Plot
4.6. Correlation Analysis of Codon Usage Bias Indices
4.7. The Relationship between CUB and Gene Expression Level
4.8. Determination of Optimal Codons
4.9. Phylogenetic and Clustering Analysis
4.10. Principal Component Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Jia, J.; Xue, Q. Codon usage biases of transposable elements and host nuclear genes in Arabidopsis thaliana and Oryza sativa. Genom. Proteom. Bioinform. 2009, 7, 175–184. [Google Scholar] [CrossRef]
- Chakraborty, S.; Yengkhom, S.; Uddin, A. Analysis of codon usage bias of chloroplast genes in Oryza species. Planta 2020, 252, 67. [Google Scholar] [CrossRef]
- Tyagi, N.; Sardar, R.; Gupta, D. Natural selection plays a significant role in governing the codon usage bias in the novel SARS-CoV-2 variants of concern (VOC). PeerJ 2022, 10, e13562. [Google Scholar] [CrossRef]
- Li, N.; Sun, M.-H.; Jiang, Z.-S.; Shu, H.-R.; Zhang, S.-Z. Genome-wide analysis of the synonymous codon usage patterns in apple. J. Integr. Agric. 2016, 15, 983–991. [Google Scholar] [CrossRef]
- Murray, E.E.; Lotzer, J.; Eberle, M. Codon usage in plant genes. Nucleic Acids Res. 1989, 17, 477–498. [Google Scholar] [CrossRef]
- Wang, L.; Xing, H.; Yuan, Y.; Wang, X.; Saeed, M.; Tao, J.; Feng, W.; Zhang, G.; Song, X.; Sun, X. Genome-wide analysis of codon usage bias in four sequenced cotton species. PLoS ONE 2018, 13, e0194372. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhang, Q.; Xiao, S.; Si, F. Deep decoding of codon usage strategies and host adaption preferences of soybean mosaic virus. Int. J. Biol. Macromol. 2022, 222, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Zhou, J.; Huo, Z.; Wu, T.; Li, Y.; Li, Y.; Wang, Y.; Wang, M. The Shift in Synonymous Codon Usage Reveals Similar Genomic Variation during Domestication of Asian and African Rice. Int. J. Mol. Sci. 2022, 23, 2860. [Google Scholar] [CrossRef] [PubMed]
- Henry, I.; Sharp, P.M. Predicting gene expression level from codon usage bias. Mol. Biol. Evol. 2007, 24, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Li, Y.; Ji, K.K.; Zhu, J.; Ling, P.; Zhou, T.; Fan, L.Y.; Xie, S.Q. Genome-wide codon usage pattern analysis reveals the correlation between codon usage bias and gene expression in Cuscuta australis. Genomics 2020, 112, 2695–2702. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Zhang, J.; Wang, Y.; Ge, W.; Jing, Y.; Cao, X.; Huo, Y.; Fu, Y. A codon-based live-cell biomonitoring system for assessing intracellular phenylalanine bioavailability in cyanobacteria. Biosens. Bioelectron. 2024, 244, 115792. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Kabade, P.G.; Gnanapragasam, N.; Singh, U.M.; Gurjar, A.K.S.; Rai, A.; Sinha, P.; Kumar, A.; Singh, V.K. Codon Usage Provide Insights into the Adaptation of Rice Genes under Stress Condition. Int. J. Mol. Sci. 2023, 24, 1098. [Google Scholar] [CrossRef]
- Bu, Y.; Wu, X.; Sun, N.; Man, Y.; Jing, Y. Codon usage bias predicts the functional MYB10 gene in Populus. J. Plant Physiol. 2021, 265, 153491. [Google Scholar] [CrossRef] [PubMed]
- Komar, A.A.; Samatova, E.; Rodnina, M.V. Translation Rates and Protein Folding. J. Mol. Biol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Nasrullah, I.; Butt, A.M.; Tahir, S.; Idrees, M.; Tong, Y. Genomic analysis of codon usage shows influence of mutation pressure, natural selection, and host features on Marburg virus evolution. BMC Evol. Biol. 2015, 15, 174. [Google Scholar] [CrossRef]
- Wu, P.; Xiao, W.; Luo, Y.; Xiong, Z.; Chen, X.; He, J.; Sha, A.; Gui, M.; Li, Q. Comprehensive analysis of codon bias in 13 Ganoderma mitochondrial genomes. Front. Microbiol. 2023, 14, 1170790. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Li, X. Analysis of synonymous codon usage patterns in different plant mitochondrial genomes. Mol. Biol. Rep. 2009, 36, 2039–2046. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, B.; Gu, G.; Yuan, J.; Shen, S.; Jin, L.; Lin, Z.; Lin, J.; Xie, X. Genome-wide identification and expression analysis of the R2R3-MYB gene family in tobacco (Nicotiana tabacum L.). BMC Genom. 2022, 23, 432. [Google Scholar] [CrossRef]
- Feng, P.M.; Chen, W.; Lin, H.; Chou, K.C. iHSP-PseRAAAC: Identifying the heat shock protein families using pseudo reduced amino acid alphabet composition. Anal. Biochem. 2013, 442, 118–125. [Google Scholar] [CrossRef]
- Deng, Y.; Zheng, H.; Yan, Z.; Liao, D.; Li, C.; Zhou, J.; Liao, H. Full-Length Transcriptome Survey and Expression Analysis of Cassia obtusifolia to Discover Putative Genes Related to Aurantio-Obtusin Biosynthesis, Seed Formation and Development, and Stress Response. Int. J. Mol. Sci. 2018, 19, 2476. [Google Scholar] [CrossRef]
- Zhong, L.; Zhou, W.; Wang, H.; Ding, S.; Lu, Q.; Wen, X.; Peng, L.; Zhang, L.; Lu, C. Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell 2013, 25, 2925–2943. [Google Scholar] [CrossRef]
- Huang, J.; Hai, Z.; Wang, R.; Yu, Y.; Chen, X.; Liang, W.; Wang, H. Genome-wide analysis of HSP20 gene family and expression patterns under heat stress in cucumber (Cucumis sativus L.). Front. Plant Sci. 2022, 13, 968418. [Google Scholar] [CrossRef]
- Hua, Y.; Liu, Q.; Zhai, Y.; Zhao, L.; Zhu, J.; Zhang, X.; Jia, Q.; Liang, Z.; Wang, D. Genome-wide analysis of the HSP20 gene family and its response to heat and drought stress in Coix (Coix lacryma-jobi L.). BMC Genom. 2023, 24, 478. [Google Scholar] [CrossRef]
- Lopes-Caitar, V.S.; de Carvalho, M.C.; Darben, L.M.; Kuwahara, M.K.; Nepomuceno, A.L.; Dias, W.P.; Abdelnoor, R.V.; Marcelino-Guimaraes, F.C. Genome-wide analysis of the Hsp20 gene family in soybean: Comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genom. 2013, 14, 577. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Cheng, Y.; Feng, K.; Ruan, M.; Ye, Q.; Wang, R.; Li, Z.; Zhou, G.; Yao, Z.; Yang, Y.; et al. Genome-Wide Identification and Expression Profiling of Tomato Hsp20 Gene Family in Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2016, 7, 1215. [Google Scholar] [CrossRef]
- Centurion, V.B.; Lacerda-Junior, G.V.; Duarte, A.W.F.; Silva, T.R.; Silva, L.J.; Rosa, L.H.; Oliveira, V.M. Dynamics of microbial stress responses driven by abiotic changes along a temporal gradient in Deception Island, Maritime Antarctica. Sci. Total Environ. 2021, 758, 143671. [Google Scholar] [CrossRef] [PubMed]
- Mu, C.; Zhang, S.; Yu, G.; Chen, N.; Li, X.; Liu, H. Overexpression of small heat shock protein LimHSP16.45 in Arabidopsis enhances tolerance to abiotic stresses. PLoS ONE 2013, 8, e82264. [Google Scholar] [CrossRef]
- Sun, X.; Sun, C.; Li, Z.; Hu, Q.; Han, L.; Luo, H. AsHSP17, a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA-dependent and independent signalling to attenuate plant response to abiotic stress. Plant Cell Environ. 2016, 39, 1320–1337. [Google Scholar] [CrossRef]
- Arias, T.; Beilstein, M.A.; Tang, M.; McKain, M.R.; Pires, J.C. Diversification times among Brassica (Brassicaceae) crops suggest hybrid formation after 20 million years of divergence. Am. J. Bot. 2014, 101, 86–91. [Google Scholar] [CrossRef]
- Fang, C.; Hamilton, J.P.; Vaillancourt, B.; Wang, Y.W.; Wood, J.C.; Deans, N.C.; Scroggs, T.; Carlton, L.; Mailloux, K.; Douches, D.S.; et al. Cold stress induces differential gene expression of retained homeologs in Camelina sativa cv Suneson. Front. Plant Sci. 2023, 14, 1271625. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Oh, J.M.; Luan, S.; Carlson, J.E.; Ahn, S.J. Cold stress causes rapid but differential changes in properties of plasma membrane H(+)-ATPase of camelina and rapeseed. J. Plant Physiol. 2013, 170, 828–837. [Google Scholar] [CrossRef]
- Qin, Y.; Li, Q.; An, Q.; Li, D.; Huang, S.; Zhao, Y.; Chen, W.; Zhou, J.; Liao, H. A phenylalanine ammonia lyase from Fritillaria unibracteata promotes drought tolerance by regulating lignin biosynthesis and SA signaling pathway. Int. J. Biol. Macromol. 2022, 213, 574–588. [Google Scholar] [CrossRef]
- Mazumdar, P.; Binti Othman, R.; Mebus, K.; Ramakrishnan, N.; Ann Harikrishna, J. Codon usage and codon pair patterns in non-grass monocot genomes. Ann. Bot. 2017, 120, 893–909. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Ma, B.; Shen, J.; Zhao, S.; Ma, X.; Wang, Z.; Fan, Y.; Tang, Q.; Wei, D. The evolution of the expansin gene family in Brassica species. Plant Physiol. Biochem. 2021, 167, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Gan, Z.; Zhang, F.; Yi, X.; Zhang, J.; Wan, X. Analysis of codon usage patterns in citrus based on coding sequence data. BMC Genom. 2020, 21, 234. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Zhang, F.; Dong, Q.; Chen, S. Insights into Comparative Genomics, Codon Usage Bias, and Phylogenetic Relationship of Species from Biebersteiniaceae and Nitrariaceae Based on Complete Chloroplast Genomes. Plants 2020, 9, 1605. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shen, Z.; Meng, X.; Zhang, L.; Liu, Z.; Liu, M.; Zhang, F.; Zhao, J. Codon usage patterns across seven Rosales species. BMC Plant Biol. 2022, 22, 65. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Quan, H.; Huang, B.; Ji, H.; Zhang, T.; Chen, J.; Zhou, J. Integrated transcriptomic and metabolomic analysis reveals the molecular basis of tissue-specific accumulation of bioactive steroidal alkaloids in Fritillaria unibracteata. Phytochemistry 2023, 214, 113831. [Google Scholar] [CrossRef]
- He, B.; Dong, H.; Jiang, C.; Cao, F.; Tao, S.; Xu, L.A. Analysis of codon usage patterns in Ginkgo biloba reveals codon usage tendency from A/U-ending to G/C-ending. Sci. Rep. 2016, 6, 35927. [Google Scholar] [CrossRef] [PubMed]
- Palanisamy, N.; Osman, N.; Ohnona, F.; Xu, H.T.; Brenner, B.; Mesplede, T.; Wainberg, M.A. Does antiretroviral treatment change HIV-1 codon usage patterns in its genes: A preliminary bioinformatics study. AIDS Res. Ther. 2017, 14, 2. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, W.; Li, H.; Wang, Y.; Li, D.; Xue, C.; Liu, Z.; Liu, M.; Zhao, J. Genome-wide analysis of the bZIP gene family in Chinese jujube (Ziziphus jujuba Mill.). BMC Genom. 2020, 21, 483. [Google Scholar] [CrossRef]
- Chamani Mohasses, F.; Solouki, M.; Ghareyazie, B.; Fahmideh, L.; Mohsenpour, M. Correlation between gene expression levels under drought stress and synonymous codon usage in rice plant by in-silico study. PLoS ONE 2020, 15, e0237334. [Google Scholar] [CrossRef]
- Sharp, P.M.; Tuohy, T.M.; Mosurski, K.R. Codon usage in yeast: Cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986, 14, 5125–5143. [Google Scholar] [CrossRef]
- Jiang, R.; Zou, M.; Qin, Y.; Tan, G.; Huang, S.; Quan, H.; Zhou, J.; Liao, H. Modeling of the Potential Geographical Distribution of Three Fritillaria Species under Climate Change. Front. Plant Sci. 2021, 12, 749838. [Google Scholar] [CrossRef]
- Waters, E.R.; Vierling, E. Plant small heat shock proteins—Evolutionary and functional diversity. New Phytol. 2020, 227, 24–37. [Google Scholar] [CrossRef]
- Hsiao, Y.Y.; Lin, C.H.; Liu, J.K.; Wong, T.Y.; Kuo, J. Analysis of Codon Usage Patterns in Toxic Dinoflagellate Alexandrium tamarense through Expressed Sequence Tag Data. Comp. Funct. Genom. 2010, 2010, 138538. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Zhou, J.; Li, Z.F.; Wang, L.; Gu, X.; Zhong, Y. Comparative Analysis of Codon Usage Patterns among Mitochondrion, Chloroplast and Nuclear Genes in Triticum aestivum L. J. Integr. Plant Biol. 2007, 49, 246–254. [Google Scholar] [CrossRef]
- Zhanjun, W.; Ziqi, W.; Zhaoxia, W.; Zulan, O.; Jie, L.; Qianwen, C.; Zhongdong, X.; Zhaoliang, Z. A comparative study of the evolution and codon usage bias in WOX gene family of three Camellia sinensis cultivars. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2022, 46, 71–80. [Google Scholar] [CrossRef]
- Kawabe, A.; Miyashita, N.T. Patterns of codon usage bias in three dicot and four monocot plant species. Genes Genet. Syst. 2003, 78, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Nag, D.; Mazumder, T.H.; Uddin, A. Codon usage pattern and prediction of gene expression level in Bungarus species. Gene 2017, 604, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Luo, Y.; Sha, A.; Xiao, W.; Xiong, Z.; Chen, X.; He, J.; Peng, L.; Zou, L. Analysis of synonymous codon usage patterns in mitochondrial genomes of nine Amanita species. Front. Microbiol. 2023, 14, 1134228. [Google Scholar] [CrossRef]
- Sun, W.; Bernard, C.; van de Cotte, B.; Van Montagu, M.; Verbruggen, N. At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J. 2001, 27, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Duret, L.; Mouchiroud, D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. USA 1999, 96, 4482–4487. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Drummond, D.A.; Wilke, C.O. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 2008, 134, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Cannarozzi, G.; Schraudolph, N.N.; Faty, M.; von Rohr, P.; Friberg, M.T.; Roth, A.C.; Gonnet, P.; Gonnet, G.; Barral, Y. A role for codon order in translation dynamics. Cell 2010, 141, 355–367. [Google Scholar] [CrossRef]
- Xiong, B.; Wang, T.; Huang, S.; Liao, L.; Wang, X.; Deng, H.; Zhang, M.; He, J.; Sun, G.; He, S.; et al. Analysis of Codon Usage Bias in Xyloglucan Endotransglycosylase (XET) Genes. Int. J. Mol. Sci. 2023, 24, 6108. [Google Scholar] [CrossRef]
- Tian, C.; Quan, H.; Jiang, R.; Zheng, Q.; Huang, S.; Tan, G.; Yan, C.; Zhou, J.; Liao, H. Differential roles of Cassia tora 1-deoxy-D-xylulose-5-phosphate synthase and 1-deoxy-D-xylulose-5-phosphate reductoisomerase in trade-off between plant growth and drought tolerance. Front. Plant Sci. 2023, 14, 1270396. [Google Scholar] [CrossRef]
- Yang, J.; Chu, Q.; Meng, G.; Kong, W. The complete chloroplast genome sequences of three Broussonetia species and comparative analysis within the Moraceae. PeerJ 2022, 10, e14293. [Google Scholar] [CrossRef]
- Jiang, W.; Lv, B.; Wu, X.; Wang, J.; Wu, G.; Shi, C.; Tang, X. Analysis of synonymous codon usage patterns in the edible fungus Volvariella volvacea. Biotechnol. Appl. Biochem. 2017, 64, 218–224. [Google Scholar] [CrossRef]
- Li, G.; Pan, Z.; Gao, S.; He, Y.; Xia, Q.; Jin, Y.; Yao, H. Analysis of synonymous codon usage of chloroplast genome in Porphyra umbilicalis. Genes Genom. 2019, 41, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Akturk Dizman, Y. Codon usage bias analysis of the gene encoding NAD(+)-dependent DNA ligase protein of Invertebrate iridescent virus 6. Arch. Microbiol. 2023, 205, 352. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Huang, D.; Fu, L.; Qin, Y.; Tang, J.; Lv, S.; Liao, H.; Zhou, J. Characterization of Arabidopsis thaliana Hsp20 gene family and its expression analysis under drought and salt stress. Life Sci. Res. 2023, 27, 162–169. [Google Scholar]
- Song, S.; Chen, A.; Zhu, J.; Yan, Z.; An, Q.; Zhou, J.; Liao, H.; Yu, Y. Structure basis of the caffeic acid O-methyltransferase from Ligusiticum chuanxiong to understand its selective mechanism. Int. J. Biol. Macromol. 2022, 194, 317–330. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, H.; Liu, J.; Chen, Y.; Yu, X.; Luo, C.; Sang, L.; Zhou, J.; Liao, H. Bioinformatic Analysis of Codon Usage Bias of HSP20 Genes in Four Cruciferous Species. Plants 2024, 13, 468. https://doi.org/10.3390/plants13040468
Ji H, Liu J, Chen Y, Yu X, Luo C, Sang L, Zhou J, Liao H. Bioinformatic Analysis of Codon Usage Bias of HSP20 Genes in Four Cruciferous Species. Plants. 2024; 13(4):468. https://doi.org/10.3390/plants13040468
Chicago/Turabian StyleJi, Huiyue, Junnan Liu, Yineng Chen, Xinyi Yu, Chenlu Luo, Luxi Sang, Jiayu Zhou, and Hai Liao. 2024. "Bioinformatic Analysis of Codon Usage Bias of HSP20 Genes in Four Cruciferous Species" Plants 13, no. 4: 468. https://doi.org/10.3390/plants13040468
APA StyleJi, H., Liu, J., Chen, Y., Yu, X., Luo, C., Sang, L., Zhou, J., & Liao, H. (2024). Bioinformatic Analysis of Codon Usage Bias of HSP20 Genes in Four Cruciferous Species. Plants, 13(4), 468. https://doi.org/10.3390/plants13040468








