Abstract
Ran GTPases play essential roles in plant growth and development. Our previous studies revealed the nuclear localization of DlRan3A and DlRan3B proteins and proposed their functional redundancy and distinction in Dimocarpus longan somatic embryogenesis, hormone, and abiotic stress responses. To further explore the possible roles of DlRan3A and DlRan3B, gene expression analysis by qPCR showed that their transcripts were both more abundant in the early embryo and pulp in longan. Heterologous expression of DlRan3A driven by its own previously cloned promoter led to stunted growth, increased root hair density, abnormal fruits, bigger seeds, and enhanced abiotic stress tolerance. Conversely, constitutive promoter CaMV 35S (35S)-driven expression of DlRan3A, 35S, or DlRan3B promoter-controlled expression of DlRan3B did not induce the alterations in growth phenotype, while they rendered different hypersensitivities to abiotic stresses. Based on the transcriptome profiling of longan Ran overexpression in tobacco plants, we propose new mechanisms of the Ran-mediated regulation of genes associated with cell wall biosynthesis and expansion. Also, the transgenic plants expressing DlRan3A or DlRan3B genes controlled by 35S or by their own promoter all exhibited altered mRNA levels of stress-related and transcription factor genes. Moreover, DlRan3A overexpressors were more tolerant to salinity, osmotic, and heat stresses, accompanied by upregulation of oxidation-related genes, possibly involving the Ran-RBOH-CIPK network. Analysis of a subset of selected genes from the Ran transcriptome identified possible cold stress-related roles of brassinosteroid (BR)-responsive genes. The marked presence of genes related to cell wall biosynthesis and expansion, hormone, and defense responses highlighted their close regulatory association with Ran.
1. Introduction
The longan tree (Dimocarpus longan Lour., Sapindaceae), a tropical evergreen fruit tree in southern China, is of great edible value and exhibits medicinal properties. However, there are certain challenges such as lacking aborted-seed varieties and the influence of environmental stresses on vegetative growth, fruit yield, and quality [1]. Being a Sapindaceae family of fruit crops, thicker arils and abortive seeds are important quality traits. In recent years, therefore, there have been several reports on the mechanisms of longan fruit and seed development which are controlled by various regulators [2,3].
Ras-related nuclear protein (Ran) GTPases, also known as molecular switches, play a universal role in nucleocytoplasmic transport, specifically expressed during embryogenesis, and are essential for cell division in animal development [4,5]. Plant Ran GTPases, along with their cofactors and nucleoporins, are involved in diverse biological processes including plant growth and root development [6,7], regulation of hormone (such as auxin and abscisic acid, ABA) sensitivity [6,8,9], enhancement of disease resistance [10], and stress responses, such as those to drought, salinity, osmotic stress, and cold and oxidative stresses [7,8,11]. This indicates that plant Ran may have a remarkable impact on plant growth and, hence, its economic traits when overexpressed.
Previous studies by our research group have cloned DlRan3A and DlRan3B, two Ran genes from somatic embryos in D. longan, and revealed their involvement in longan somatic embryogenesis [12]. DlRan3A and DlRan3B promoters (hereafter, referred to as ‘pDlRan3A’ and ‘pDlRan3B’, respectively), 1256 bp and 1569 bp in length, were cloned from embryogenic callus (EC) [13,14]. The deletion experiments showed that pDlRan3A and pDlRan3B might be involved in transcriptional control of plant hormones and specific defense reactions [14,15]. However, the molecular mechanism of longan Ran and its relationship with plant growth (especially seed and fruit development) and responses to the external environment need further investigation.
In this study, expression profiling of DlRan3A and DlRan3B was analyzed in different longan tissues, during zygotic embryo development, and in the pulp of ripening fruits, to reveal the potential role of Ran in embryo and fruit development in woody fruit trees. Currently, the molecular basis of the Ran gene and its promoter for the regulation in plant embryo and fruit development is poorly understood, in part due to the fact that few studies have aimed to investigate global changes in the gene expression elicited by overexpression of the plant Ran gene. For selection and further application of Ran genes in molecular breeding using transgenic technology, we generated transgenic tobacco plants overexpressing DlRan3A and DlRan3B, using both CaMV 35S (35S) and their own promoters. Based on the analysis of plant phenotype and environmental stress response, transcriptome analysis of transgenic lines was subsequently performed to identify the possible downstream genes responsible for the specific plant growth and stress tolerance phenotypes among different transgenic lines. The selected genes shared or were distinct between transgenic lines, respectively, will be useful to examine the effect and underlying molecular mechanism of longan Ran overexpression, and for genetic engineering to enhance the stress tolerance of woody fruit trees, including longan.
2. Results
2.1. Longan DlRan3A and DRan3B Are Highly Expressed in Early Embryos and Pulp (Aril)
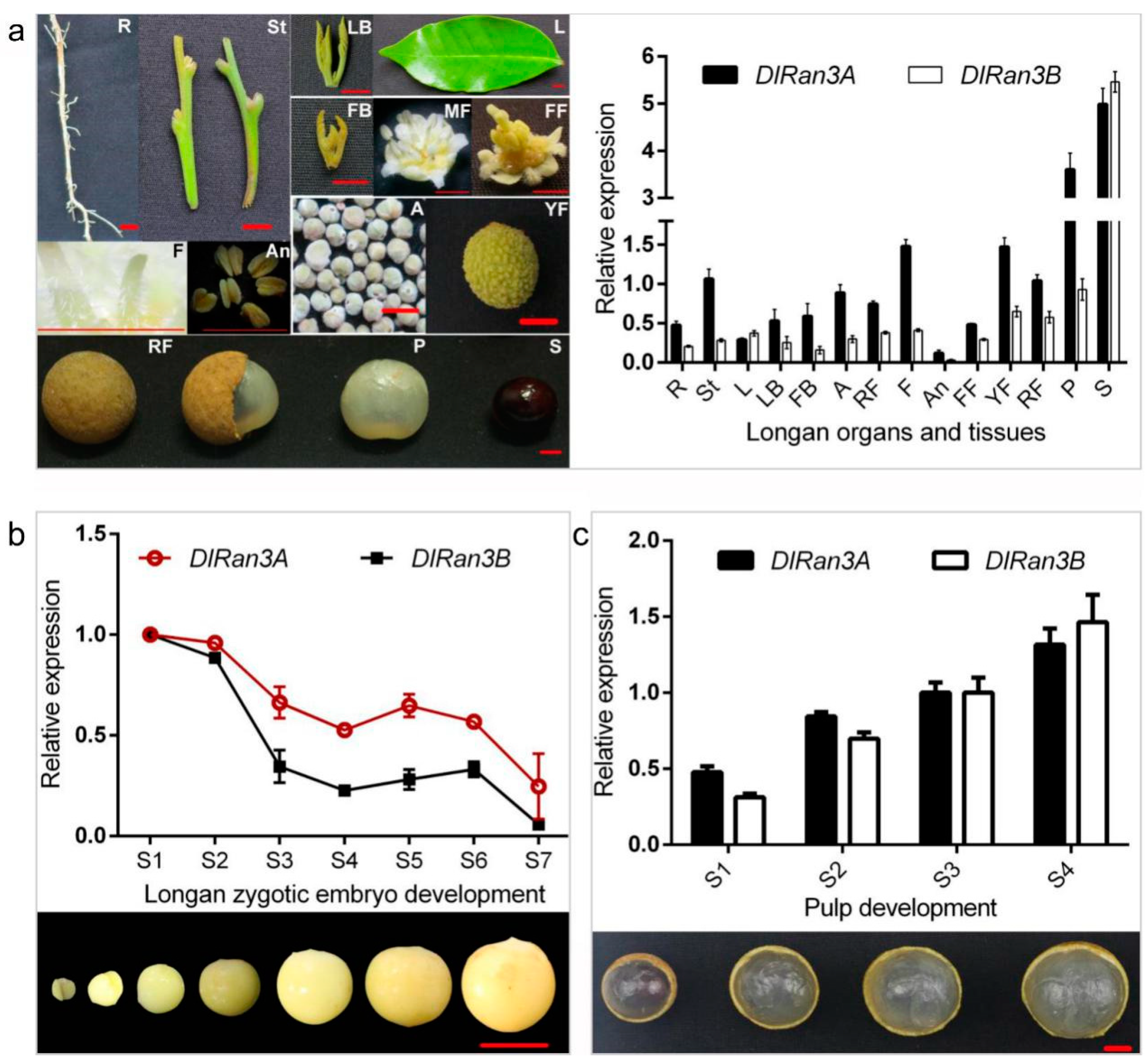
To explore the potential developmental role of DlRan3A and DlRan3B, we first investigated their expression patterns in D. longan Lour. cv. Honghezi in diverse tissues (Figure 1a). We found that, apart from the fundamental expressions in longan tissues, DlRan3A and DlRan3B were both predominantly expressed in the seeds and then in the pulp, but displayed the lowest levels in the anthers (Figure 1a). This indicates that Ran is indispensable to longan growth and development while DlRan3A and DlRan3B genes might play more significant roles in seed and pulp development.
Figure 1.
Relative expression levels of DlRan3A and DlRan3B in different tissues, developing zygotic embryos and pulp in longan. (a) Relative expression of DlRan3A and DlRan3B in different tissues (updated from Chen [16]). R: root; St: stem; L: leaf; LB: leaf bud; FB: floral bud; A: alabastrum; MF: male flower; F: filament; An: anther; FF: female flower; YF: young fruit; RF: ripe fruit; P: pulp; S: seed; bar = 5 mm. (b) Relative expression of DlRan3A and DlRan3B during zygotic embryo development. Longan zygotic embryos from stage S1 to S7 were collected from 16 June to 12 July in 2015, every four or every five days; bar = 10 mm. (c) Relative expression of DlRan3A and DlRan3B in different sizes of longan pulp. Longan pulp from stage S1 to S4 were collected in August 2015; bar = 10 mm.
To further characterize DlRan3A and DlRan3B expression patterns during longan seed (embryo) and pulp development, we next examined their expressions during ‘Honghezi’ zygotic embryo development and in the pulp of ripening fruits. In the early stages (S1 and S2), both genes showed high expression with a fluctuating trend that declined during the zygotic embryo development (Figure 1b). However, both gene expressions showed an increase in the pulp of ripening fruits (Figure 1c).
2.2. Characterization of DlRan3A and DlRan3B Genes and the Use of Their Own Promoters by Heterologous Expression in Tobacco Plants
To further investigate the function of DlRan3A and DlRan3B, we cloned each gene into pCAMBIA1301 driven by 35S or Ran promoters and transformed into Agrobacterium tumefaciens strain EHA105. The T2 line 35S-driven expression of DlRan3A or DlRan3B (henceforth, referred to as ‘35S_A’ or ‘35S_B’, respectively) and those driven by the Ran promoter (hereinafter, referred to as ‘PA_A’ or ‘PB_B’, respectively) were employed for the following experiments.
At the same planting time, excluding PA_A, there were no significant differences in plant height and blade generation rate among the transgenic tobaccos (Figure S3). Notably, PA_A showed a dwarf phenotype with reduced height and thick stems, along with phenotypes of late flowering and reduced axillary buds (Figure 2a). After vertical cultivation for 7 d, 14 d, and 21 d, excluding PA_A, there were no significant differences in root growth among the transgenic tobaccos. Notably, PA_A exhibited a sturdier root system along with increased numbers of root hairs (Figure 2b and Figure S4). Even after 50–60 d of growth, PA_A maintained distinct phenotypes in fruit and seed development, showing abnormal fruits, larger seeds, decreased seed setting rate, and increased seed weight (Figure 2c and Figure S5). This indicates that pDlRan3A-driven expression of DlRan3A caused stunted growth and significantly affected the development of root, fruit, and seed. Moreover, the histochemical staining assay conducted to assess pDlRan3A- or pDlRan3B-driven expression of the GUS gene in transgenic tobaccos (hereafter, referred to as ‘PA_GUS’ or ‘PB_GUS’, respectively) revealed that both the promoters led to GUS expression in roots, leaves, flowers, fruits, and seeds. However, the DlRan3A promoter could drive greater GUS accumulation in petals (Figures S6 and S7).

Figure 2.
Phenotype of transgenic tobaccos. (a) The entire phenotypes of transgenic tobaccos. WT and T2 lines of P35S_A, PA_A, P35S_B, and PB_B (45 d) are displayed in the graph from left to right. (b) The phenotype of transgenic tobacco roots (21 d). WT and T2 lines of P35S_A and PA_A are displayed in the graph from left to right. The upper images illustrate the transgenic tobaccos germinated and vertically cultivated on MS medium for 21 d, and the lower images illustrate the 21-day-cultivated tobacco roots stained with propidium iodide, as observed under a confocal microscope (bar = 100 μm). (c) The phenotypes of flowers, fruits, and seeds of transgenic tobaccos. The WT and T2 lines of P35S_A and PA_A are displayed in the graph from left to right (bar = 5 mm).
2.3. Heterologous Expression in Tobaccos Reveals Shared and Distinct Roles of DlRan3A and DlRan3B in Various Stresses
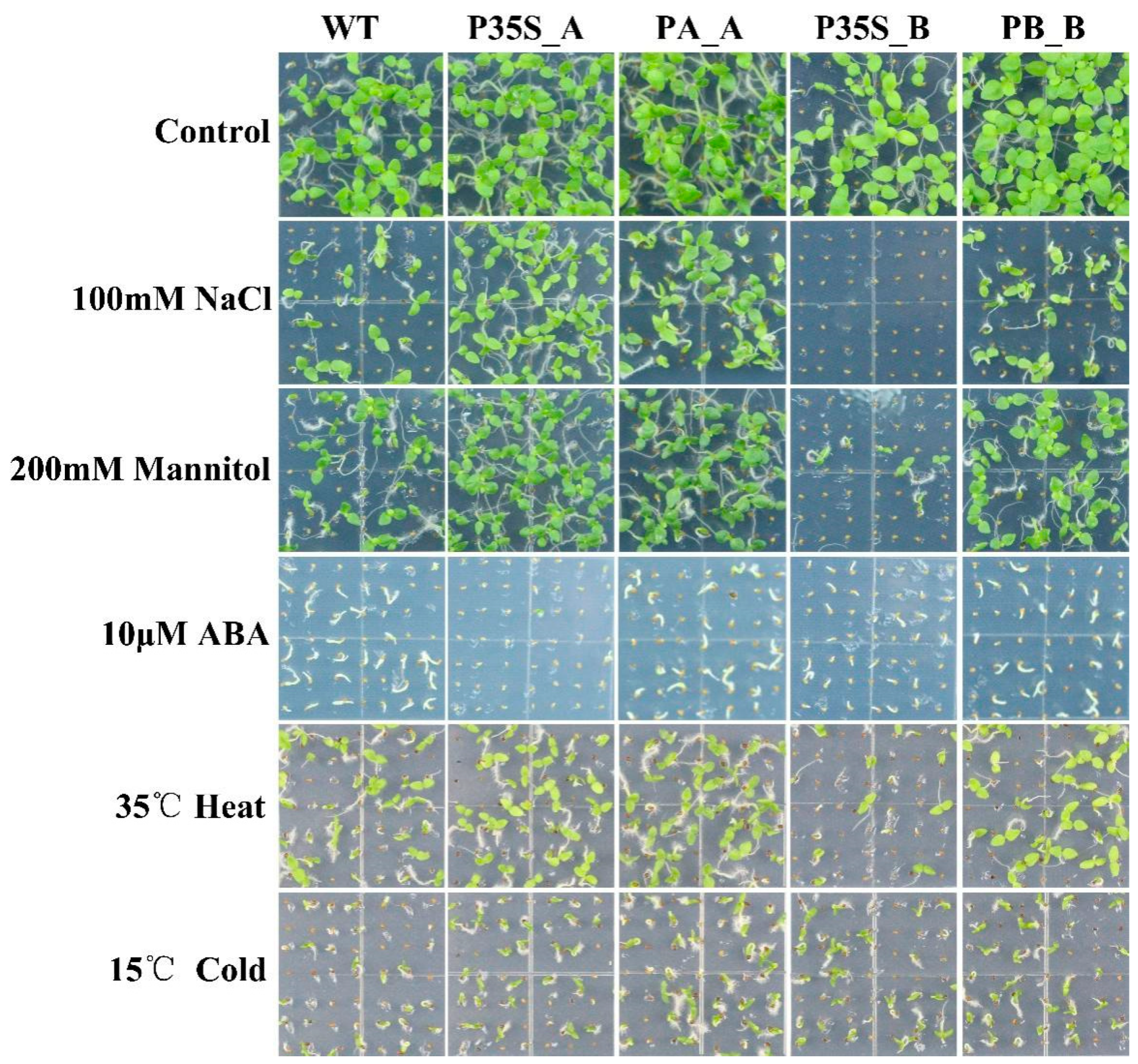
Under 100 mM NaCl stress, 200 mM mannitol stress, 10 μM ABA, 35 °C heat stress, and 15 °C cold stress, tobaccos expressing DlRan3A (i.e., P35S_A and PA_A) exhibited stronger tolerance to salinity, osmotic, and heat stresses. In contrast, tobaccos expressing DlRan3B (i.e., P35S_B, and PB_B) showed relatively weaker tolerance against abiotic stress. Remarkably, P35S_B tobaccos were hypersensitive to salinity, osmotic, and heat stresses, while P35S_A tobaccos were hypersensitive to ABA. Additionally, tobaccos expressing DlRan3A or DlRan3B showed mild tolerance against cold stress (Figure 3).
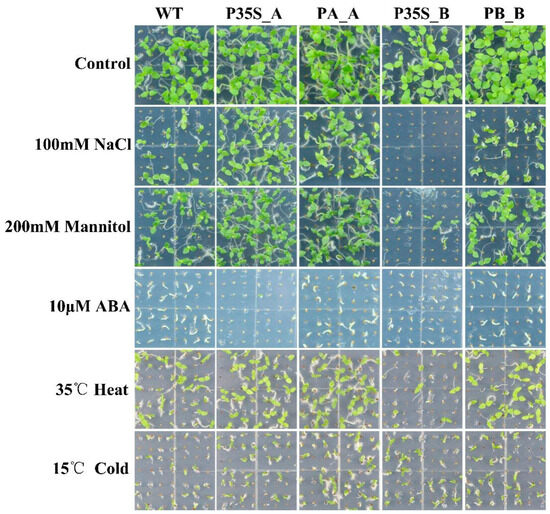
Figure 3.
Abiotic stress tolerance of transgenic tobaccos. Graphics related to the WT and T2 lines of P35S_A, PA_A, P35S_B, and PB_B are displayed from left to right; a control group without any treatment and the treatment groups belonging to 100 mM NaCl, 200 mM mannitol, 10 μM ABA, 35 °C heat, and 15 °C cold stresses are displayed from top to bottom.
2.4. Transcriptome Profiling of Transgenic Tobaccos Revealed Alteration in Genes Involved in Biosynthetic Processes Related to the Cell Wall, Hormone Signaling, and Stress Responses
To understand the molecular functions of longan Ran GTPases, RNA-sequencing (RNA-Seq) profiling was conducted to estimate the effects on the entire transcriptome. Sequencing libraries were generated from WT, P35S_A, P35S_B, PA_A, and PB_B transgenic tobacco plants cultivated in Murashige and Skoog (MS) medium for 21 d. Reads alignment to the reference genome is shown in Table S5. The differentially expressed genes (DEGs) with higher transcript levels in transgenic lines than the corresponding control sample were denoted “up-regulated genes”, whereas those with lower transcript levels were defined as “down-regulated genes”. As shown in Table S6, the PB_B versus WT group (hereinafter, referred to as ‘PB_B/WT’) showed the largest number of DEGs, followed by PA_A versus WT groups (hereinafter, referred to as ‘PA_A/WT’); the DEGs in the PA_A versus P35S_A groups or PB_B versus P35S_B groups (hereinafter, referred to as ‘PA_A/P35S_A’ or ‘PB_B/P35S_B’, respectively) were less than any transgenic lines versus the WT groups.
Notably, not many DEGs in P35S_A versus WT groups (hereinafter, referred to as ‘35S_A/WT’) were shared with the P35S_B versus WT groups (hereinafter, referred to as ‘35S_B/WT’) (Figure S8a and Table S7). Furthermore, functional annotation revealed that the shared DEGs are mainly involved in cell wall organization or biogenesis (such as cellulose synthase, CESA) and plant resistance (such as asparagine synthetase, AS, and heat shock protein, HSP). Importantly, PA_A/WT and 35S_A/WT shared a certain number of DEGs (Figure S8b and Table S8). These are also mainly involved in cell wall organization or biogenesis (such as cellulose synthase) and plant resistance, such as AS, HSP, pleiotropic drug resistance protein (PDR), calmodulin-binding protein (CBP), peroxide (PER), and respiratory burst oxidase homolog (RBOH). Among the DEGs, most DEGs were up-regulated, except some like CYP450 (cytochrome P450) and HSP. The shared DEGs between PB_B/WT and 35S_B/WT are listed in Figure S8c and Table S9, including calcium-binding protein, CESA, expansin, and genes involved in plant resistance (such as AS, NAC, ERF, WRKY, bHLH, C3H, GRAS, and HSP). Among these, most DEGs associated with plant resistance were down-regulated.
Additionally, we also analyzed the expression profiles of transcription factor genes, cell wall biosynthesis and expansin/extensin genes, hormone-related genes, and stress-related genes between non-transgenic and transgenic tobaccos. As shown in Figure S9, the heat map indicated that certain transcription factors (MYB, NAC, WRKY, ERF, GRAS, bHLH, etc.) were down-regulated in the transgenic lines, especially in the 35S_B and PB_B lines. Regarding hormone-related genes, auxin-related DEGs were up- or down-regulated; ethylene-related ERFs were mostly down-regulated, especially in the 35S_B and PB_B lines. Also, all of the brassinosteroid (BR)-related EXORDIUMs were up-regulated in the transgenic lines, along with down-regulated MYC2s (jasmonic acid-related) and up-regulated ARR9 (cytokinin-related). Notably, most extensin genes were up-regulated in PA_A along with three extensin genes (Niben101Scf02042g00001, Niben101Scf02191g03003, and Niben101Scf03036g00006). The two expansins (Niben101Scf03913g01045 and Niben101Scf20887g00008) were significantly up-regulated or down-regulated in the PA_A/P35S_A comparisons, respectively (Figure S10). The expression profiles of stress-related genes are shown in Figures S11 and S12.
2.5. Functional Classification, and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analyses of DEGs
Gene Ontology (GO) enrichment analysis was performed for global functional analysis of DEGs related to longan Ran expression. Compared to WT, PA_A showed the greatest number of significantly enriched pathways and related DEGs (Table S10).
In P35S_A (compared to WT), the oxidoreductase activity pathway acting on peroxide as an acceptor was significantly enriched for four DEGs: the peroxide gene, RBOH, and Novel00358, were up-regulated, while CAT1 (catalase isozyme 1) was down-regulated. In PA_A (compared to WT), among the sixteen significantly enriched pathways, “cell wall organization or biogenesis” or related pathways were predominant along with six up-regulated DEGs (mainly extensin gene). Other pathways were mainly enriched for upregulated CESAs, XET/XTH (xyloglucan endotransglucosylase/hydrolase), and UGE (Bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase), while UGT73C3 (UDP-glycosyltransferase 73C3), GT (Anthocyanidin 3-O-glucosyltransferase), and CIPK (Calcineurin B-like-interacting serine/threonine-protein kinase 11) were down-regulated.
Further comparative analysis of PA_A/P35S_A or PB_B/P35S_B illuminated the biological function of longan Ran promoters. For PA_A/P35S_A or PB_B/P35S_B, all the listed cell wall biogenesis pathways were significantly enriched for the three extensin genes mentioned above. However, these were up-regulated in PA_A/P35S_A but down-regulated in PB_B/P35S_B.
Likewise, in PA_A/PB_B, the significantly enriched pathways were mainly related to cell wall and oxidative stress. Notably, the oxidative stress-related pathways were significantly enriched for four up-regulated peroxide genes (Niben101Scf03990g00010, Niben101Scf00416g01009, Niben101Scf02709g01005, and Niben101Scf02349g03001) and a GPX (glutathione peroxidase) gene (Niben101Scf06369g03006) (also see Figure S13).
Moreover, the KEGG biochemical pathways analysis of related DEGs was implemented. Compared to WT, the pathways related to alanine, aspartate, and glutamate metabolism were significantly enriched in P35S_A, P35S_B, or PA_A, lines, respectively (Table S11), suggesting the key role of longan Ran in these processes. Also, we found five significantly enriched pathways in PA_A (compared to PB_B). Most of these pathways were related to up-regulated DEGs, including five GST (glutathione S-transferase) (Niben101Scf00069g02013, Niben101Scf10316g03004, Niben101Scf03147g10010, Niben101Scf03482g0101, and Niben101Scf11037g00006) and a GPX (mentioned above in the GO analysis of PA_A/PB_B) genes.
2.6. Verification qPCR Validation of Candidate DEGs
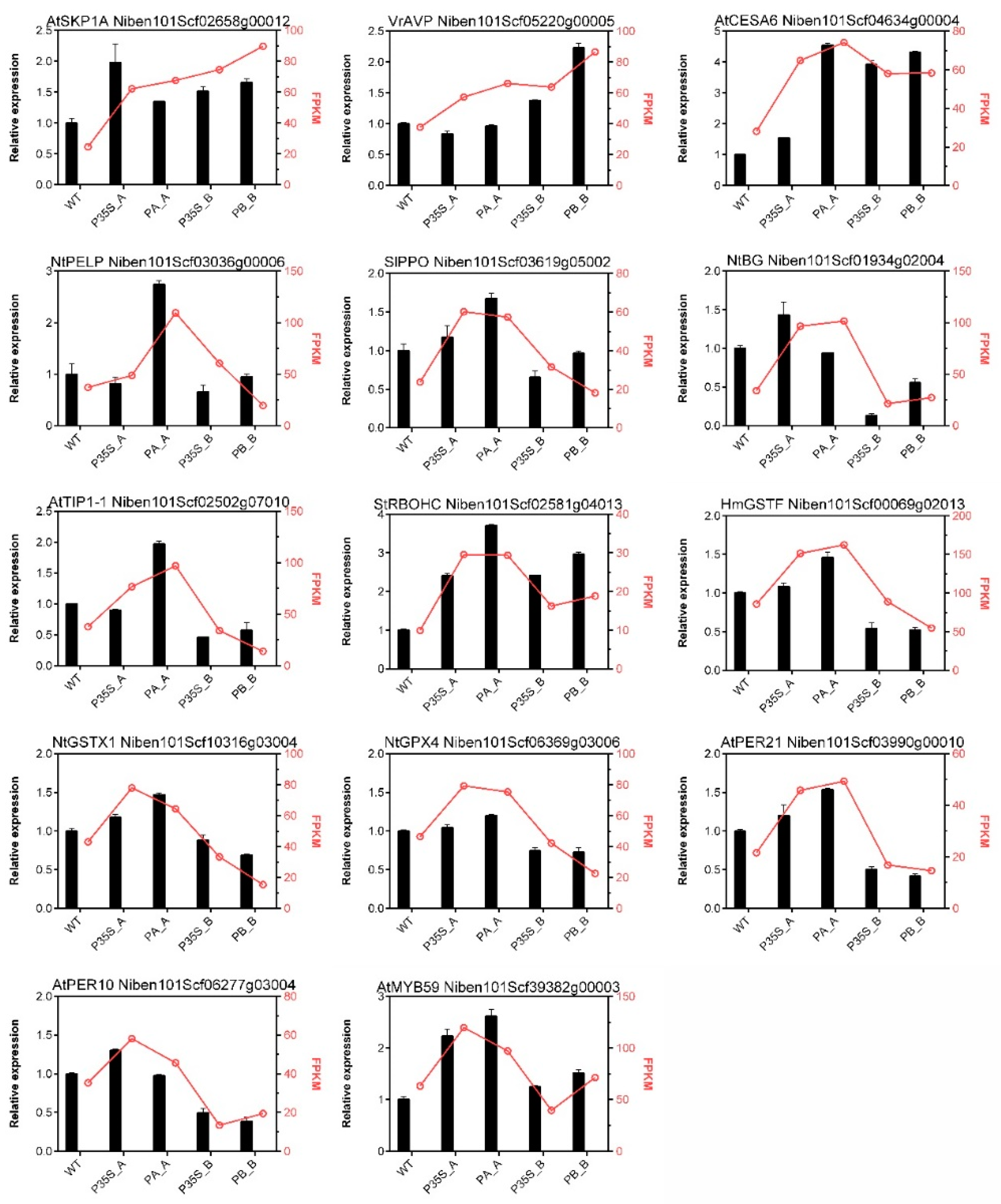
To validate the RNA-Seq results, 14 DEGs were selected for qPCR assay. The gene symbols, FPKM values, and relative expression levels are shown in Figure 4. Although the fold changes in transcript levels detected by RNA-Seq and qPCR were not perfectly matched, most of the candidate DEGs showed similar trends of change in gene expression as in the RNA-Seq results. In general, the expression patterns measured by the two methods were consistent.
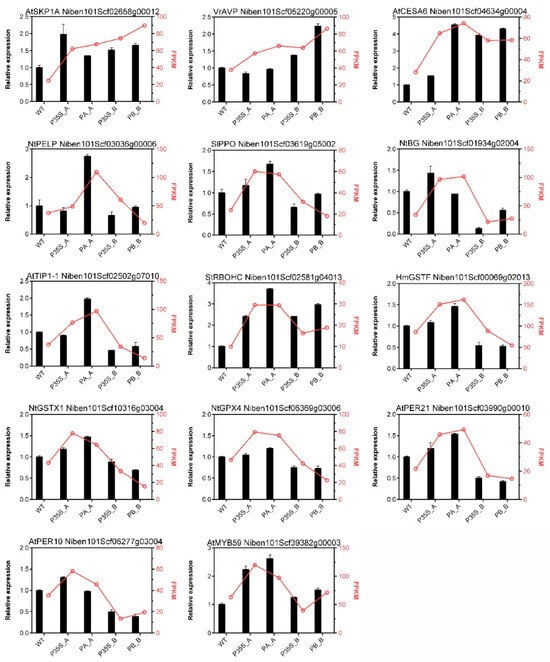
Figure 4.
Validation of RNA-seq results by qPCR. The corresponding gene ID is mentioned at the top of each graph. The columns represent the relative expression levels measured by qPCR using NbEF1a as a reference gene, and the red lines portray the FPKM change from RNA-seq.
3. Discussion
3.1. DlRan3A and DlRan3B Up-Regulate Cell Wall Biosynthesis and Expansion Genes to Regulate Early Embryo and Pulp Development in Longan
Studies have proved that plant Ran is ubiquitously expressed in all tissues, such as in tomato [17], Arabidopsis [18], tall fescue [19], sugarcane [20], etc. In longan, DlRan3A and DlRan3B also exhibited a certain fundamental expression in almost the whole tree, suggesting their essential roles in development. Ran is more expressed in meristematic tissues, embryos, or roots, which provides a clue to its functional diversity in plants [18,19,20]. Arabidopsis RAN1 interaction with DEM1 (defective embryo and meristems 1) is required for organized cell divisions during embryonic and post-embryonic growth in tomato [21]. Moreover, it was shown to mediate seed development by affecting the onset of endosperm cellularization [22]. A previous study revealed that, akin to its homolog AtRan3, longan Ran might have similar roles in cell proliferation during somatic embryogenesis [12]. Notably, DlRan3A and DlRan3B indicated functional similarity and specificity in transcriptional regulation during longan somatic embryogenesis [14,15]. Here, based on expression profiling of DlRan3A and DlRan3B at different development stages of seeds, fruits, and other tissues, we found that both genes were highly expressed in seeds and pulp (aril), as well as in the young embryos of early-stage zygotic embryo development. Additionally, they may positively regulate fruit pulp swelling. Importantly, based on the previous similar findings of their roles in late embryogenesis (cotyledon embryos), DlRan3A and DlRan3B might regulate early embryo development, especially the formation of cotyledons.
Proteins involved in cell wall synthesis and expansion in plants are closely related to plant development including embryo and fruit development [23]. For instance, XET/XTH, an important enzyme for cell wall loosening, has a key role in plant somatic embryogenesis [24]. Additionally, the genes encoding for cellulose synthase-like protein AtCSLA7, hydroxyproline-rich glycoprotein RSH (root-shoot-hypocotyl-defective), and AtEXT3 (extensin 3), are essential for cell wall structure and normal embryo development in Arabidopsis [25,26]. The current study suggests that longan Ran expression led to up-regulation of numerous cell wall biosynthesis and expansin/extensin genes, indicating the role of Ran in cell wall development. Additionally, during the fruit expansion stage, the cell wall needs to turn loose. We found that DlRan3A and DlRan3B were highly expressed in longan pulp, and their expression levels increased with the expansion of longan pulp (aril), along with several up-regulated genes related to cell wall loosening or fruit ripening (encoding expansins, XETs, XTHs, etc.). This indicates the potential role of Ran in fruit development by enhancing the accumulation of cell wall-loosening proteins. The current study provides significant new insights for Ran function during embryo and pulp (aril) development. However, the potential mechanism(s) of Ran-mediated transcript accumulation of cell wall-related genes during early embryo and pulp (aril) development needs to be further revealed.
3.2. pDlRan3A Driven Expression of DlRan3A Led to Stunted Plant Growth, Higher Root Hair Density, Abnormal Fruits, and Bigger Seeds, Potentially via Partial Regulation of Expansin- and Extensin-like Genes
Ran GTPase is closely related to both vegetative and reproductive growth in plants. Expression of wheat TaRAN1 prolongs the life cycle, elevates mitotic index, decreases lateral roots, increases primordial tissue, and enhances auxin hypersensitivity [6]. Similarly, ectopic expression of FaRan in Arabidopsis increases axillary buds and reduces apical dominance, suggesting its potential role in the initiation of meristem and subsequent growth and development [19]. In this study, pDlRan3A-controlled expression of DlRan3A led to stunted plant growth, increased root hair density, abnormal fruits, and larger seeds, suggesting the important role of Ran in certain organs. It seems that an excessive or ectopic presence of Ran, beyond its optimal concentration, may significantly affect plant development.
The agronomic characteristic dwarfism, such as reduced height and thick stems, is often desirable to bolster lodging resistance [27]. The current results suggest that pDlRan3A-driven expression of DlRan3A can be an additional functional factor for stunted plant growth. Importantly, stunted or dwarfed plant growth has also been linked to transcriptional regulation of genes related to cell wall synthesis and expansion [28], which is consistent with our finding that two expansin genes, AtEXLA2 Niben101Scf03913g01045 and OsEXPA4 Niben101Scf20887g00008, were, respectively, up-regulated and down-regulated in the PA_A/P35S_A comparison. Despite the fact that the suppressed expression of the expansin gene is known to reduce plant growth while the overexpression does not necessarily promote it [28,29,30], our finding highlights the role of DlRan3A in the regulation of stunted plant growth and provides a novel clue for understanding the molecular mechanism of Ran in the activation or suppression of expansin genes.
Root hairs, offering the plant a competitive advantage in the acquisition of water and nutrients from the soil, can enhance plant tolerance to abiotic stress [31]. Though previous studies have revealed the functional involvement of small GTPases in root hair growth [32,33], detailed investigations are required to understand the underlying mechanism(s). In particular, the role of Ran GTPases has not yet been reported in root hair growth. Here, we found that three extensin-like genes were up-regulated in PA_A (compared to P35S_A), and down-regulated in PB_B (compared to P35S_B), while both types were significantly enriched in pathways of cell wall organization or biogenesis. Based on the phenotypes of transgenic plants, we propose that DlRan3A and DlRan3B, driven by their native promoters, might play different roles in cell wall organization or biogenesis in longan tissues, especially via regulation of extensin-like genes. Emerging evidence suggests that extensin- and extensin-like proteins are essential for cell wall self-assembly and, hence, root hair development, such as in tomato [34], barley [35], Arabidopsis [36], etc. Here, we showed that DlRan3A plays a very specific role in root hair development via up-regulation of extensin-like genes, but further studies are required to discover and validate the downstream effectors of Ran in root hair development.
In addition, one of the three extensin-like genes was annotated as pistil-specific extensin-like protein gene ‘PELP’ (Niben101Scf03036g00006). Notably, extensins have been reported in reproductive development in some plants. For instance, the class III pistil-specific extensin-like proteins (PELPIIIs) of Nicotiana were associated with interspecific incompatibility, specific inhibition of pollen tube, and growth [37,38]. pDlRan3A-driven expression of DlRan3A led to abnormal fruits, larger seeds, a reduced seed setting rate, and increased seed weight, coinciding with increased and abnormal expression of PELP in PA_A. Thus, DlRan3A and PELP might be closely interrelated and involved in reproductive development, providing new perspectives on Ran-mediated transcriptional regulation of plant reproductive processes.
3.3. DlRan3A and DlRan3B Function in Stress Tolerance by Regulating Different Stress-Responsive Genes
A close association between Ran and plant responses to environmental stresses, such as salinity [39], osmotic stress [8], drought [7], low temperatures [9,40], aluminum toxicity [41], and oxidative stress [11], has been elucidated in numerous studies. However, the understanding of the underlying mechanisms is slowly improving, and there have been rare reports about the role of longan Ran in defense responses. In this study, using transcriptome analysis, we revealed numerous DEGs related to cell wall biosynthesis and stress response (PER, aquaporin, AS, GST, HSP, DNAJ, etc.), as well as stress-related transcription factor (MYB, NAC, WRKY, ERF, GRAS, bHLH, etc.) DEGs. The data strongly suggest the important roles of Ran in a complex stress regulatory network. We found that the tobacco overexpressing DlRan3A (P35S_A and PA_A) were more tolerant to abiotic stress (salinity, osmotic, and heat stress). Conversely, tobacco overexpressing DlRan3B (P35S_B and PB_B) exhibited relatively poor tolerance compared to the P35S_A and PA_A lines. In particular, compared to WT and DlRan3B overexpressing tobaccos, P35S_A tobaccos showed a little more sensitivity to ABA, which plays a critical role in various stress responses. More studies are needed to further elucidate the role of DlRan3A in relationship with ABA accumulation and stress (salinity, osmotic, and heat stress) tolerance.
Previous studies in rice suggesting that RAN overexpression improved cold tolerance are partially supported by our results. Despite Ran’s associations with cell division, the hormone BR might also participate in Ran-regulated cold tolerance. Notably, many previous studies have shown BR-induced cold tolerance in plants, such as in Arabidopsis and Brassica napus [42], Cucumis sativus [43], Chorispora bungeana [44], Elymus nutans [45], Medicago truncatula [46], etc. Importantly, plants overexpressing longan Ran showed increased accumulation of EXORDIUM transcripts than the WT (Figure S7), and well-characterized BR-responsive genes, such as expansins, XETs, aquaporins, and ASs [47,48,49], were significantly up-regulated in plants with increased longan Ran expression. The phenotypic observations, together with the transcriptome analysis of the BR-responsive genes, suggest a possible role of longan Ran in activated BR signaling and BR-mediated plant defense mechanisms, especially in cold tolerance.
Despite the similarities, the functional divergence between the two members of the longan Ran family in stress responses (salinity, osmotic, and heat) might be partially attributed to the inhibition of stress-regulated transcription factor (WRKY, ERF, GRAS, bHLH, C3H, C2H2, etc.) genes in P35S_B or PB_B. Moreover, the activation of oxidative stress-related genes (PER, GST, GPX, PPO, RBOH, etc.) can stimulate the antioxidant defense system in P35S_vA and PA_A, resulting in the differences between DlRan3A overexpressing tobaccos and DlRan3B overexpressing ones in terms of their stress responses (Figure S13). Plant RBOHs are the key enzymes that catalyze the generation of reactive oxygen species (ROS) during plant defense responses and are involved in the modulation of root growth [50]. Plant CBL (calcineurin B-like) and CIPK (CBL-interacting protein kinase) proteins, form one of the important Ca2+ decoding complexes to decipher Ca2+ signals elicited by environmental challenges [51]. Rac/Rop GTPases and CBL-CIPKs involve integration of calcium signaling into ROS regulation via direct interaction with RBOH [52]. Using GO analysis, we showed that CIPK was significantly down-regulated in PA_A, while the RBOH was up-regulated. Considering the fact that RBOH acts as a convergence point targeted by a complex regulatory network, these data endorse the notion that the resistant phenotypes and associated molecular changes in plants overexpressing DlRan3A are at least partly due to the Ran–RBOH–CIPK regulatory network. This new evidence suggests the involvement of Ran in plant defense responses via calcium signaling and ROS regulation.
4. Materials and Methods
4.1. Plant Materials
The different tissues of longan were collected from Fujian Agriculture and Forestry University in Fuzhou. All materials were mixed samples from at least six rootstock longan plants, collected and stored at −80 °C for further studies. Nicotiana benthamiana tobaccos were used for stable genetic transformation.
4.2. Gene Expression Analysis
Total RNA was extracted using the RNAprep Pure Plant Kit (TIANGEN Code, DP441, Beijing, China) or total plant RNA extraction kit (BioTeke Code, RP3312, Beijing, China) following the manufacturer’s protocol. cDNAs were synthesized using the PrimeScriptTM Perfect Real-Time RT Reagent Kit (TaKaRa Code, RR037A (Dalian, China)). Quantitative real-time PCR analysis (qPCR) was performed to evaluate the transcript levels of the DlRan3A and DlRan3B genes in longan tissues, during zygotic embryo and pulp developments. Typical reactions were prepared using the SYBR Premix Ex Taq kit (Takara) and all the qPCR reactions were performed in triplicate. QPCR assays were implemented using the LightCycler 480 qPCR instrument (Roche Applied Science, Basel, Switzerland) and cycling conditions were chosen according to the manufacturer’s protocol. The expression profiles of DlRan3A and DlRan3B in longan tissues were quantified using three pre-microRNAs (pre-miR167f3p, pre-miR171f, and pre-miR394a) as the reference genes; and the expression levels during zygotic embryo development were quantified using the 2−ΔΔCT method, with longan Fe-SOD as a reference gene. The expression levels of DlRan3A and DlRan3B in other longan samples were quantified using the internal standards as described previously [53]. Primer names and sequences are provided in Table S1.
4.3. Vector Construction
The coding sequences of DlRan3A (JQ775539) and DlRan3B (JQ279697) were PCR-amplified from longan cDNA using primers with BamH I/Sal I restriction sites at the 5′/3′ ends, respectively. To generate expression constructs pCAMBIA1301-35S-DlRan3A (hereinafter, referred to as ‘35S_A’) and pCAMBIA1301-35S-DlRan3B (hereinafter, referred to as ‘35S_B’), the PCR-amplified products were in-fusion cloned into the corresponding sites of pCAMBIA1301SN vector (modified by Feng [54]) having a 35S promoter and nos terminator, respectively. pDlRan3A and pDlRan3B were amplified from longan DNA, using primers with EcoR I/Kpn I restriction enzyme sites at the 5′/3′ ends. The 35S promoter was removed from the construct 35S_A and 35S_B by EcoR I and Kpn I digestion, and the amplified products were in-fusion cloned into the corresponding sites of 35S-removed vectors, respectively, to generate expression constructs pCAMBIA1301-pDlRan3A(1256bp)-DlRan3A (hereinafter, referred to as ‘PA_A’) and pCAMBIA1301-pDlRan3B (1569bp)-DlRan3B (hereinafter, referred to as ‘PB_B’). To construct pCAMBIA1301-pDlRan3A(1256bp)-GUS (hereinafter, referred to as ‘PA_GUS’) and pCAMBIA1301-pDlRan3A(1256bp)-GUS (hereinafter, referred to as ‘PB_GUS’), pDlRan3A and pDlRan3B were amplified using primers with Hind III/Nco I restriction sites, and then in-fusion cloned into pCAMBIA1301 with removed 35S. These expression constructs were transformed into N. benthamiana using Agrobacterium strain EHA105. Primer sequences for the isolation of the DlRan3A and DlRan3B genes, as well as their promoter sequences, are provided in Supplementary data Table S2. The design of these constructs is depicted in Supplementary data Figure S1.
4.4. Generation of T2 Transgenic Tobacco Plants
The Agrobacterium-mediated transformation of tobacco leaf segments and regeneration of T0 transgenic tobaccos were performed following the method by Feng [54]. To generate T1 tobacco plants, the T0 lines of P35S_A, PA_A, P35S_B, and PB_B plants were grown in a growth chamber until flowering, and then the self-pollination stage. The resulting generations were selected against hygromycin pressure (40 mg/L), PCR verified, and validated with GUS staining by using GUS histochemical assays kit (Real-Times, Beijing, China). Similarly, the final T2 lines, used for further analysis, were obtained through self-pollination of T1 plants and then subjected to similar screening. The single T2 line of P35S_A or PA_A was obtained from T1 lines exhibiting the highest expression levels of DlRan3A. Similarly, the single T2 line of P35S_B or PB_B was obtained from T1 lines exhibiting the highest expression levels of DlRan3B (Figure S2). The used primer sequences are provided in Table S3.
4.5. Phenotypic Analysis
The effects of DlRan3A or DlRan3B overexpression were analyzed by scoring a range of specific plant phenotypes: flowering time, plant height, blade generation rate, and the development of roots, flowers, fruits, and seeds. The data for phenotype analysis were acquired from a minimum of fifteen independent plants. The tobacco root tips were stained by propidium iodide as described previously and observed by laser scanning confocal microscopy (Olympus, Tokyo, Japan; FV1200) [40].
4.6. Analysis of Environmental Stress Response
Seeds of T1 lines of P35S_A, PA_A, P35S_B, and PB_B tobacco plants (T2 lines) were germinated on MS medium with 100 mM NaCl, 200 mM mannitol, or 10 μM ABA. All seedlings were cultivated for 10 d in a chamber of 25 °C. For the heat and cold treatments, the tobacco seeds germinated on MS medium were exposed to 35 or 15 °C, respectively, for 6 h and then cultivated in the same condition as mentioned above. The control transgenic and non-transgenic tobaccos groups were cultivated without any treatments.
4.7. RNA-Sequencing
A cDNA library was generated from a pool of equal quantities of total RNA from 21 d cultivated tobacco seedlings of non-transgenic WT and transgenic T2 lines, 35S_A, PA_A, P35S_B, and PB_B, respectively. Plant samples were sent to the Novogene company (Beijing, China) for RNA extraction, cDNA library preparation, sequencing, quality control, reads mapping to the reference genome, and the gene expression quantification. RNA-seq of the mRNA and libraries was performed on an Illumina® HiSeq™ 2000 platform according to Cui’s method [55]. DEGs were screened according to their expression profiles meeting the following criteria: |log2| (transgenic lines/corresponding control) > 1 and the adjusted p value < 0.005. Expression profiles of the screened DEGs were visualized by heatmap using the TBtools software (v0.6673) [56]. The iTAK database was used to define the transcription factor family according to relative rules [57,58]. Hormone-related genes and stress-related genes were analyzed based on annotations in the TAIR (The Arabidopsis Information Resource) database. The GO and KEGG enrichment analyses of the DEGs were conducted following the method by Zhao [59]. The GO terms and KEGG pathways with corrected p-values < 0.05 were considered significantly enriched.
4.8. qPCR Validation
Total RNAs were isolated from transgenic tobaccos using the TriPure Isolation Reagent (Roche) following the manufacturer’s protocol. cDNA synthesis and qPCR analysis were conducted as above described, using the 2−ΔΔCT method with NbEF1a as a reference gene [16]. Primer sequences are provided in Table S4.
5. Conclusions
In this study, we investigated the expression and function of longan Ran GTPase genes DlRan3A and DlRan3B. The results of the study indicated that DlRan3A and DlRan3B have important roles in longan early embryo and pulp development. Heterologous expression of DlRan3A and DlRan3B by employing 35S or Ran promoters, combined with Ran-overexpression transcriptome of transgenic plants, showed that DlRan3A and DlRan3B regulate cell wall-related genes to affect plant growth. Particularly, pDlRan3A-driven expression of DlRan3A led to stunted plant growth, higher root hair density, abnormal fruits, and bigger seeds, potentially via partial regulation of the expansin- and extensin-like genes. Furthermore, DlRan3A and DlRan3B might function via shared molecular mechanisms in cold stress response but differed in conferring plants with salinity, osmotic, and heat stress tolerance or sensitivity, by up-regulated or down-regulated different stress- or hormone-related genes. Overall, the present study provided a widespread characterization on longan Ran GTPase genes DlRan3A and DlRan3B, highlighting the associated gene expression mechanisms incorporated by Ran.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants13040480/s1, Figure S1: Constructed vectors of DlRan3A and DlRan3B; Figure S2: Gene-relative expression in transgenic modified tobacco (T1 lines); Figure S3: Height (a) and leaf number (b) of transgenic tobacco; Figure S4: Phenotype of transgenic tobacco root (7 d and 14 d); Figure S5: Hundred-grain weight of transgenic tobacco; Figure S6: GUS staining of transgenic tobacco seedlings (T1 lines); Figure S7: GUS staining of flowers, fruits, and seeds of transgenic tobacco; Figure S8: Expression profiles of the shared DEGs in transgenic lines; Figure S9: Expression profiles of DEGs classified as genes encoding transcription factors (TFs) (a) and hormone-related genes (HRGs) (b); Figure S10: Expression profiles of DEGs involved in cell wall development; Figure S11: Expression profiles of stress-related DEGs; Figure S12: Mapman software (v3.6.0) visualization of stress-related genes; Figure S13: Expression profiles of the antioxidant-defense-related DEGs; Table S1: Sequences of primers used for qPCR analysis of DlRan3A and DlRan3B in longan; Table S2: Primer sequences used for DlRan3A and DlRan3B constructs for genetic transformation of Nicotiana benthamiana; Table S3: Sequences of primers used for qPCR detection in transgenic tobaccos; Table S4: Primer sequences used for qPCR verification; Table S5: Comparison of reads by transcriptome sequencing to reference genome; Table S6: Differentially expressed genes (DEGs) of the GM N. benthamiana lines; Table S7: Shared DEGs between the P35S_A and P35S_B lines; Table S8: Shared DEGs between the P35S_A and PA_A lines; Table S9: Shared DEGs between the P35S_B and PB_B lines; Table S10: Significant GO enrichment pathways of the genetically modified (GM) N. benthamiana lines; Table S11: Significant KEGG enrichment pathways of the GM N. benthamiana lines.
Author Contributions
Conceptualization, Z.L. and X.X. (Xuhan Xu); methodology, Q.T. and X.X. (Xiying Xie); validation, Q.T., X.X. (Xiying Xie) and R.L.; resources, Z.Z.; data curation, Y.C.; writing—original draft preparation, Q.T.; writing—review and editing, Y.L.; supervision, C.C.; funding acquisition, Z.L. and Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31572088, 31672127), the Special fund for scientific and technological innovation of Fujian Agricultural and Forestry University Applied Basic Research (CXZX2017189 and CXZX2017314), the Natural Science Foundation of Fujian Province (2020N0061 and 2021J05199), and the startup fund of Minnan Normal University (4206-L21715).
Data Availability Statement
The RNA sequencing data have been uploaded to the NCBI (National Center for Biotechnology Information) SRA (Sequence Read Archive) under accession number PRJNA607013.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lin, Y.; Min, J.; Lai, R.; Wu, Z.; Chen, Y.; Yu, L.; Cheng, C.; Jin, Y.; Tian, Q.; Liu, Q.; et al. Genome-wide sequencing of longan (Dimocarpus longan Lour.) provides insights into molecular basis of its polyphenol-rich characteristics. Gigascience 2017, 6, gix023. [Google Scholar] [CrossRef] [PubMed]
- Jue, D.; Liu, L.; Sang, X.; Shu, B.; Wang, J.; Wang, Y.; Zhang, C.; Shi, S. SNP-based high-density genetic map construction and candidate gene identification for fruit quality traits of Dimocarpus longan Lour. Sci. Hortic. 2021, 284, 110086. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, C.; Xu, X.; Cai, R.; Guan, Q.; Chen, X.; Chen, Y.; Zhang, Z.; XuHan, X.; Lin, Y.; et al. Riboflavin mediates m6A modification targeted by miR408, promoting early somatic embryogenesis in longan. Plant Physiol. 2023, 192, 1799–1820. [Google Scholar] [CrossRef] [PubMed]
- Salus, S.S.; Demeter, J.; Sazer, S. The Ran GTPase system in fission yeast affects microtubules and cytokinesis in cells that are competent for nucleocytoplasmic protein transport. Mol. Cell. Biol. 2002, 22, 8491–8505. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scrofani, J.; Sardon, T.; Meunier, S.; Vernos, I. Microtubule Nucleation in Mitosis by a RanGTP-Dependent Protein Complex. Curr. Biol. 2015, 25, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, Y.; Han, Y.; Bao, S.; Du, J.; Yuan, M.; Xu, Z.; Chong, K. Overexpression of RAN1 in rice and Arabidopsis alters primordial meristem, mitotic progress, and sensitivity to auxin. Plant Physiol. 2006, 140, 91–101. [Google Scholar] [CrossRef]
- Malambane, G.; Madumane, K.; Sewelo, L.T.; Batlang, U. Drought stress tolerance mechanisms and their potential common indicators to salinity, insights from the wild watermelon (Citrullus lanatus): A review. Front. Plant Sci. 2022, 13, 1074395. [Google Scholar] [CrossRef]
- Zang, A.; Xu, X.; Neill, S.; Cai, W. Overexpression of OsRAN2 in rice and Arabidopsis renders transgenic plants hypersensitive to salinity and osmotic stress. J. Exp. Bot. 2010, 61, 777–789. [Google Scholar] [CrossRef]
- Chen, N.; Xu, Y.; Wang, X.; Du, C.; Du, J.; Yuan, M.; Xu, Z.; Chong, K. OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress. Plant Cell Environ. 2011, 34, 52–64. [Google Scholar] [CrossRef]
- Xu, P.; Ma, W.; Liu, J.; Hu, J.; Cai, W. Overexpression of a small GTP-binding protein Ran1 in Arabidopsis leads to promoted elongation growth and enhanced disease resistance against P. syringae DC3000. Plant J. 2021, 108, 977–991. [Google Scholar] [CrossRef]
- Yan, J.J.; Xie, B.; Zhang, L.; Li, S.J.; van Peer, A.F.; Wu, T.J.; Chen, B.Z.; Xie, B.G. Small GTPases and Stress Responses of vvran1 in the Straw Mushroom Volvariella volvacea. Int. J. Mol. Sci. 2016, 17, 1527. [Google Scholar] [CrossRef]
- Fang, Z.-Z.; Zhang, Y.-L.; Lai, C.C.; Lai, Z. Developmental regulation of Ran 3untranslated region during somatic embryogenesis in Dimocarpus longan Lour. Sci. Hortic. 2014, 176, 297–302. [Google Scholar] [CrossRef]
- Tian, Q.; Lin, Y.; Lai, Z.; Wang, T. Cloning and Bioinformatic Analysis of the Promoters of DlRan3A and DlRan3B from Embryogenic Callus in Dimocarpus longan. Chin. J. Trop. Crops 2014, 35, 82–89. [Google Scholar]
- Tian, Q.; Lin, Y.; Yang, M.; Zhang, D.; Lai, R.; Lai, Z. DlRan3A is involved in hormone, light, and abiotic stress responses in embryogenic callus of Dimocarpus longan Lour. Gene 2015, 569, 267–275. [Google Scholar] [CrossRef]
- Tian, Q.; Lin, Y.; Zhang, D.; Lai, R.; Lai, Z. Ras-Related Nuclear Protein Ran3B Gene Is Involved in Hormone Responses in the Embryogenic Callus of Dimocarpus longan Lour. Int. J. Mol. Sci. 2016, 17, 873. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, X.; Chen, X.; Chen, Y.; Zhang, Z.; Xuhan, X.; Lin, Y.; Lai, Z. Seed-Specific Gene MOTHER of FT and TFL1(MFT) Involved in Embryogenesis, Hormones and Stress Responses in Dimocarpus longan Lour. Int. J. Mol. Sci. 2018, 19, 2403. [Google Scholar] [CrossRef] [PubMed]
- Ach, R.A.; Gruissem, W. A small nuclear GTP-binding protein from tomato suppresses a Schizosaccharomyces pombe cell-cycle mutant. Proc. Natl. Acad. Sci. USA 1994, 91, 5863–5867. [Google Scholar] [CrossRef] [PubMed]
- Haizel, T.; Merkle, T.; Pay, A.; Fejes, E.; Nagy, F. Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J. 1997, 11, 93–103. [Google Scholar] [CrossRef]
- Lü, S.H.; Fan, Y.L.; Jin, C.X. Overexpression of a Ran GTPase homologous gene, FaRan from tall fescue, in transgenic Arabidopsis. Biol. Plant. 2011, 55, 331–334. [Google Scholar] [CrossRef]
- Huang, N.; Ling, H.; Zhang, X.; Mao, H.; Su, Y.; Su, W.; Liu, F.; Xu, L.; Chen, R.; Que, Y. A Small GTP-Binding Gene ScRan from Sugarcane is Involved in Responses to Various Hormone Stresses and Sporisirium scitamineum Challenge. Sugar Technol. 2018, 20, 669–680. [Google Scholar] [CrossRef]
- Reynolds, J.P. Characterisation of the Defective Embryo and Meristems (DEM) Genes of Plants. Ph.D. Thesis, The University of Queensland, Townsville, QLD, Australia, 2013. [Google Scholar]
- Liu, P.; Qi, M.; Wang, Y.; Chang, M.; Liu, C.; Sun, M.; Yang, W.; Ren, H. Arabidopsis RAN1 mediates seed development through its parental ratio by affecting the onset of endosperm cellularization. Mol. Plant 2014, 7, 1316–1328. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wolf, S.; Hématy, K.; Höfte, H. Growth control and cell wall signaling in plants. Annu. Rev. Plant Biol. 2012, 63, 381–407. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, R.; Filipecki, M.; Tagashira, N.; Wiśniewska, A.; Gaj, P.; Plader, W.; Malepszy, S. Xyloglucan endotransglucosylase/hydrolase genes in cucumber (Cucumis sativus)-differential expression during somatic embryogenesis. Physiol. Plant. 2004, 120, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Hall, Q.; Cannon, M.C. The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell 2002, 14, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Cannon, M.C.; Terneus, K.; Hall, Q.; Tan, L.; Wang, Y.; Wegenhart, B.L.; Chen, L.; Lamport, D.T.; Chen, Y.; Kieliszewski, M.J. Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. USA 2008, 105, 2226–2231. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wu, K.; Harberd, N.P.; Fu, X. Green Revolution DELLAs: From translational reinitiation to future sustainable agriculture. Mol. Plant 2021, 14, 547–549. [Google Scholar] [CrossRef] [PubMed]
- Budot, B.O.; Encabo, J.R.; Ambita, I.D.; Atienza-Grande, G.A.; Satoh, K.; Kondoh, H.; Ulat, V.J.; Mauleon, R.; Kikuchi, S.; Choi, I.R. Suppression of cell wall-related genes associated with stunting of Oryza glaberrima infected with Rice tungro spherical virus. Front. Microbiol. 2014, 5, 26. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Lee, Y.; Cho, H.T.; Kende, H. Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell 2003, 15, 1386–1398. [Google Scholar] [CrossRef]
- Ilias, I.A.; Negishi, K.; Yasue, K.; Jomura, N.; Morohashi, K.; Baharum, S.N.; Goh, H.H. Transcriptome-wide effects of expansin gene manipulation in etiolated Arabidopsis seedling. J. Plant Res. 2019, 132, 159–172. [Google Scholar] [CrossRef]
- Koevoets, I.T.; Venema, J.H.; Elzenga, J.T.; Testerink, C. Roots Withstanding their Environment: Exploiting Root System Architecture Responses to Abiotic Stress to Improve Crop Tolerance. Front. Plant Sci. 2016, 7, 1335. [Google Scholar] [CrossRef]
- Lei, M.J.; Wang, Q.; Li, X.; Chen, A.; Luo, L.; Xie, Y.; Li, G.; Luo, D.; Mysore, K.S.; Wen, J.; et al. The small GTPase ROP10 of Medicago truncatula is required for both tip growth of root hairs and nod factor-induced root hair deformation. Plant Cell 2015, 27, 806–822. [Google Scholar] [CrossRef]
- Kang, E.; Zheng, M.; Zhang, Y.; Yuan, M.; Yalovsky, S.; Zhu, L.; Fu, Y. The Microtubule-Associated Protein MAP18 Affects ROP2 GTPase Activity during Root Hair Growth. Plant Physiol. 2017, 174, 202–222. [Google Scholar] [CrossRef]
- Bucher, M.; Brunner, S.; Zimmermann, P.; Zardi, G.I.; Amrhein, N.; Willmitzer, L.; Riesmeier, J.W. The expression of an extensin-like protein correlates with cellular tip growth in tomato. Plant Physiol. 2002, 128, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Kwasniewski, M.; Janiak, A.; Mueller-Roeber, B.; Szarejko, I. Global analysis of the root hair morphogenesis transcriptome reveals new candidate genes involved in root hair formation in barley. J. Plant Physiol. 2010, 167, 1076–1083. [Google Scholar] [CrossRef] [PubMed]
- Velasquez, S.M.; Ricardi, M.M.; Dorosz, J.G.; Fernandez, P.V.; Nadra, A.D.; Pol-Fachin, L.; Egelund, J.; Gille, S.; Harholt, J.; Ciancia, M.; et al. O-glycosylated cell wall proteins are essential in root hair growth. Science 2011, 332, 1401–1403. [Google Scholar] [CrossRef] [PubMed]
- Eberle, C.A.; Anderson, N.O.; Clasen, B.M.; Hegeman, A.D.; Smith, A.G. PELPIII: The class III pistil-specific extensin-like Nicotiana tabacum proteins are essential for interspecific incompatibility. Plant J. 2013, 74, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.M.L.; Noyszewski, A.K.; Smith, A.G. Structure and function of class III pistil-specific extensin-like protein in interspecific reproductive barriers. BMC Plant Biol. 2019, 19, 118. [Google Scholar] [CrossRef] [PubMed]
- Ralevski, A. The Interaction of Early Salt Stress-Induced 2 (ESI2) and the Ran G Protein in Arabidopsis. Master’s Thesis, Concordia University, Montreal, QC, Canada, 2013. [Google Scholar]
- Xu, P.; Cai, W. RAN1 is involved in plant cold resistance and development in rice (Oryza sativa). J. Exp. Bot. 2014, 65, 3277–3287. [Google Scholar] [CrossRef] [PubMed]
- Karuppanapandian, T.; Rhee, S.-J.; Kim, E.-J.; Han, B.K.; Hoekenga, O.A.; Lee, G.P. Proteomic analysis of differentially expressed proteins in the roots of Columbia-0 and Landsberg erecta ecotypes of Arabidopsis thaliana in response to aluminum toxicity. Can. J. Plant Sci. 2012, 92, 1267–1282. [Google Scholar] [CrossRef]
- Kagale, S.; Divi, U.K.; Krochko, J.E.; Keller, W.A.; Krishna, P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 2007, 225, 353–364. [Google Scholar] [CrossRef]
- Xia, X.-J.; Wang, Y.-J.; Zhou, Y.-H.; Tao, Y.; Mao, W.-H.; Shi, K.; Asami, T.; Chen, Z.; Yu, J.-Q. Reactive Oxygen Species Are Involved in Brassinosteroid-Induced Stress Tolerance in Cucumber. Plant Physiol. 2009, 150, 801–814. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, H.; Zhao, Z.; An, L. Abscisic acid is involved in brassinosteroids-induced chilling tolerance in the suspension cultured cells from Chorispora bungeana. J. Plant Physiol. 2011, 168, 853–862. [Google Scholar] [CrossRef]
- Fu, J.; Sun, P.; Luo, Y.; Zhou, H.; Gao, J.; Zhao, D.; Pubu, Z.; Liu, J.; Hu, T. Brassinosteroids enhance cold tolerance in Elymus nutans via mediating redox homeostasis and proline biosynthesis. Environ. Exp. Bot. 2019, 167, 103831. [Google Scholar] [CrossRef]
- Arfan, M.; Zhang, D.W.; Zou, L.J.; Luo, S.S.; Tan, W.R.; Zhu, T.; Lin, H.H. Hydrogen Peroxide and Nitric Oxide Crosstalk Mediates Brassinosteroids Induced Cold Stress Tolerance in Medicago truncatula. Int. J. Mol. Sci. 2019, 20, 144. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.A.; Mateos, I.; Labrador, E.; Dopico, B. Brassinolides and IAA induce the transcription of four alpha-expansin genes related to development in Cicer arietinum. Plant Physiol. Biochem. 2004, 42, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Lisso, J.; Steinhauser, D.; Altmann, T.; Kopka, J.; Müssig, C. Identification of brassinosteroid-related genes by means of transcript co-response analyses. Nucleic Acids Res. 2005, 33, 2685–2696. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Divi, U.K.; Rahman, T.; Krishna, P. Gene expression and functional analyses in brassinosteroid-mediated stress tolerance. Plant Biotechnol. J. 2016, 14, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Orman-Ligeza, B.; Parizot, B.; de Rycke, R.; Fernandez, A.; Himschoot, E.; Van Breusegem, F.; Bennett, M.J.; Perilleux, C.; Beeckman, T.; Draye, X. RBOH-mediated ROS production facilitates lateral root emergence in Arabidopsis. Development 2016, 143, 3328–3339. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Sanyal, S.K.; Pandey, G.K. Ca2+–CBL–CIPK: A modulator system for efficient nutrient acquisition. Plant Cell Rep. 2021, 40, 2111–2122. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Yan, M.; Zhang, Q. Functional regulation of plant NADPH oxidase and its role in signaling. Plant Signal. Behav. 2017, 12, 1356970. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.L.; Lai, Z.X. Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci. 2010, 178, 359–365. [Google Scholar] [CrossRef]
- Feng, X. Genome-Wide Identification and Function Analysis of the Superoxide Dismutase Gene Family in Musa spp. Ph.D. Thesis, Fujian Agriculture and Forestry University, Fuzhou, China, 2016. [Google Scholar]
- Cui, Y.; Zhu, Y.; Lin, Y.; Chen, L.; Feng, Q.; Wang, W.; Xiang, H. New insight into the mechanism underlying the silk gland biological process by knocking out fibroin heavy chain in the silkworm. BMC Genome 2018, 19, 215. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, P.; Riaño-Pachón, D.M.; Corrêa, L.G.; Rensing, S.A.; Kersten, B.; Mueller-Roeber, B. PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010, 38, D822–D827. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef]
- Zhao, W.; Yang, X.; Yu, H.; Jiang, W.; Sun, N.; Liu, X.; Liu, X.; Zhang, X.; Wang, Y.; Gu, X. RNA-Seq-based transcriptome profiling of early nitrogen deficiency response in cucumber seedlings provides new insight into the putative nitrogen regulatory network. Plant Cell Physiol. 2015, 56, 455–467. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).