Physiological Responses to Salt Stress at the Seedling Stage in Wild (Oryza rufipogon Griff.) and Cultivated (Oryza sativa L.) Rice
Abstract
1. Introduction
2. Results
2.1. Plant Survival and Growth Capacity
2.2. Na+ and K+ Homeostasis in Roots and Leaves
2.3. Carbon and Nitrogen Content in Leaves
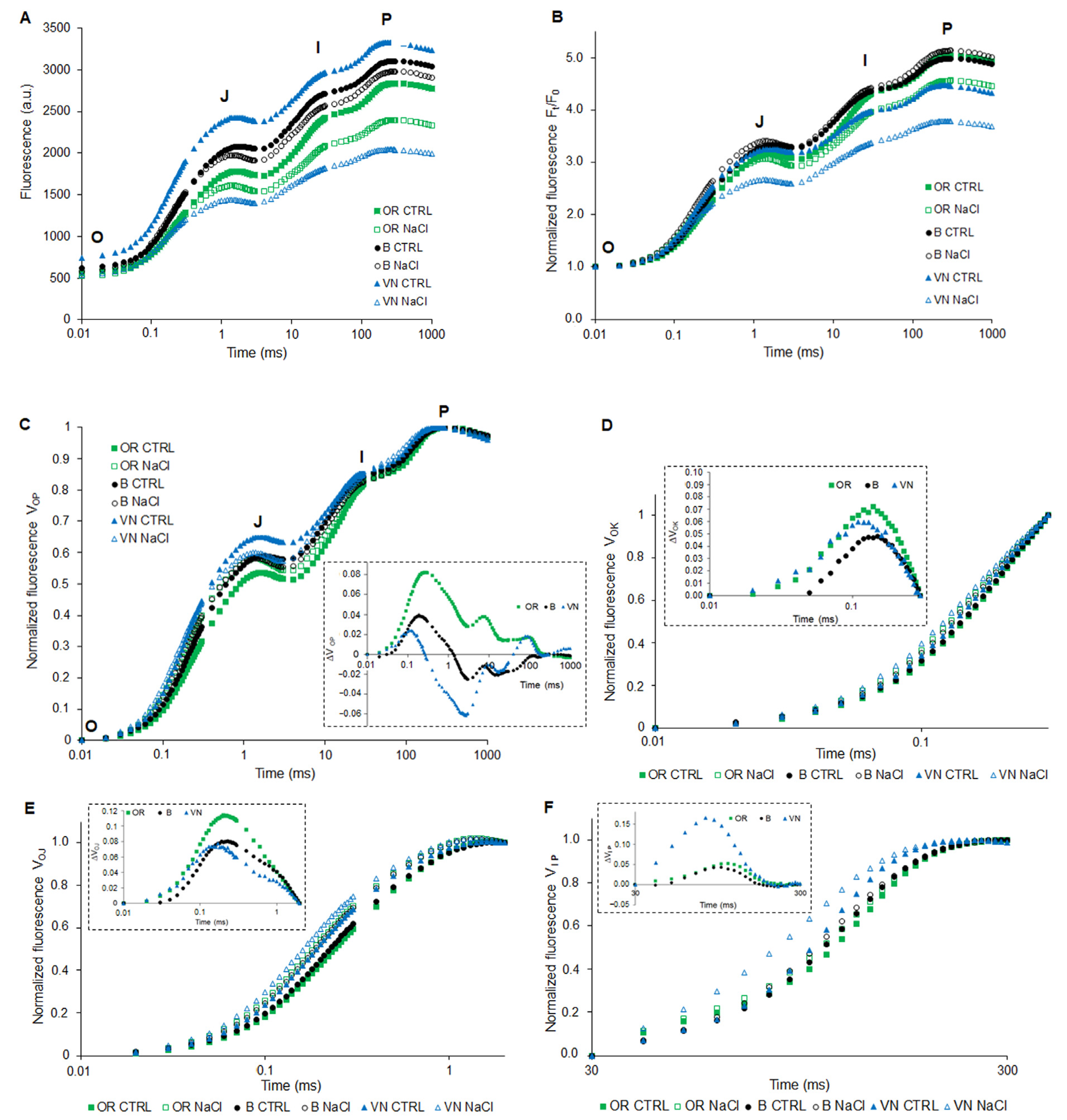
2.4. Pigments Content and Photosythetic Performance by JIP-Test
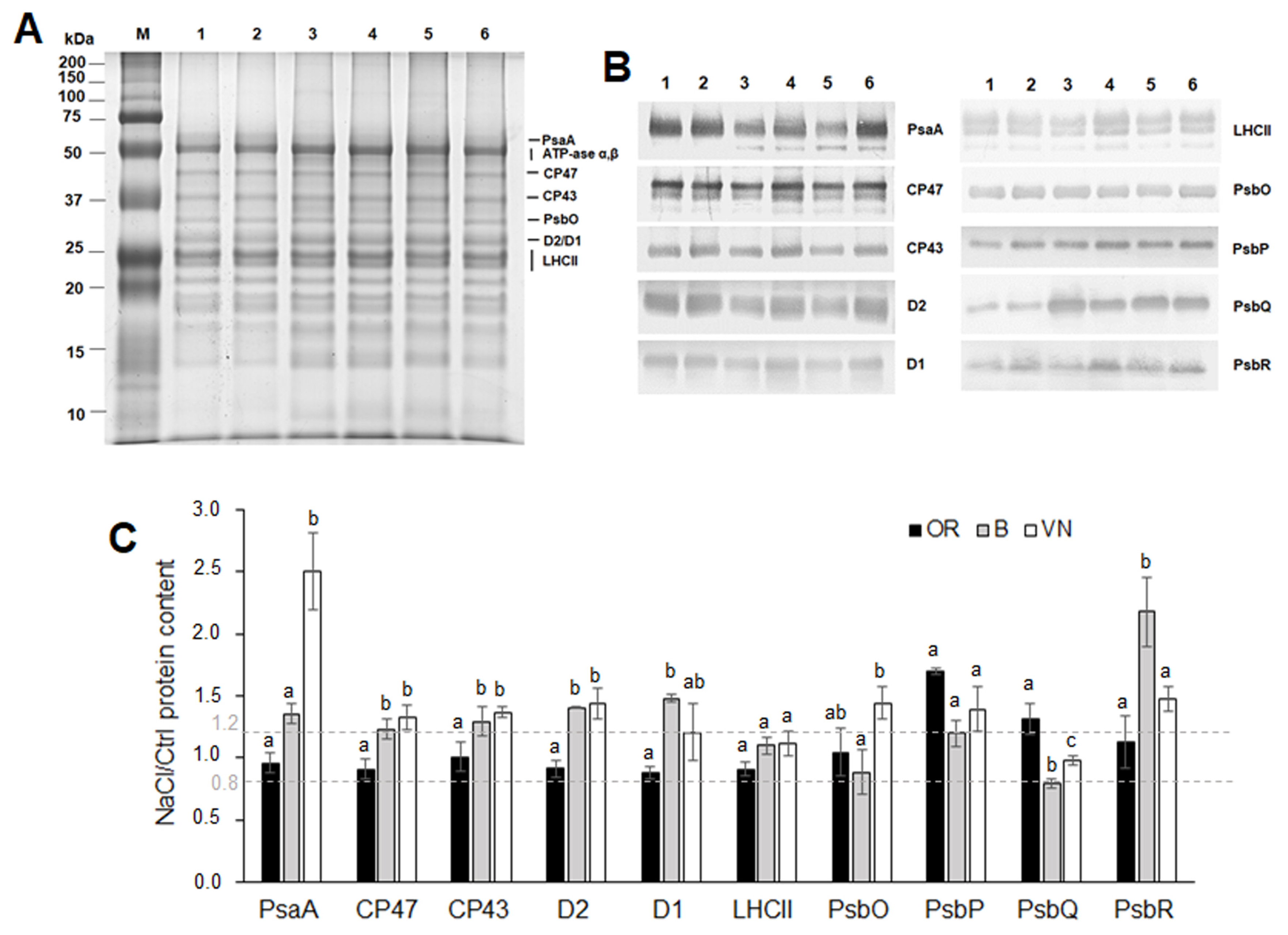
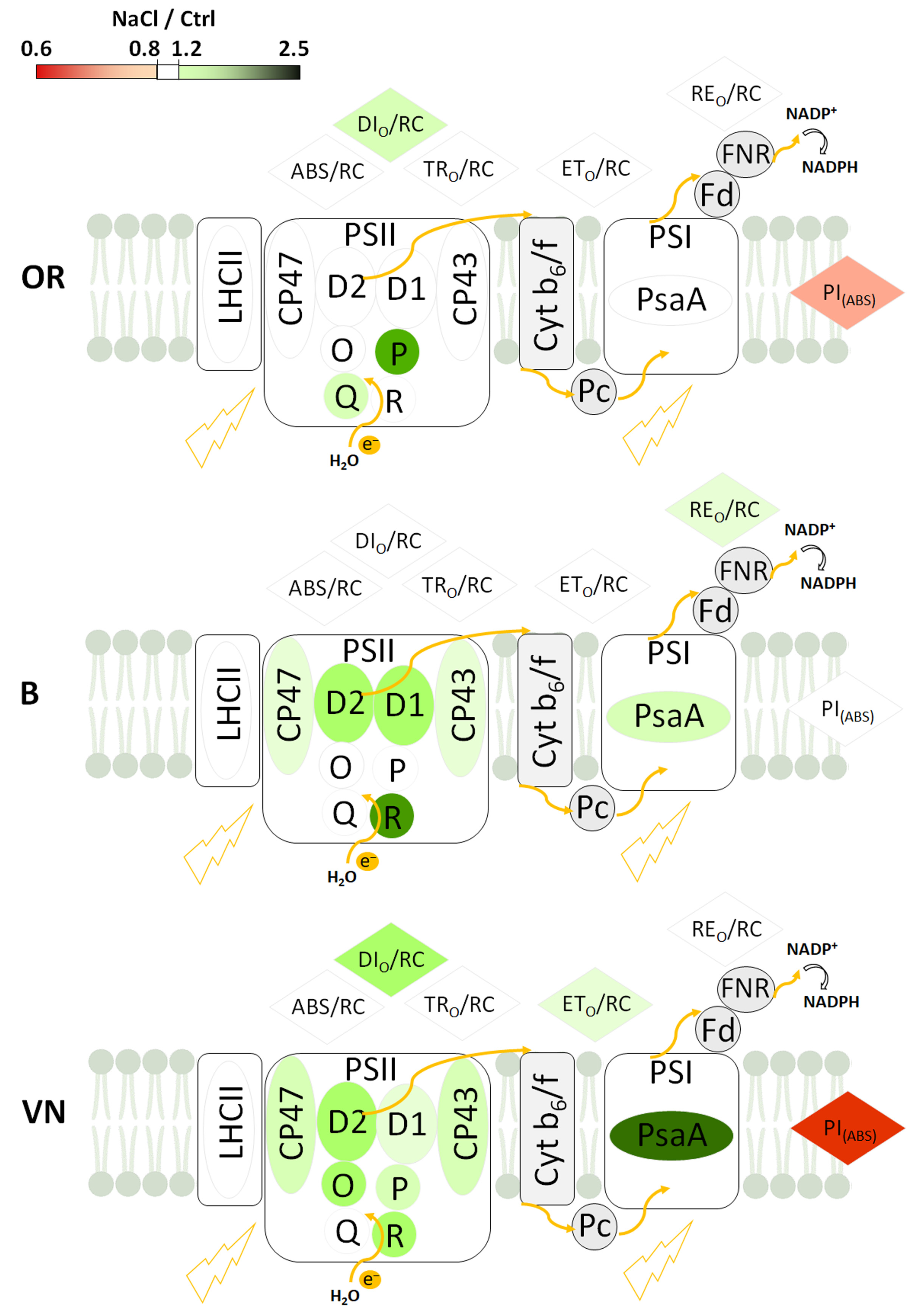
2.5. Thylakoid Membranes and Photosystem I and Photosystem II Protein Quantification
3. Discussion
3.1. Effects of Salinity on Plant Survival
3.2. Effects of Salinity on Plant Growth, Water Status and Na+ and K+ Ion Homeostasis
3.3. Effects of Salinity on Plant Photosynthetic Apparatus Integrity and Performance
4. Materials and Methods
4.1. Plant Material
4.2. SES Evaluation and Plant Selection
4.3. Biomass and Morphometric Measurements
4.4. Relative Water Content
4.5. Determination of Ion Contents in Roots and Leaves
4.6. Carbon and Nitrogen Determination in Leaves
4.7. Chlorophyll and Carotenoid Contents
4.8. Chlorophyll a Fluorescence Analysis and JIP-Test
| Fluorescence Parameters | Description |
|---|---|
| Basic parameters calculated from the extracted data | |
| F0 ≅ F50μs or ≅ F20μs | Fluorescence when all PSII RCs are open (≅to the minimal reliable recorded fluorescence) |
| Fm (=FP) | Maximal fluorescence, when all PSII RCs are closed (=FP when the actinic light intensity is above 500 μmol(photon) m−2 s1 and provided that all RCs are active as QA-reducing) |
| FV = Fm − F0 | Maximal variable fluorescence |
| Area | Total complementary area between the fluorescence induction curve and the line F = FP (meaningful only when FP = Fm), relates to the pool size of PSII electron transport acceptors |
| Fv/F0 = (Fm − F0)/F0 | Maximum efficiency of water-splitting reaction on the donor side of PSII |
| Vt = (Ft − F0)/(Fm − F0) | Relative variable fluorescence at time t |
| VI = (FI − F0)/(Fm − F0) | Relative variable fluorescence at the I-step (at 30 ms) |
| VJ = (FJ − F0)/(Fm − F0) | Relative variable fluorescence at the J-step (at 2 ms) |
| M0 = [(∆F/∆t)0]/(Fm − F0) = 4 × (F300μs − F0)/(Fm − F0) | Approximated initial slope (in ms−1) of the fluorescence transient normalised on the maximal variable fluorescence Fm − F0 = FV; equivalently, initial slope (50 to 300 μs; in ms−1) of the Vt = f(t) kinetics |
| Sm = Area/(Fm − F0) = Area/FV | Normalised Area (reflecting multiple turnover of QA reduction events and representing energy necessary for the closure of all RCs) |
| N = Sm × (M0/VJ) | Turnover number (expresses how many times QA is reduced in the time interval from 0 to tFm) |
| Specific energy fluxes (per active PSII reaction center) | |
| ABS/RC = M0 × (1/VJ) × (1/ϕPo) | Absorption flux (exciting PSII antenna Chl a molecules) per RC (also used as a unit-less measure of PSII apparent antenna size) |
| RC/ABS = [(F2ms − F0)/4(F300μs − F0)] × (FV/Fm) = φP0 × (VJ/M0) | Density of RCs per chlorophyll (reciprocal of ABS/RC) |
| TR0/RC = M0 × (1/VJ) | Trapped energy flux (leading to QA reduction) per RC (at t = 0) |
| ET0/RC = M0 × (1/VJ) × (1 − VJ) | Electron transport flux (further than QA−) per RC (at t = 0) |
| RE0/RC = M0 × (1/VJ) × (1 − VI) | Electron flux reducing end electron acceptors at the PSI acceptor side per RC (at t = 0) |
| DI0/RC = (ABS/RC) − (TR0/RC) | Dissipated energy flux per RC (at t = 0) |
| Quantum yields and efficiencies | |
| Ψ0 = ET0/TR0 = (1 − VJ) | Efficiency/probability that an electron moves further than QA− (at t = 0) |
| φP0 = TR0/ABS = [1 − (F0/Fm)] = Fv/Fm | Maximum quantum yield for primary PSII photochemistry (maximum efficiency at which light absorbed by PSII is used for reduction of QA) (at t = 0) |
| φE0 = ET0/ABS = [1 − (F0/Fm)] × Ψ0 | Probability that an absorbed photon moves an electron further than QA− (at t = 0) |
| Performance index | |
| PI(ABS) = (RC/ABS) × [φP0/(1 − φP0)] × [Ψ0/(1 − Ψ0)] | PSII Performance index on absorption basis |
4.9. Isolation of Thylakoid Membranes, Gel Electrophoresis and Western Blotting
4.10. Statistical Analyses
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fukagawa, N.K.; Ziska, L.H. Rice: Importance for global nutrition. J. Nutr. Sci. Vitaminol. 2019, 65, S2–S3. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Shannon, M.C. Salinity effects on seedling growth and yield components of rice. Crop Sci. 2000, 40, 996–1003. [Google Scholar] [CrossRef]
- Balasubramaniam, T.; Shen, G.; Esmaeili, N.; Zhang, H. Plants’ response mechanisms to salinity stress. Plants 2023, 12, 2253. [Google Scholar] [CrossRef] [PubMed]
- FAO. Handbook for Saline Soil Management; Food and Agriculture Organization: Rome, Italy, 2018; ISBN 978-92-5-130141-8. [Google Scholar]
- Sparks, D.L. The chemistry of saline and sodic soils. In Environmental Soil Chemistry, 2nd ed.; Sparks, D.L., Ed.; Academic Press: Burlington, NJ, USA, 2003; pp. 285–300. ISBN 978-0-12-656446-4. [Google Scholar]
- Asch, F.; Wopereis, M.C.S. Responses of field-grown irrigated rice cultivars to varying levels of floodwater salinity in a semi-arid environment. Field Crops Res. 2001, 70, 127–137. [Google Scholar] [CrossRef]
- Reddy, I.N.B.L.; Kim, B.K.; Yoon, I.S.; Kim, K.H.; Kwon, T.R. Salt tolerance in rice: Focus on mechanisms and approaches. Rice Sci. 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Singh, A. Soil salinity: A global threat to sustainable development. Soil Use Manag. 2022, 38, 39–67. [Google Scholar] [CrossRef]
- Rodríguez Coca, L.I.; García González, M.T.; Gil Unday, Z.; Jiménez Hernández, J.; Rodríguez Jáuregui, M.M.; Fernández Cancio, Y. Effects of sodium salinity on rice (Oryza sativa L.) cultivation: A review. Sustainability 2023, 15, 1804. [Google Scholar] [CrossRef]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Bressan, R.A. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Moradi, F.; Ismail, A.M. Responses of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice. Ann. Bot. 2007, 99, 1161–1173. [Google Scholar] [CrossRef]
- Xu, J.; Huang, X.; Lan, H.; Zhang, H.; Huang, J. Rearrangement of nitrogen metabolism in rice (Oryza sativa L.) under salt stress. Plant Signal. Behav. 2016, 11, e1138194. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Lu, Y.; Bao, S.; Zhang, Q.; Geng, Y.; Shao, X. Carbon and nitrogen metabolism in rice cultivars affected by salt-alkaline stress. Crop Pasture Sci. 2021, 72, 372–382. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Kim, D.; Paek, N.-C. Salt treatments and induction of senescence. In Methods in Molecular Biology; Guo, Y., Ed.; Springer: New York, NY, USA, 2018; pp. 141–149. ISBN 978-1-4939-7672-0. [Google Scholar]
- Sade, N.; Del Mar Rubio-Wilhelmi, M.; Umnajkitikorn, K.; Blumwald, E. Stress-induced senescence and plant tolerance to abiotic stress. J. Exp. Bot. 2018, 69, 845–853. [Google Scholar] [CrossRef]
- Negrão, S.; Courtois, B.; Ahmadi, N.; Abreu, I.; Saibo, N.; Oliveira, M.M. Recent updates on salinity stress in rice: From physiological to molecular responses. Crit. Rev. Plant Sci. 2011, 30, 329–377. [Google Scholar] [CrossRef]
- Khush, G.S. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 1997, 35, 25–34. [Google Scholar] [CrossRef]
- Lee, K.-S.; Choi, W.-Y.; Ko, J.-C.; Kim, T.-S.; Gregorio, G.B. Salinity tolerance of japonica and indica rice (Oryza sativa L.) at the seedling stage. Planta 2003, 216, 1043–1046. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Li, Y.; Huang, R. Advances and challenges in the breeding of salt-tolerant rice. Int. J. Mol. Sci. 2020, 21, 8385. [Google Scholar] [CrossRef]
- Song, Z.; Li, B.; Chen, J.; Lu, B.R. Genetic diversity and conservation of common wild rice (Oryza rufipogon) in China. Plant Species Biol. 2005, 20, 83–92. [Google Scholar] [CrossRef]
- Solis, C.A.; Yong, M.T.; Vinarao, R.; Jena, K.; Holford, P.; Shabala, L.; Zhou, M.; Shabala, S.; Chen, Z.H. Back to the wild: On a quest for donors toward salinity tolerant rice. Front. Plant Sci. 2020, 11, 323. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, S.; Fu, Y.; Su, Z.; Wang, X.; Sun, C. Identification of a drought tolerant introgression line derived from Dongxiang common wild rice (O. rufipogon Griff.). Plant Mol. Biol. 2006, 62, 247–259. [Google Scholar] [CrossRef]
- Tian, L.; Tan, L.; Liu, F.; Cai, H.; Sun, C. Identification of quantitative trait loci associated with salt tolerance at seedling stage from Oryza rufipogon. J. Genet. Genom. 2011, 38, 593–601. [Google Scholar] [CrossRef]
- Wang, S.; Cao, M.; Ma, X.; Chen, W.; Zhao, J.; Sun, C.; Tan, L.; Liu, F. Integrated RNA sequencing and QTL mapping to identify candidate genes from Oryza rufipogon associated with salt tolerance at the seedling stage. Front. Plant Sci. 2017, 8, 1427. [Google Scholar] [CrossRef]
- Brar, D.S.; Khush, G.S. Wild relatives of rice: A valuable genetic resource for genomics and breeding research. In The Wild Oryza Genomes; Mondal, T.K., Henry, R.J., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–25. ISBN 978-3-319-71997-9. [Google Scholar]
- Senadhira, D.; Zapata-Arias, F.J.; Gregorio, G.B.; Alejar, M.S.; de la Cruz, H.C.; Padolina, T.F.; Galvez, A.M. Development of the first salt-tolerant rice cultivar through indica/indica anther culture. Field Crops Res. 2002, 76, 103–110. [Google Scholar] [CrossRef]
- Ganie, S.A.; Wani, S.H.; Henry, R.; Hensel, G. Improving rice salt tolerance by precision breeding in a new era. Curr. Opin. Plant Biol. 2021, 60, 101996. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, A.; Tinarelli, A. Rice cultivation in the E.U. Ecological conditions and agronomical practices. In Pesticide Risk Assessment in Rice Paddies: Theory and Practice; Elsevier: Amsterdam, The Netherlands, 2008; pp. 1–24. [Google Scholar] [CrossRef]
- Bertazzini, M.; Sacchi, G.A.; Forlani, G. A differential tolerance to mild salt stress conditions among six Italian rice genotypes does not rely on Na+ exclusion from shoots. J. Plant Physiol. 2018, 226, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, P.; Cui, F.; Zhang, F.; Luo, X.; Xie, J. Transcriptome analysis of salt stress responsiveness in the seedlings of dongxiang wild rice (Oryza rufipogon Griff.). PLoS ONE 2016, 11, e0146242. [Google Scholar] [CrossRef] [PubMed]
- Solis, C.A.; Yong, M.T.; Venkataraman, G.; Milham, P.; Zhou, M.; Shabala, L.; Holford, P.; Shabala, S.; Chen, Z.H. Sodium sequestration confers salinity tolerance in an ancestral wild rice. Physiol. Plant. 2021, 172, 1594–1608. [Google Scholar] [CrossRef] [PubMed]
- Marè, C.; Zampieri, E.; Cavallaro, V.; Frouin, J.; Grenier, C.; Courtois, B.; Brottier, L.; Tacconi, G.; Finocchiaro, F.; Serrat, X.; et al. Marker-assisted introgression of the salinity tolerance locus Saltol in temperate Japonica rice. Rice 2023, 16, 2. [Google Scholar] [CrossRef]
- IRRI. Standard Evaluation System for Rice (SES), 3rd ed.; International Rice Research Institute: Manila, Philippines, 2013; ISBN 9781612099897. [Google Scholar]
- Gregorio, G.B.; Senadhira, D.; Mendoza, R.D. Screening rice for salinity tolerance. In IRRI Discussion Paper Series No. 22; International Rice Research Institute: Manila, Philippines, 1997; pp. 1–30. [Google Scholar]
- Tsimilli-Michael, M. Revisiting JIP-test: An educative review on concepts, assumptions, approximations, definitions and terminology. Photosynthetica 2020, 58, 275–292. [Google Scholar] [CrossRef]
- Barera, S.; Pagliano, C.; Pape, T.; Saracco, G.; Barber, J. Characterization of PSII-LHCII supercomplexes isolated from pea thylakoid membrane by one-step treatment with α- and β-dodecyl-D-maltoside. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 3389–3399. [Google Scholar] [CrossRef]
- Eynard, A.; Lal, R.; Wiebe, K. Crop response in salt-affected soils. J. Sustain. Agric. 2005, 27, 5–50. [Google Scholar] [CrossRef]
- Vázquez-Glaría, A.; Eichler-Löbermann, B.; Loiret, F.G.; Ortega, E.; Kavka, M. Root-system architectures of two cuban rice cultivars with salt stress at early development stages. Plants 2021, 10, 1194. [Google Scholar] [CrossRef]
- Samarajeewa, P.K.; Barrero, R.A.; Umeda-Hara, C.; Kawai, M.; Uchimiya, H. Cortical cell death, cell proliferation, macromolecular movements and rTip1 expression pattern in roots of rice (Oryza sativa L.) under NaCl stress. Planta 1999, 207, 354–361. [Google Scholar] [CrossRef]
- Toyofuku, K.; Matsunami, M.; Ogawa, A. Genotypic variation in osmotic stress tolerance among rice cultivars and its association with L-type lateral root development. Plant Prod. Sci. 2015, 18, 246–253. [Google Scholar] [CrossRef]
- Nibau, C.; Gibbs, D.J.; Coates, J.C. Branching out in new directions: The control of root architecture by lateral root formation. New Phytol. 2008, 179, 595–614. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Albacete, A.; Ghanem, M.E.; Martínez-Andújar, C.; Acosta, M.; Sánchez-Bravo, J.; Martínez, V.; Lutts, S.; Dodd, I.C.; Pérez-Alfocea, F. Hormonal changes in relation to biomass partitioning and shoot growth impairment in salinized tomato (Solanum lycopersicum L.) plants. J. Exp. Bot. 2008, 59, 4119–4131. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, T.C.; Xu, L.K. Sensitivity of growth of roots versus leaves to water stress: Biophysical analysis and relation to water transport. J. Exp. Bot. 2000, 51, 1595–1616. [Google Scholar] [CrossRef] [PubMed]
- Formentin, E.; Sudiro, C.; Perin, G.; Riccadonna, S.; Barizza, E.; Baldoni, E.; Lavezzo, E.; Stevanato, P.; Sacchi, G.A.; Fontana, P.; et al. Transcriptome and cell physiological analyses in different rice cultivars provide new insights into adaptive and salinity stress responses. Front. Plant Sci. 2018, 9, 204. [Google Scholar] [CrossRef] [PubMed]
- Yeo, A.R.; Lee, À.S.; Izard, P.; Boursier, P.J.; Flowers, T.J. Short- and long-term effects of salinity on leaf growth in rice (Oryza sativa L.). J. Exp. Bot. 1991, 42, 881–889. [Google Scholar] [CrossRef]
- Yeo, A.R.; Yeo, M.E.; Flowers, T.J. The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions. J. Exp. Bot. 1987, 38, 1141–1153. [Google Scholar] [CrossRef]
- Ochiai, K.; Matoh, T. Characterization of the Na+ delivery from roots to shoots in rice under saline stress: Excessive salt enhances apoplastic transport in rice plants. Soil Sci. Plant Nutr. 2002, 48, 371–378. [Google Scholar] [CrossRef]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef]
- Kobayashi, N.I.; Yamaji, N.; Yamamoto, H.; Okubo, K.; Ueno, H.; Costa, A.; Tanoi, K.; Matsumura, H.; Fujii-Kashino, M.; Horiuchi, T.; et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017, 91, 657–670. [Google Scholar] [CrossRef]
- Ismail, A.M.; Horie, T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef]
- Farooq, M.; Park, J.R.; Jang, Y.H.; Kim, E.G.; Kim, K.M. Rice cultivars under salt stress show differential expression of genes related to the regulation of Na+/K+ balance. Front. Plant Sci. 2021, 12, 680131. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, B.; Yun, P.; Shabala, L.; Zhou, M.; Sellamuthu, G.; Venkataraman, G.; Chen, Z.H.; Shabala, S. Unravelling the physiological basis of salinity stress tolerance in cultivated and wild rice species. Funct. Plant Biol. 2022, 49, 351–364. [Google Scholar] [CrossRef]
- Rewald, B.; Shelef, O.; Ephrath, J.E.; Rachmilevitch, S. Adaptive plasticity of salt-stressed root systems. In Ecophysiology and Responses of Plants under Salt Stress; Ahmad, P., Azooz, M.M., Prasad, M.N.V., Eds.; Springer: New York, NY, USA, 2013; pp. 169–201. ISBN 978-1-4614-4747-4. [Google Scholar]
- Mcallister, C.H.; Beatty, P.H.; Good, A.G. Engineering nitrogen use efficient crop plants: The current status. Plant Biotechnol. J. 2012, 10, 1011–1025. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, M.; Guo, R.; Shi, D.; Liu, B.; Lin, X.; Yang, C. Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Farquar, G.D.; Ehleringer, J.R.; Hubick, K.T. Carbon isotope discrimination and photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989, 40, 503–537. [Google Scholar] [CrossRef]
- Sarabi, B.; Fresneau, C.; Ghaderi, N.; Bolandnazar, S.; Streb, P.; Badeck, F.W.; Citerne, S.; Tangama, M.; David, A.; Ghashghaie, J. Stomatal and non-stomatal limitations are responsible in down-regulation of photosynthesis in melon plants grown under the saline condition: Application of carbon isotope discrimination as a reliable proxy. Plant Physiol. Biochem. 2019, 141, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Badeck, F.W.; Tcherkez, G.; Nogués, S.; Piel, C.; Ghashghaie, J. Post-photosynthetic fractionation of stable carbon isotopes between plant organs—A widespread phenomenon. Rapid Commun. Mass Spectrom. 2005, 19, 1381–1391. [Google Scholar] [CrossRef] [PubMed]
- Tcherkez, G.; Mahé, A.; Hodges, M. 12C/13C fractionations in plant primary metabolism. Trends Plant Sci. 2011, 16, 499–506. [Google Scholar] [CrossRef]
- Hartman, G.; Danin, A. Isotopic values of plants in relation to water availability in the Eastern Mediterranean region. Oecologia 2010, 162, 837–852. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.P.; Sarkar, R.K. Distinction and characterisation of salinity tolerant and sensitive rice cultivars as probed by the chlorophyll fluorescence characteristics and growth parameters. Funct. Plant Biol. 2014, 41, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Kalaji, H.M.; Jajoo, A.; Oukarroum, A.; Brestic, M.; Zivcak, M.; Samborska, I.A.; Cetner, M.D.; Łukasik, I.; Goltsev, V.; Ladle, R.J. Chlorophyll a fluorescence as a tool to monitor physiological status of plants under abiotic stress conditions. Acta Physiol. Plant. 2016, 38, 102. [Google Scholar] [CrossRef]
- Oukarroum, A.; Bussotti, F.; Goltsev, V.; Kalaji, H.M. Correlation between reactive oxygen species production and photochemistry of photosystems I and II in Lemna gibba L. plants under salt stress. Environ. Exp. Bot. 2015, 109, 80–88. [Google Scholar] [CrossRef]
- Mehta, P.; Jajoo, A.; Mathur, S.; Bharti, S. Chlorophyll a fluorescence study revealing effects of high salt stress on Photosystem II in wheat leaves. Plant Physiol. Biochem. 2010, 48, 16–20. [Google Scholar] [CrossRef]
- Strasser, R.J.; Srivastava, A.; Tsimilli-Michael, M. Probing Photosynthesis; Yunus, M., Pathre, U., Mohanty, P., Eds.; CRC Press: Boca Raton, FL, USA, 2000; ISBN 9780429179549. [Google Scholar]
- Faseela, P.; Sinisha, A.K.; Brestič, M.; Puthur, J.T. Chlorophyll a fluorescence parameters as indicators of a particular abiotistress in rice. Photosynthetica 2020, 58, 293–300. [Google Scholar] [CrossRef]
- Masarmi, A.G.; Solouki, M.; Fakheri, B.; Kalaji, H.M.; Mahgdingad, N.; Golkari, S.; Telesiński, A.; Lamlom, S.F.; Kociel, H.; Yousef, A.F. Comparing the salinity tolerance of twenty different wheat genotypes on the basis of their physiological and biochemical parameters under NaCl stress. PLoS ONE 2023, 18, e0282606. [Google Scholar] [CrossRef]
- Roose, J.L.; Wegener, K.M.; Pakrasi, H.B. The extrinsic proteins of Photosystem II. Photosynth. Res. 2007, 92, 369–387. [Google Scholar] [CrossRef]
- Pagliano, C.; Saracco, G.; Barber, J. Structural, functional and auxiliary proteins of photosystem II. Photosynth. Res. 2013, 116, 167–188. [Google Scholar] [CrossRef] [PubMed]
- Ceppi, M.G.; Oukarroum, A.; Çiçek, N.; Strasser, R.J.; Schansker, G. The IP amplitude of the fluorescence rise OJIP is sensitive to changes in the photosystem I content of leaves: A study on plants exposed to magnesium and sulfate deficiencies, drought stress and salt stress. Physiol. Plant. 2012, 144, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lan, H.; Fang, H.; Huang, X.; Zhang, H.; Huang, J. Quantitative proteomic analysis of the rice (Oryza sativa L.) salt response. PLoS ONE 2015, 10, e0120978. [Google Scholar] [CrossRef] [PubMed]
- Lekklar, C.; Suriya-arunroj, D.; Pongpanich, M.; Comai, L.; Kositsup, B.; Chadchawan, S.; Buaboocha, T. Comparative genomic analysis of rice with contrasting photosynthesis and grain production under salt stress. Genes 2019, 10, 562. [Google Scholar] [CrossRef]
- Golding, A.J.; Johnson, G.N. Down-regulation of linear and activation of cyclic electron transport during drought. Planta 2003, 218, 107–114. [Google Scholar] [CrossRef]
- Ozakca, D.U. Effect of abiotic stress on Photosystem I-related gene transcription in photosynthetic organisms. In Photosynthesis; Dubinsky, Z., Ed.; IntechOpen: Rijeka, Croatia, 2013. [Google Scholar]
- Mancarella, S.; Orsini, F.; Van Oosten, M.J.; Sanoubar, R.; Stanghellini, C.; Kondo, S.; Gianquinto, G.; Maggio, A. Leaf sodium accumulation facilitates salt stress adaptation and preserves photosystem functionality in salt stressed Ocimum basilicum. Environ. Exp. Bot. 2016, 130, 162–173. [Google Scholar] [CrossRef]
- Liu, Z.; Zou, L.; Chen, C.; Zhao, H.; Yan, Y.; Wang, C.; Liu, X. iTRAQ-based quantitative proteomic analysis of salt stress in Spica Prunellae. Sci. Rep. 2019, 9, 9590. [Google Scholar] [CrossRef]
- Hewitt, E.J.; Eden, A. Sand and Water Culture Methods Used in the Study of Plant Nutrition. In Commonwealth Agricultural Bureaux; Commonwealth Agricultural Bureaux: Farnham Royal, UK, 1953; Volume 78. [Google Scholar]
- Barrs, H.; Weatherley, P. A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust. J. Biol. Sci. 1962, 15, 413–428. [Google Scholar] [CrossRef]
- Calà, E.; Fracchia, A.; Robotti, E.; Gulino, F.; Gullo, F.; Oddone, M.; Massacane, M.; Cordone, G.; Aceto, M. On the traceability of the hazelnut production chain by means of trace elements. Molecules 2022, 27, 3854. [Google Scholar] [CrossRef] [PubMed]
- Pagliano, C.; Raviolo, M.; Dalla Vecchia, F.; Gabbrielli, R.; Gonnelli, C.; Rascio, N.; Barbato, R.; La Rocca, N. Evidence for PSII donor-side damage and photoinhibition induced by cadmium treatment on rice (Oryza sativa L.). J. Photochem. Photobiol. B Biol. 2006, 84, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Porra, R.; Thompson, W.; Kriedemann, P. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 1999, 975, 384–394. [Google Scholar] [CrossRef]
- Wellburn, A.R. The spectral determination of Chlorophylls a and b, as well as total Carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- Strasser, R.J.; Srivastava, A. Govindjee Polyphasic chlorophyll a fluorescence transient in plants and cyanobacteria. Photochem. Photobiol. 1995, 61, 32–42. [Google Scholar] [CrossRef]
- Stirbet, A. Govindjee On the relation between the Kautsky effect (chlorophyll a fluorescence induction) and Photosystem II: Basics and applications of the OJIP fluorescence transient. J. Photochem. Photobiol. B Biol. 2011, 104, 236–257. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and Carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Barbato, R.; Friso, G.; Rigoni, F.; Dalla Vecchia, F.; Giacometti, G.M. Structural changes and lateral redistribution of Photosystem II during donor side photoinhibition of thylakoids. J. Cell Biol. 1992, 119, 325–335. [Google Scholar] [CrossRef]
- Bohra, A.; Kilian, B.; Sivasankar, S.; Caccamo, M.; Mba, C.; McCouch, S.R.; Varshney, R.K. Reap the crop wild relatives for breeding future crops. Trends Biotechnol. 2022, 40, 412–431. [Google Scholar] [CrossRef]
- Brar, D.S.; Khush, G.S. Utilization of wild species of genus Oryza in rice improvement. In Monograph on Genus Oryza; Nanda, J.S., Sharma, S.D., Eds.; Science Publishers: Enfield, UK, 2003; pp. 283–309. [Google Scholar]
- Huang, X.; Kurata, N.; Wei, X.; Wang, Z.X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W.; et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef]
- Zhang, B.; Ma, L.; Wu, B.; Xing, Y.; Qiu, X. Introgression lines: Valuable resources for functional genomics research and breeding in rice (Oryza sativa L.). Front. Plant Sci. 2022, 13, 863789. [Google Scholar] [CrossRef] [PubMed]
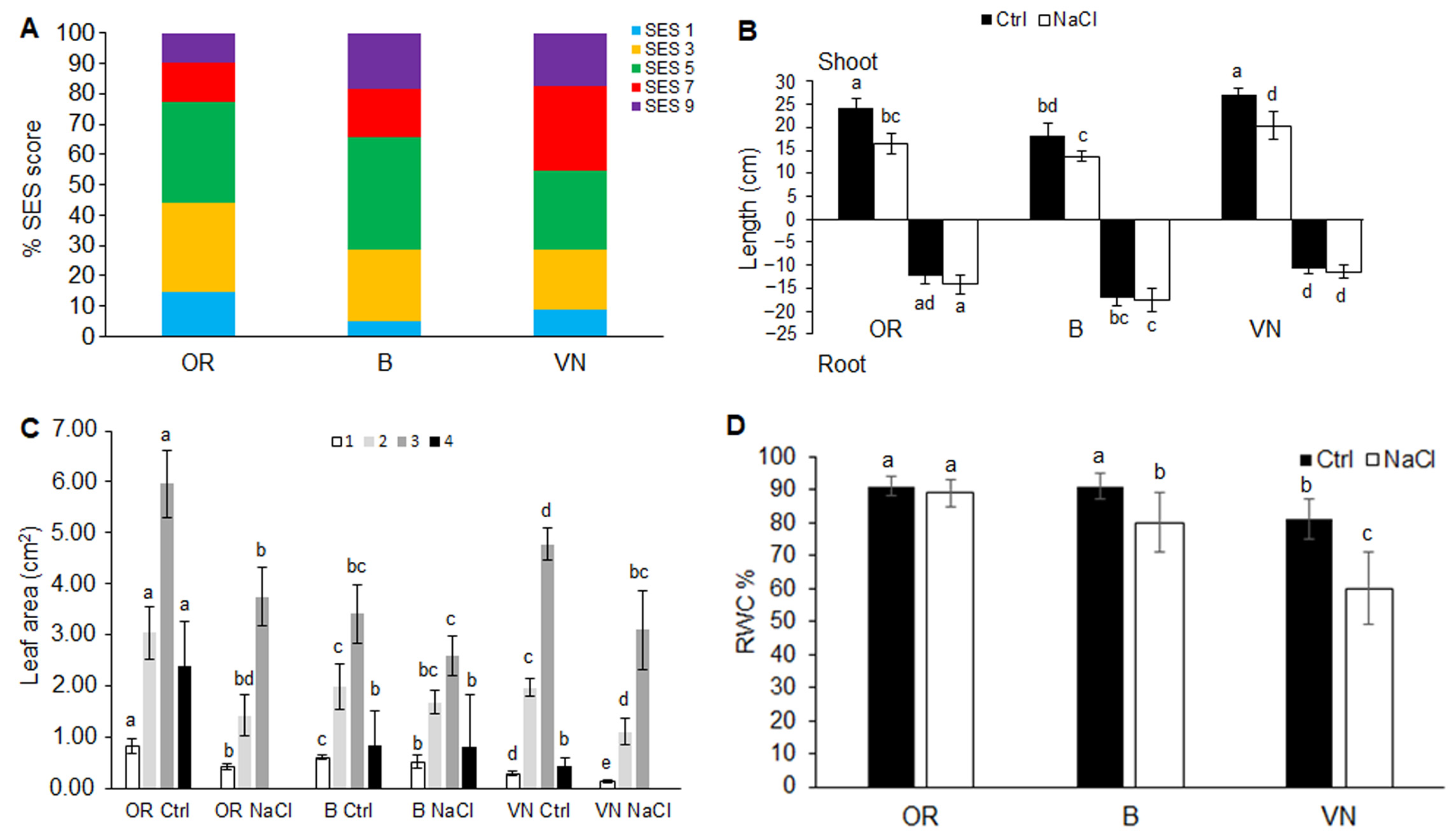
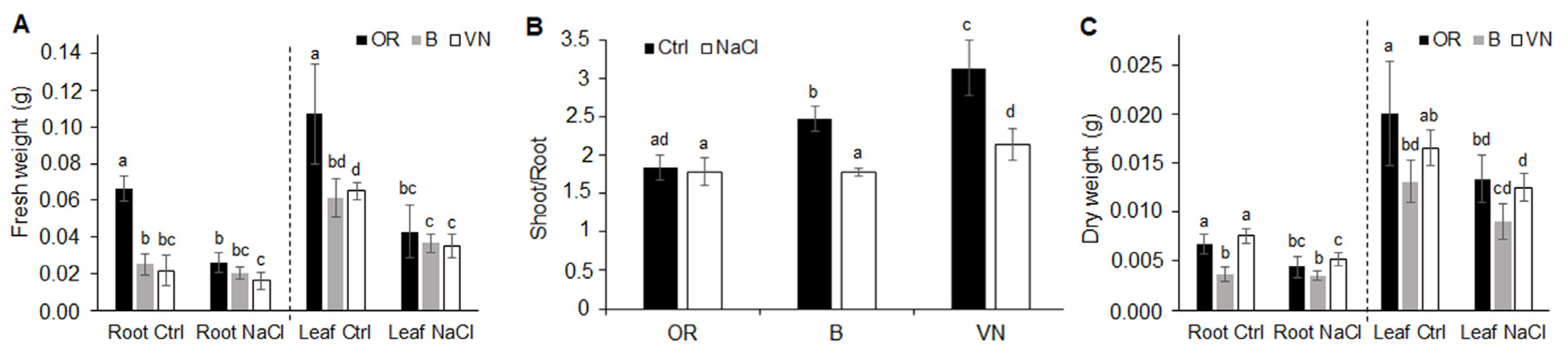
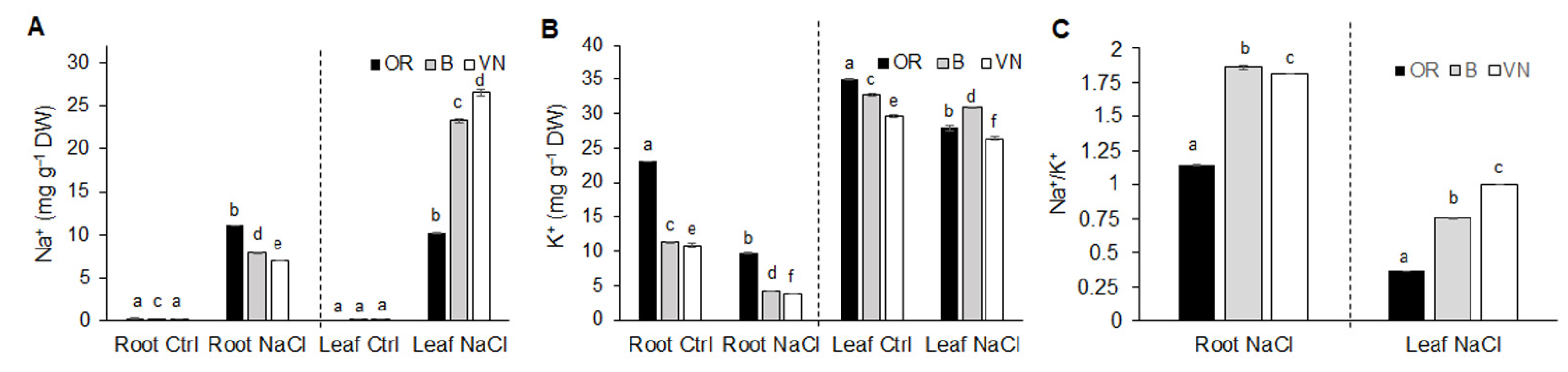
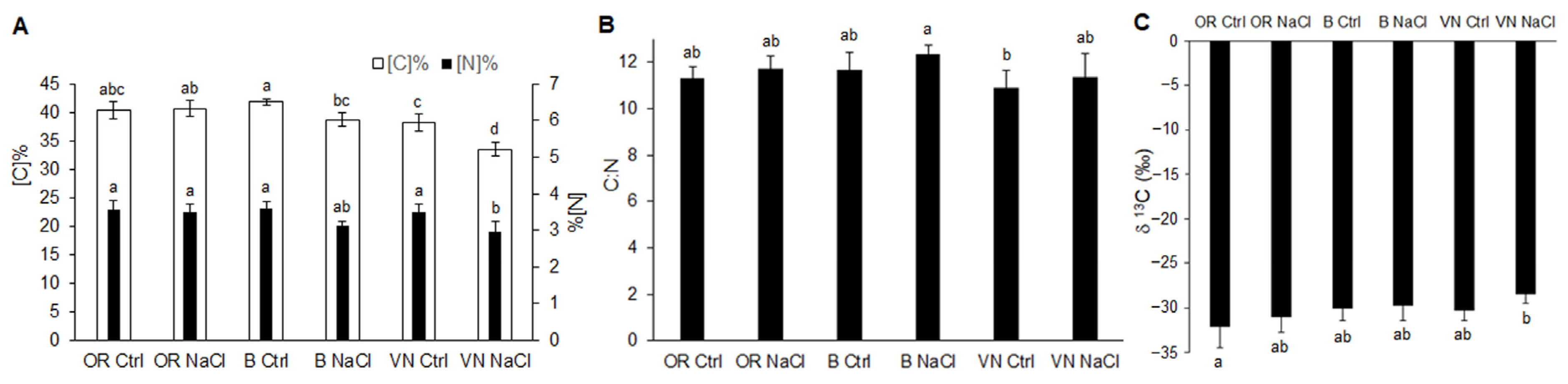

| Plant | Length (cm) | Volume (cm3) | Average Diameter (mm) | Surface Area (cm2) | Tips Number | Branching Degree |
|---|---|---|---|---|---|---|
| OR Ctrl | 295.2 ± 39.6 a | 0.26 ± 0.04 a | 0.348 ± 0.030 a | 29.05 ± 6.06 a | 520 ±170 a | 2.14 ± 0.42 a |
| OR NaCl | 139.7 ± 32.1 b | 0.18 ± 0.03 b | 0.395 ± 0.046 b | 17.43 ± 3.04 b | 319 ± 94 b | 2.25 ± 0.22 a |
| B Ctrl | 143.8 ± 25.1 b | 0.16 ± 0.03 b | 0.372 ± 0.022 ab | 17.17 ± 4.3 b | 375 ± 118 b | 2.38 ± 0.23 a |
| B NaCl | 147.9 ± 24.8 b | 0.14 ± 0.03 b | 0.357 ± 0.020 a | 17.87 ± 3.4 b | 353 ± 47 b | 2.41 ± 0.29 a |
| VN Ctrl | 240.2 ± 27.9 a | 0.24 ± 0.03 ac | 0.357 ± 0.022 a | 26.04 ± 2.63 ac | 551 ± 62 a | 2.30 ± 0.28 a |
| VN NaCl | 161.0 ± 27.8 b | 0.17 ± 0.03 bc | 0.373 ± 0.025 ab | 21.04 ± 3.25 bc | 363 ± 78 b | 2.37 ± 0.13 a |
| Plant | Chl a (mg g−1 FW) | Chl b (mg g−1 FW) | Carotenoids (mg g−1 FW) | Chl a/b | Chl/Carotenoids |
|---|---|---|---|---|---|
| OR Ctrl | 3.16 ± 0.35 a | 0.75 ± 0.05 a | 0.64 ± 0.07 a | 4.26 ± 0.32 a | 6.11 ± 0.18 ac |
| OR NaCl | 2.10 ± 0.26 bc | 0.50 ± 0.07 bc | 0.41 ± 0.05 bc | 4.25 ± 0.24 a | 6.39 ± 0.28 a |
| B Ctrl | 2.01 ± 0.27 bc | 0.45 ± 0.06 bc | 0.46 ± 0.05 bc | 4.45 ± 0.26 ab | 5.42 ± 0.26 b |
| B NaCl | 2.31 ± 0.20 b | 0.52 ± 0.05 b | 0.48 ± 0.04 c | 4.47 ± 0.18 ab | 5.84 ± 0.21 c |
| VN Ctrl | 1.96 ± 0.18 bc | 0.42 ± 0.06 c | 0.46 ± 0.04 bc | 4.72 ± 0.14 b | 5.18 ± 0.24 b |
| VN NaCl | 1.89 ± 0.15 c | 0.43 ± 0.04 c | 0.40 ± 0.03 b | 4.45 ± 0.18 ab | 5.77 ± 0.20 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trotti, J.; Trapani, I.; Gulino, F.; Aceto, M.; Minio, M.; Gerotto, C.; Mica, E.; Valè, G.; Barbato, R.; Pagliano, C. Physiological Responses to Salt Stress at the Seedling Stage in Wild (Oryza rufipogon Griff.) and Cultivated (Oryza sativa L.) Rice. Plants 2024, 13, 369. https://doi.org/10.3390/plants13030369
Trotti J, Trapani I, Gulino F, Aceto M, Minio M, Gerotto C, Mica E, Valè G, Barbato R, Pagliano C. Physiological Responses to Salt Stress at the Seedling Stage in Wild (Oryza rufipogon Griff.) and Cultivated (Oryza sativa L.) Rice. Plants. 2024; 13(3):369. https://doi.org/10.3390/plants13030369
Chicago/Turabian StyleTrotti, Jacopo, Isabella Trapani, Federica Gulino, Maurizio Aceto, Miles Minio, Caterina Gerotto, Erica Mica, Giampiero Valè, Roberto Barbato, and Cristina Pagliano. 2024. "Physiological Responses to Salt Stress at the Seedling Stage in Wild (Oryza rufipogon Griff.) and Cultivated (Oryza sativa L.) Rice" Plants 13, no. 3: 369. https://doi.org/10.3390/plants13030369
APA StyleTrotti, J., Trapani, I., Gulino, F., Aceto, M., Minio, M., Gerotto, C., Mica, E., Valè, G., Barbato, R., & Pagliano, C. (2024). Physiological Responses to Salt Stress at the Seedling Stage in Wild (Oryza rufipogon Griff.) and Cultivated (Oryza sativa L.) Rice. Plants, 13(3), 369. https://doi.org/10.3390/plants13030369








