Abstract
The focus on increasing wheat (Triticum aestivum L.) grain yield at the expense of grain quality and nutrient accumulation can lead to shortages in macronutrient minerals, which are dangerous for human health. This is important, especially in nations where bread wheat is used in most daily dietary regimens. One efficient way to guarantee nutritional security is through biofortification. A genome-wide association mapping approach was used to investigate the genetic basis of the differences in macronutrient mineral accumulation in wheat grains. N, P, K, Na, Ca, and Mg concentrations were measured after a panel of 200 spring wheat advanced lines from the Wheat Association Mapping Initiative were cultivated in the field. The population exhibited a wide range of natural variations in macronutrient minerals. The minerals were found to have strong positive correlations except for magnesium, which had negative correlation patterns with N, P, and K. Furthermore, there were negative correlations between N and each of Ca and Na. Remarkably, genotypes with large yields contained moderate levels of critical metals. Of the 148 significant SNPs above −log10(P) = 3, 29 had −log10(P) values greater than 4. Four, one, and nineteen significant SNPs with a −log10(P) between 4 and 5.8 were associated with N and mapped on chromosomes 1A, 1B, and 1D, respectively. Three significant SNPs on chromosome A3 were associated with K. Two significant SNPs were associated with Ca and Na and mapped on chromosomes B3 and A4, respectively. Our findings offer crucial information about the genetic underpinnings of nutritional mineral concentration augmentation, which can guide future breeding research to enhance human nutrition.
1. Introduction
Wheat (Triticum aestivum L.) is the second most widely grown crop, with 790.42 million tons produced in 2024 [1]. Global consumption is expected to increase by 132 million tons by 2050 [2], necessitating more significant research into boosting output and quality. Wheat is essential for human health as it contains carbohydrates, proteins, and inorganic metals. Nonetheless, wheat grain has a naturally low nutrient content, including various minerals and vitamins essential to various biological processes. Maintaining mineral uptake and accumulation in wheat is critical for maximizing wheat growth, productivity, and nutritional quality [3,4], particularly for countries that rely heavily on wheat in their daily diet.
Similar to the depth of knowledge regarding the impacts of heavy metals [5,6,7,8,9], the overabundance of the necessary minerals has received attention [3,10,11]. While excessive uptake and accumulation of these minerals can interfere with growth, affect nutrient uptake, and cause mineral toxicity, nitrogen (N+), phosphorus (P+), potassium (K+), calcium (Ca++), and magnesium (Mg++) all play essential roles in various physiological processes [3,12,13,14].
Numerous biological substances, such as coenzymes, amino acids, nucleic acids, and chlorophyll, require N+. However, a high-N+ diet may cause adverse metabolic alterations that disrupt lipid and N metabolism in wheat, lowering grain filling [15,16]. In addition, P+ functions as a substrate in a range of physiological processes, including photosynthesis, respiration, signal transduction, and energy metabolism. It is a structural element of numerous biological molecules, including DNA, RNA, ATP, phospholipids, and carbohydrates [17]. On the other hand, excessive P+ consumption may inhibit meristematic activity and primary root growth, leading to deficiencies in other plant minerals such as iron (Fe+++) and zinc (Zn++) [18,19]. K+ improves photosynthesis, C− and N+ metabolizing enzymes, and nitrate transport and assimilation, improving nitrogen utilization efficiency. However, high K+ consumption can hinder photosynthesis [20].
Na+ can accumulate in high quantities and be beneficial in replacing K+ in vacuolar osmoticum, where it acts as an osmotic agent. Except for a few halophytes, most plants do not require Na+ [21]. Ca+ deficit increases a plant’s susceptibility to biotic and abiotic stressors, as it plays a significant role in cell wall development, plant design, quality, and yield formation [22,23]. Mg+ is a cofactor for enzymes that support carbon fixation, metabolism, and chlorophyll [24]. Excessive magnesium intake can lead to a 54–67% decline in rice growth [25].
Developing stable varieties that balance vital mineral contents through traditional and modern plant breeding is a long-term method for maintaining mineral homeostasis [26,27]. However, this requires a better understanding of the genetic determinants controlling such quantitative features of mineral concentration [7,28]. Genome-wide association mapping (GWAM) effectively identifies genetic regions contributing to observed trait variation in a population. GWAM investigates genetic variations across the entire genome to discover genetic markers, such as single nucleotide polymorphisms (SNPs) statistically related to phenotypic variation [29].
Genomic prediction (GP) predicts phenotypes or breeding values of genotypes on unobserved individuals by connecting a set of markers to variability in cultivar phenotypes that have been observed [30]. In breeding programmes, GP relies on genome-wide marker data to forecast the breeding value of complex traits, offering an alternative to handle complex traits governed by numerous genes with minor impacts [7,30,31]. GP uses various techniques such as ridge-regression best linear unbiased prediction (rrBLUP), which enables effective prediction using unreplicated training data [32].
Our study was conducted to unravel the genetic architecture underlying five essential minerals and to identify candidate genes for these traits in spring wheat. In addition, we identified several genotypes with high contents of each of the measured minerals. Finally, we performed GP for the studied macronutrient minerals.
2. Results
2.1. Statistical Analysis
All traits exhibited a wide range of genetic variation (Table 1). The least observed variance was for Na, with minimum and maximum values of 0.06 and 0.12, respectively. Mg showed the second most minor variance, with a mean of 0.17, a minimum of 0.03, and a maximum of 0.38 mg kg−1. The P concentration ranged from 0.20 to 0.58, with an average of 0.34 mg kg−1. The minimum and the maximum of K were 0.21 and 0.62 mg kg−1, respectively. The most significant variations were observed for N and Ca, with a minimum of 0.58 and 0.11 and a maximum of 3.37 and 0.78 mg kg−1, respectively.

Table 1.
Population performance of the measured macronutrient minerals: nitrogen (N), phosphorus (P), potassium (K), calcium (Ca), and magnesium (Mg). Minimum (min), maximum (max), and mean values in mg kg−1.
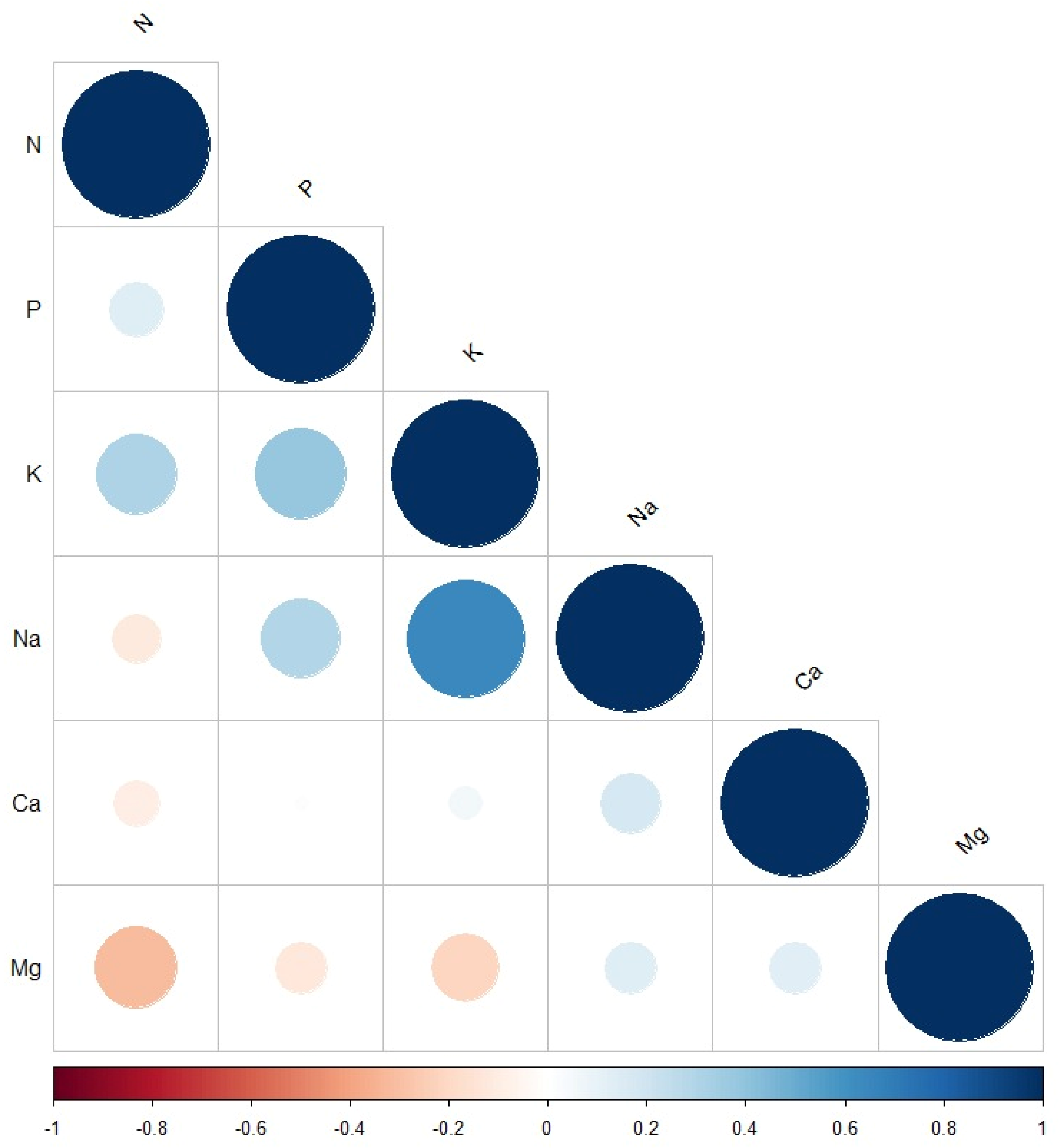
The correlation plot (Figure 1) revealed a positive relationship between all evaluated parameters except between N and each of Na, Ca, and Mg. The strongest positive correlation was between Na and K, followed by the correlation between P and K. The strongest negative correlation was observed between N and Mg, followed by the correlation between K and Mg. No correlation was observed between P and Ca.
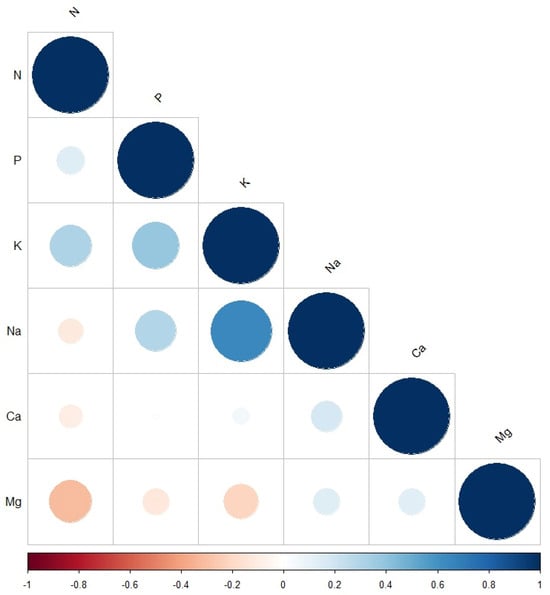
Figure 1.
Correlation plot between the measured macroelement minerals: N, P, K, Na, Ca, and Mg. The size and the colour of the circles indicate the significance level. The blue and red indicate positive and negative correlations between the measured minerals.
Checking the five high-yielding genotypes from Said et al. [33] for their macronutrient mineral concentrations revealed moderate concentrations of the six minerals (Table 2). We observed overlapping genotypes between the highest N, P, K, Na, Ca, and Mg contents. For example, genotype 80836 had the highest N content and the third-highest Ca content. Genotype 369673 showed the highest P and K contents.

Table 2.
Macronutrient mineral contents in mg kg−1 for genotypes with the highest yield [33]: N, P, K, Na, Ca, and Mg concentrations. GY represents grain yield data from Said et al. [33].
2.2. Genome-Wide Association Mapping
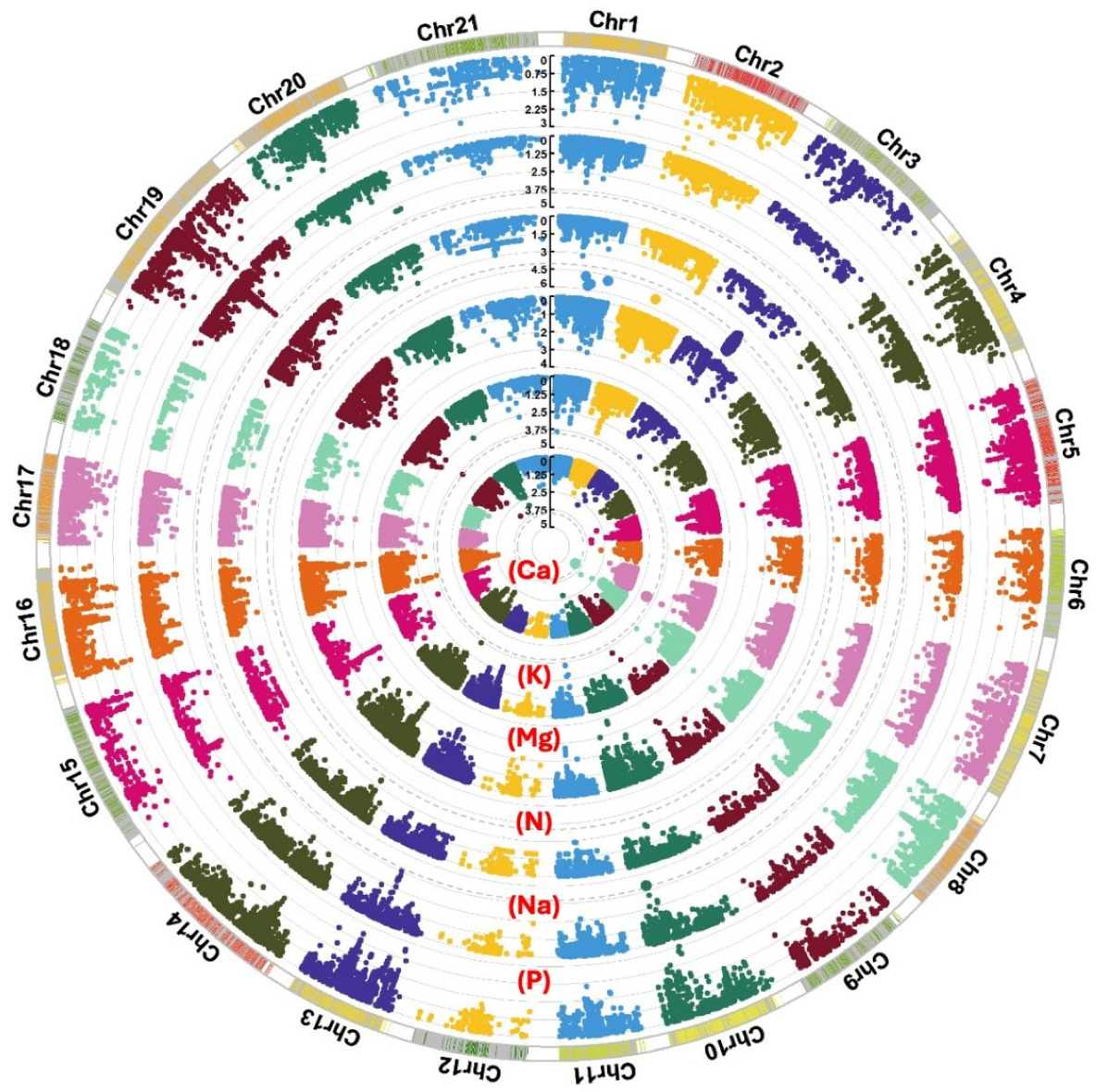
A total of 148 significant SNPs, above −log10(P) = 3, were associated with the measured macroelement minerals (Figure 2 and Table S1), of which 29 SNPs were above −log10(P) = 4 (Table 3). For example, 24 SNPs between −log10(P) = 4 and 5.8 were associated with N and mapped on 1A, 1B, and 1D. Only one SNP, BS00021657_51, on 7A, was significantly linked to P. Among the 28 SNPs associated with K, one QTL harboured six SNPs linked to four genes at 177 cM on 3A. Three of the six SNPs were significant at −log10(P) = 4.3. Ten additional SNPs were linked to Na, four SNPs were mapped to 4A, and six SNPs were on 5A. One QTL sheltered 36 SNPs, significant at −log10(P) = 4.3, on 2B at 134 cM that were associated with Ca and contained 20 genes. A total of 11 SNPs showed a significant link to Mg, of which three were on 2B, three were on 4A, and five were on 6A.
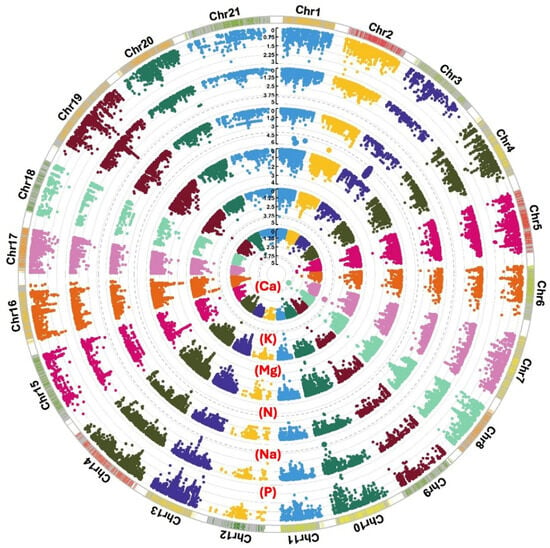
Figure 2.
Circular Manhattan plots displaying significant SNPs associated with the wheat content of macronutrient minerals Ca, K, Mg, N, Na, and P at a −log10(P) value ≥ 3. Genome-wide association mapping was carried out for the 200 wheat genotypes using 26,814 SNPs distributed over the 21 wheat chromosomes. The significance level is denoted by dashed-grey circles.

Table 3.
Selected significant SNPs associated with macronutrient mineral contents. The positions (Pos) in centimorgan on chromosomes (Chr.) are indicated. The −log10(P) values indicate the significance value. The explained phenotypic variance (R2) and the effect of each allele are given. Names of candidate genes are presented according to the second version of the International Wheat Genome Sequencing (IWGS2).
2.3. Genomic Prediction
The predictability values for our wheat panel, which ranged from 0.036 to 0.14, were low. A prediction of 0.059 for N, 0.039 for P, 0.14 for K, 0.098 for Na, 0.078 for Ca, and 0.10 for Mg was obtained using the rrBLUP and a training population of 130 genotypes.
3. Discussion
The expansion of the world population places enormous pressure on agricultural systems to increase food production. However, this focus on quantity often ignores essential quality factors, including mineral imbalances [34]. It becomes obvious how nutritional imbalances interact, demonstrating how an excess or limited availability of one element can reduce the intake of other essential nutrients, ultimately affecting plant health and development [35,36]. Therefore, a balanced approach considering mineral buildup is required to overcome this difficulty.
Exploring natural phenotypic variation in wheat is the first step in boosting the levels of essential macronutrients in wheat grains using genomic breeding techniques [3,7]. We found significant natural variation in the grain concentrations of N, P, K, Na, Ca, and Mg using our panel of 200 advanced spring wheat lines, consistent with previous findings [3,5].
Choosing high-yielding genotypes with acceptable amounts of mineral accumulation is the foundation of biofortification [5,7,27,37]. We examined a variety of approaches to determine which genotypes performed best. First, we examined the five most productive genotypes from the previous study [33] that used the same population. Those genotypes showed moderate nutritional values of N, P, K, Ca, and Mg and low Na contents. Interestingly, those genotypes had some of the lowest Ni and Cd contents and some of the highest Mn, Fe, Cu, and Zn contents [7]. Our results imply that those genotypes could be good candidates for further breeding efforts to increase their content of the macronutrient minerals. The pyramiding of proteins and micro- and macronutrients into high-yielding wheat genotypes can alleviate malnutrition, affecting about one-third of the global population [38].
According to an earlier investigation [39], increasing nitrogen availability in the soil via a high input level enhanced wheat grain N content. At 238 kg ha−1 of ammonium nitrate (33.5% N) employed in our study, the five most productive genotypes [33] had a moderate level of N content. Instead, another five genotypes showed double N content. N is necessary to structure many biological molecules, including coenzymes, amino acids, and proteins. Therefore, it can indicate protein content (PC) using nitrogen-to-protein conversion factors [40,41,42]. Using wheat grains in dietary regimes can meet up to 25% of the protein needs [43]. However, breeding wheat to enhance grain yield and PC is still tricky due to the established negative correlation between the two characteristics [44,45]. Our results showed similar trends: high-yielding genotypes showed moderate N content, and genotypes with the highest seed N contents had low grain yield. Therefore, more breeding efforts should investigate how to improve wheat nutritional quality, particularly protein content.
High-quality wheat genome sequencing provides the base for identifying genetic loci and genes controlling wheat macronutrient content [46]. We used 200 advanced spring wheat lines from the WAMI population, genotyped with 26,814 SNPs [47,48], and mapped 148 significant SNPs above −log10(P) = 3, of which 29 SNPs were above −log10(P) = 4. The GAWM approach benefits from ancestral recombination, giving a higher resolution than the traditional QTL mapping using biparental populations [29]. The threshold −log10(P) = 3 is commonly used in wheat research [3,7]. However, the population structure of the GWAM population results in a high probability of false-positive associations. Therefore, we discuss SNPs with −log10(P) ≥ 4, multiple significant SNPs at the same position, or those that collocate with earlier findings.
A QTL associated with Ca content harboured 32 SNPs on Chr. 2B at 134 cM and two more SNPs at 136 and 141 cM. This QTL collocated with the SNP Kukri_c900_1334 [3] and the SNP IACX6292 [12] associated with Ca content. Another colocation observed for Ca was the SNP RAC875_rep_c78007_349, which was mapped here on Chr. 7B and collocated with wsnp_BE445506B_Ta_2_1 and wsnp_BE445506B_Ta_2_4 [12]. The SNP Kukri_c9895_1325 on Chr. 3B was linked to Ca and collocated with a previous SNP associated with Ca accumulation in wheat [14]. One more SNP, wsnp_Ku_c28104_38042857, was reported to be associated with Ca on Chr. 7A [49], collocated with our SNP wsnp_Ex_rep_c67593_66232317 mapped here.
The SNP BS00068508_51 mapped on Chr. 3A was linked to K [3], collocated with the six SNPs mapped here for the same trait (Table 3 and Table S1). One more SNP, BS00047691_51, was reported to be associated with P on Chr. 7A [49], collocated with our SNP BS00021657_51 mapped here for P. Four SNPs, wsnp_Ex_c34597_42879693, RFL_Contig6053_3082, and wsnp_Ex_c2236_4189774, on Chr. 6A and wsnp_Ex_c34597_42879718 on chromosome 6B were associated with Mg. The four SNPs were reported earlier to be significantly associated with Fe, Cu, and Cd using the same population [7]. We checked the correlation between the Mg and the three minerals, which revealed significant positive correlations of 0.6, 0.4, and 0.5, respectively, indicating the possible pleiotropic effects of the four SNPs on Mg, Fe, Cu, and Cd.
The significant positive correlations between the measured minerals reflect the presence of shared genetic mechanisms, known as pleiotropic effects, underlying the accumulation of these nutrients in wheat grains. Similar correlation trends have been previously reported in bread wheat [3,50]. In contrast, the significant negative correlations indicate possible antagonistic pleiotropic effects [51], critical in breeding programmes as breeding for one trait negatively impacts others. For example, negative correlations were observed between Mg and each of N, P, and K. Similar to our results, Alomari et al. [3] found a negative correlation between Mg and K and a moderate positive correlation between Mg and Ca. However, contradicting correlations were observed between Mg and P.
Generally, GP is a viable strategy for improving complicated features, like macronutrient metal content, mainly when studying a sizable germplasm panel with many markers. In line with earlier findings for wheat landraces evaluated in Afghanistan [52], both requirements enable more precise estimations of low breeding values for the tested metals. An individual is anticipated to perform below the reference population’s average for the trait under evaluation if their genetic breeding value is negative. Although GP values should range from 0 to 1, negative values are commonly reported. They can be explained by numerous factors, including a systematic bias of the correlation coefficient, as when the expected accuracy is low, negative prediction accuracy may be an artefact of the mathematical procedures used to calculate correlation coefficients. In addition, negative GP values can be due to statistical aberrations or remote QTL effects [53,54].
Our study offers valuable insights into the phenotypic diversity of the macronutrient minerals in the WAMI mapping population. For example, we observed that the five most high-yielding genotypes of the population have moderate concentrations of the six studied minerals. In contrast, genotypes with high seed N contents showed low grain yield. Therefore, pyramiding is required for high-yielding genotypes to increase their nutritional values. More studies are necessary to better understand the connections between the various mineral components in wheat and offer more specific knowledge that can result in practical breeding to address nutritional issues. The natural variation in the macronutrient minerals within the population facilitates GWAM investigations. Our results revealed 148 SNP markers with significant effects on the accumulation of micronutrient minerals in wheat grains. These data can be used in marker-assisted breeding programmes to improve wheat nutritional quality. In addition, it is necessary to validate those candidate genes or use genome editing technologies to edit new cultivars.
4. Materials and Methods
4.1. Plant Materials and Soil Conditions
We used 200 advanced spring wheat lines from the Wheat Association Mapping Initiative (WAMI) population and genotyped with 26,814 SNPs from the International Maize and Wheat Improvement Center (CIMMYT) [47,48]. The field study was carried out at the Agriculture Farm of Sohag University, Egypt. The soil texture is composed of two layers: sandy clay loam at depths ranging from 0 to 30 cm and sandy loam soil from 30 to 45 cm. The essential relevant chemical characteristics of the experimental soil are shown in Table 4.

Table 4.
Chemical characteristics of the experimental soil at depths ranging from 0 to 30 cm (sandy clay loam) and from 30 to 45 cm (sandy loam soil).
The experimental setup used a randomized block design with three replications. Each block of a 2 m row with a 0.10 m plant spacing represented a genotype. NPK fertilization was as follows: 238 kg ha−1 of ammonium nitrate (33.5% N), 75 kg ha−1 of calcium superphosphate (15.5% P2O), and 58 kg ha−1 of potassium sulphate (48% K2O).
4.2. Mineral Measurements
A combination of concentrated HNO3 and HCLO4 (10:4) was added to 0.5 g of crushed wheat seeds in the digesting vessel, which was then sealed and cooked in a water bath at 80 °C for 120 min. The solution was cooled to room temperature and filtered through Whatman No. 1 filter paper into a volumetric flask containing 50 mL of double-distilled deionized water. N, P, K, Na, Ca, and Mg contents were determined in the extracts using an atomic absorption spectrophotometer (AAS) [55], LAAS-A11 (Labtron Equipment Ltd, Camberley, UK).
4.3. Statistical Analysis, Genome-Wide Association Mapping, and Genomic Prediction
Statistical analysis was performed using R for Windows version 4.1.2, with the R tool corrplot used to create frequency distributions.
Using the mixed linear model (MLM), kinship matrix, and principal component, the TASSEL program, version 5.0 [56], was used to map SNP markers associated with mineral contents.
GP was assessed using the R software package ridge-regression best linear unbiased prediction (rrBLUP) [32]. The 200 investigated genotypes underwent fivefold cross-validations, and the training sets consisted of 130 randomly selected genotypes. Following 100 iterations, the mean correlation accuracy was determined based on the correlation between the observed and predicted values, which validated the prediction ability computation.
4.4. Candidate Genes Identification
We retrieved the flanking sequences of the significant SNPs [57] and then identified the candidate genes using the GrainGenes BLAST service (https://wheat.pw.usda.gov/blast/, accessed on 28 February 2022), and the wheat IWGSC RefSeq v2.1 [58]. We used the KnetMiner gene discovery tool [59] (https://knetminer.org, accessed on 28 February 2022), to find large-genome-scale knowledge graphs and show intriguing subgraphs of relevant information regarding gene biology and functions, gene networks, and phenotypes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants13243472/s1. Table S1: Significant SNPs associated with measured traits with their predicted genes.
Author Contributions
Conceptualization, M.A. and M.E.-S.; investigation, M.A. and M.E.-S.; resources, M.A.; data curation, M.E.-S.; writing—original draft preparation, M.E.-S.; writing—review and editing, M.E.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
All data are available in the manuscript or its Supplementary Materials.
Acknowledgments
The authors thank Mohamed Mosalam, Department of Plant Breeding, Hochschule Geisenheim University, 65366 Geisenheim, Germany, for creating Figure 2.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Foreign Agricultural Service, United States Department of Agriculture. Available online: https://fas.usda.gov/data/production/commodity/0410000 (accessed on 19 November 2024).
- Erenstein, O.; Jaleta, M.; Mottaleb, K.A.; Sonder, K.; Donovan, J.; Braun, H.-J. Global trends in wheat production, consumption and trade. In Wheat Improvement: Food Security in a Changing Climate; Reynolds, M.P., Braun, H.-J., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 47–66. [Google Scholar]
- Alomari, D.Z.; Alqudah, A.M.; Pillen, K.; von Wirén, N.; Röder, M.S. Toward identification of a putative candidate gene for nutrient mineral accumulation in wheat grains for human nutrition purposes. J. Exp. Bot. 2021, 72, 6305–6318. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Wu, Y.; Fan, J.; Zhang, F.; Guo, J.; Zheng, J.; Wu, L. Quantifying grain yield, protein, nutrient uptake and utilization of winter wheat under various drip fertigation regimes. Agric. Water Manag. 2022, 261, 107380. [Google Scholar] [CrossRef]
- Bhatta, M.; Baenziger, P.S.; Waters, B.M.; Poudel, R.; Belamkar, V.; Poland, J.; Morgounov, A. Genome-wide association study reveals novel genomic regions associated with 10 grain minerals in synthetic hexaploid wheat. Int. J. Mol. Sci. 2018, 19, 3237. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Huang, L.; Li, T.; Liu, Y.; Yan, Z.; Tang, G.; Zheng, Y.; Liu, D.; Wu, B. Genome-wide association study for grain micronutrient concentrations in wheat advanced lines derived from wild emmer. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- El-Soda, M.; Aljabri, M. Genome-wide association mapping of grain metal accumulation in wheat. Genes 2022, 13, 1052. [Google Scholar] [CrossRef]
- Rathan, N.D.; Krishna, H.; Ellur, R.K.; Sehgal, D.; Govindan, V.; Ahlawat, A.K.; Krishnappa, G.; Jaiswal, J.P.; Singh, J.B.; Sv, S.; et al. Genome-wide association study identifies loci and candidate genes for grain micronutrients and quality traits in wheat (Triticum aestivum L.). Sci. Rep. 2022, 12, 7037. [Google Scholar] [CrossRef]
- Sabiha, J.; Siddque, N.; Waheed, S.; Uz Zaman, Q.; Aslam, A.; Tufail, M.; Nasir, R. Uptake of heavy metal in wheat from application of different phosphorus fertilizers. J. Food Compos. Anal. 2023, 115, 104958. [Google Scholar] [CrossRef]
- Lan, Y.; Kuktaite, R.; Chawade, A.; Johansson, E. Chasing high and stable wheat grain mineral content: Mining diverse spring genotypes under induced drought stress. PLoS ONE 2024, 19, e0298350. [Google Scholar] [CrossRef]
- Sigalas, P.P.; Shewry, P.R.; Riche, A.; Wingen, L.; Feng, C.; Siluveru, A.; Chayut, N.; Burridge, A.; Uauy, C.; Castle, M.; et al. Improving wheat grain composition for human health by constructing a QTL atlas for essential minerals. Commun. Biol. 2024, 7, 1001. [Google Scholar] [CrossRef]
- Alomari, D.Z.; Eggert, K.; von Wirén, N.; Pillen, K.; Röder, M.S. Genome-wide association study of calcium accumulation in grains of European wheat cultivars. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Ahmad, A.; Aslam, Z.; Javed, T.; Hussain, S.; Raza, A.; Shabbir, R.; Mora-Poblete, F.; Saeed, T.; Zulfiqar, F.; Ali, M.M.; et al. Screening of wheat (Triticum aestivum L.) genotypes for drought tolerance through agronomic and physiological response. Agronomy 2022, 12, 287. [Google Scholar] [CrossRef]
- Shi, X.; Zhou, Z.; Li, W.; Qin, M.; Yang, P.; Hou, J.; Huang, F.; Lei, Z.; Wu, Z.; Wang, J. Genome-wide association study reveals the genetic architecture for calcium accumulation in grains of hexaploid wheat (Triticum aestivum L.). BMC Plant Biol. 2022, 22, 229. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Xie, Y.; Hu, L.; Si, J.; Wang, Z. Excessive nitrogen application dampens antioxidant capacity and grain filling in wheat as revealed by metabolic and physiological analyses. Sci. Rep. 2017, 7, 43363. [Google Scholar] [CrossRef] [PubMed]
- Elhanafi, L.; Houhou, M.; Rais, C.; Mansouri, I.; Elghadraoui, L.; Greche, H. Impact of excessive nitrogen fertilization on the biochemical quality, phenolic compounds, and antioxidant power of Sesamum indicum L seeds. J. Food Qual. 2019, 2019, 9428092. [Google Scholar] [CrossRef]
- Khan, F.; Siddique, A.B.; Shabala, S.; Zhou, M.; Zhao, C. Phosphorus plays key roles in regulating plants’ physiological responses to abiotic stresses. Plants 2023, 12, 2861. [Google Scholar] [CrossRef]
- Shukla, D.; Rinehart, C.A.; Sahi, S.V. Comprehensive study of excess phosphate response reveals ethylene mediated signaling that negatively regulates plant growth and development. Sci. Rep. 2017, 7, 3074. [Google Scholar] [CrossRef]
- Pan, Y.; Song, Y.; Zhao, L.; Chen, P.; Bu, C.; Liu, P.; Zhang, D. The genetic basis of phosphorus utilization efficiency in plants provide new insight into woody perennial plants improvement. Int. J. Mol. Sci. 2022, 23, 2353. [Google Scholar] [CrossRef]
- Xu, X.; Du, X.; Wang, F.; Sha, J.; Chen, Q.; Tian, G.; Zhu, Z.; Ge, S.; Jiang, Y. Effects of potassium levels on plant growth, accumulation and distribution of carbon, and nitrate metabolism in apple dwarf rootstock seedlings. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Nieves-Cordones, M.; Al Shiblawi, F.R.; Sentenac, H. Roles and transport of sodium and potassium in plants. In The Alkali Metal Ions: Their Role for Life; Sigel, A., Sigel, H., Sigel, R.K.O., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 291–324. [Google Scholar]
- Hepler, P.K. Calcium: A central regulator of plant growth and development. Plant Cell 2005, 17, 2142–2155. [Google Scholar] [CrossRef]
- Dayod, M.; Tyerman, S.D.; Leigh, R.A.; Gilliham, M. Calcium storage in plants and the implications for calcium biofortification. Protoplasma 2010, 247, 215–231. [Google Scholar] [CrossRef]
- Hermans, C.; Verbruggen, N. Physiological characterization of Mg deficiency in Arabidopsis thaliana. J. Exp. Bot. 2005, 56, 2153–2161. [Google Scholar] [CrossRef] [PubMed]
- Franklin, W.T.; Olsen, J.S.; Soltanpour, P.N. Effects of excessive magnesium in irrigation waters on wheat and corn growth. Commun. Soil Sci. Plant Anal. 1991, 22, 49–61. [Google Scholar] [CrossRef]
- Garg, M.; Sharma, N.; Sharma, S.; Kapoor, P.; Kumar, A.; Chunduri, V.; Arora, P. Biofortified crops generated by breeding, agronomy, and transgenic approaches are improving lives of millions of people around the world. Front. Nutr. 2018, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Ofori, K.F.; Antoniello, S.; English, M.M.; Aryee, A.N.A. Improving nutrition through biofortification—A systematic review. Front. Nutr. 2022, 9. [Google Scholar] [CrossRef]
- Kartseva, T.; Alqudah, A.M.; Aleksandrov, V.; Alomari, D.Z.; Doneva, D.; Arif, M.A.R.; Börner, A.; Misheva, S. Nutritional genomic approach for improving grain protein content in wheat. Foods 2023, 12, 1399. [Google Scholar] [CrossRef]
- El-Soda, M.; Sarhan, M.S. From gene mapping to gene editing, a guide from the Arabidopsis research. Annu. Plant Rev. 2021, 4, 733–766. [Google Scholar]
- García-Barrios, G.; Crespo-Herrera, L.; Cruz-Izquierdo, S.; Vitale, P.; Sandoval-Islas, J.S.; Gerard, G.S.; Aguilar-Rincón, V.H.; Corona-Torres, T.; Crossa, J.; Pacheco-Gil, R.A. Genomic prediction from multi-environment trials of wheat breeding. Genes 2024, 15, 417. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Endelman, J.B. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 2011, 4. [Google Scholar] [CrossRef]
- Said, A.A.; MacQueen, A.H.; Shawky, H.; Reynolds, M.; Juenger, T.E.; El-Soda, M. Genome-wide association mapping of genotype-environment interactions affecting yield-related traits of spring wheat grown in three watering regimes. Environ. Exp. Bot. 2022, 194, 104740. [Google Scholar] [CrossRef]
- Ayoubi, S.; Mehnatkesh, A.; Jalalian, A.; Sahrawat, K.L.; Gheysari, M. Relationships between grain protein, Zn, Cu, Fe and Mn contents in wheat and soil and topographic attributes. Arch. Agron. Soil Sci. 2014, 60, 625–638. [Google Scholar] [CrossRef]
- El-Soda, M.; Neris Moreira, C.; Goredema-Matongera, N.; Jamar, D.; Koornneef, M.; Aarts, M.G.M. QTL and candidate genes associated with leaf anion concentrations in response to phosphate supply in Arabidopsis thaliana. BMC Plant Biol. 2019, 19, 410. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kumar, S.; Mohapatra, T. Interaction between macro- and micro-nutrients in plants. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Shepelev, S.; Morgounov, A.; Flis, P.; Koksel, H.; Li, H.; Savin, T.; Sharma, R.; Wang, J.; Shamanin, V. Variation of macro- and microelements, and trace metals in spring wheat genetic resources in Siberia. Plants 2022, 11, 149. [Google Scholar] [CrossRef]
- Balyan, H.S.; Gupta, P.K.; Kumar, S.; Dhariwal, R.; Jaiswal, V.; Tyagi, S.; Agarwal, P.; Gahlaut, V.; Kumari, S. Genetic improvement of grain protein content and other health-related constituents of wheat grain. Plant Breed. 2013, 132, 446–457. [Google Scholar] [CrossRef]
- Debaeke, P.; Aussenac, T.; Fabre, J.L.; Hilaire, A.; Pujol, B.; Thuries, L. Grain nitrogen content of winter bread wheat (Triticum aestivum L.) as related to crop management and to the previous crop. Eur. J. Agron. 1996, 5, 273–286. [Google Scholar] [CrossRef]
- Yeoh, H.-H.; Wee, Y.-C. Leaf protein contents and nitrogen-to-protein conversion factors for 90 plant species. Food Chem. 1994, 49, 245–250. [Google Scholar] [CrossRef]
- Fujihara, S.; Sasaki, H.; Aoyagi, Y.; Sugahara, T. Nitrogen-to-protein conversion factors for some cereal products in Japan. J. Food Sci. 2008, 73, C204–C209. [Google Scholar] [CrossRef]
- Yamaguchi, M. Determination of the nitrogen-to-protein conversion factor in cereals. In Seed Analysis; Linskens, H.F., Jackson, J.F., Eds.; Springer: Berlin/Heidelberg, Germany, 1992; pp. 95–107. [Google Scholar]
- Tabbita, F.; Pearce, S.; Barneix, A.J. Breeding for increased grain protein and micronutrient content in wheat: Ten years of the GPC-B1 gene. J. Cereal Sci. 2017, 73, 183–191. [Google Scholar] [CrossRef]
- Boussakouran, A.; El Yamani, M.; Sakar, E.H.; Rharrabti, Y. Trend in yield and protein content relationship in a historical series of durum wheat varieties under rainfed and irrigated environments. Crop Environ. 2024, 3, 171–176. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, J.; Li, D.; Batchelor, W.D.; Wu, D.; Zhen, X.; Ju, H. Future climate change impacts on wheat grain yield and protein in the North China Region. Sci. Total Environ. 2023, 902, 166147. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.W.; Borrill, P. Applying genomic resources to accelerate wheat biofortification. Heredity 2020, 125, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.S.; Dreisigacker, S.; Pena, R.J.; Sukumaran, S.; Reynolds, M.P. Genetic characterization of the wheat association mapping initiative (WAMI) panel for dissection of complex traits in spring wheat. TAG. Theor. Appl. Genetics. Theor. Angew. Genet. 2015, 128, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Ahirwar, R.N.; Mishra, V.K.; Chand, R.; Budhlakoti, N.; Mishra, D.C.; Kumar, S.; Singh, S.; Joshi, A.K. Genome-wide association mapping of spot blotch resistance in wheat association mapping initiative (WAMI) panel of spring wheat (Triticum aestivum L.). PLoS ONE 2018, 13, e0208196. [Google Scholar] [CrossRef]
- Alqudah, A.M.; Elkelish, A.; Abu-Elsaoud, A.M.; Hassan, S.E.-D.; Thabet, S.G. Genome-wide association study reveals the genetic basis controlling mineral accumulation in wheat grains under potassium deficiency. Genet. Resour. Crop Evol. 2024. [Google Scholar] [CrossRef]
- Shi, R.-l.; Tong, Y.-p.; Jing, R.-l.; Zhang, F.-s.; Zou, C.-q. Characterization of quantitative trait loci for grain minerals in hexaploid wheat (Triticum aestivum L.). J. Integr. Agric. 2013, 12, 1512–1521. [Google Scholar] [CrossRef]
- El-Soda, M.; Malosetti, M.; Zwaan, B.J.; Koornneef, M.; Aarts, M.G. Genotype x environment interaction QTL mapping in plants: Lessons from Arabidopsis. Trends Plant Sci. 2014, 9, 390–398. [Google Scholar] [CrossRef]
- Manickavelu, A.; Hattori, T.; Yamaoka, S.; Yoshimura, K.; Kondou, Y.; Onogi, A.; Matsui, M.; Iwata, H.; Ban, T. Genetic nature of elemental contents in wheat grains and its genomic prediction: Toward the effective use of wheat landraces from Afghanistan. PLoS ONE 2017, 12, e0169416. [Google Scholar] [CrossRef]
- Zhou, Y.; Vales, M.I.; Wang, A.; Zhang, Z. Systematic bias of correlation coefficient may explain negative accuracy of genomic prediction. Brief. Bioinform. 2016, 18, 744–753. [Google Scholar] [CrossRef]
- Misztal, I.; Lourenco, D. Potential negative effects of genomic selection. J. Anim. Sci. 2024, 102, skae155. [Google Scholar] [CrossRef]
- Adelantado, J.V.G.; Martinez, V.P.; Garcia, A.P.; Reig, F.B. Atomic-absorption spectrometric determination of calcium, magnesium and potassium in leaf samples after decomposition with molten sodium hydroxide. Talanta 1991, 38, 959–963. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Wang, L.; Rimbert, H.; Rodriguez, J.C.; Deal, K.R.; De Oliveira, R.; Choulet, F.; Keeble-Gagnère, G.; Tibbits, J.; Rogers, J.; et al. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 2021, 107, 303–314. [Google Scholar] [CrossRef]
- Hassani-Pak, K.; Singh, A.; Brandizi, M.; Hearnshaw, J.; Parsons, J.D.; Amberkar, S.; Phillips, A.L.; Doonan, J.H.; Rawlings, C. KnetMiner: A comprehensive approach for supporting evidence-based gene discovery and complex trait analysis across species. Plant Biotechnol. J. 2021, 19, 1670–1678. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).