Abstract
Cucurbitacins, oxygenated tetracyclic triterpenoids that are found mainly in the Cucurbitaceae family, play essential roles as defensive compounds, serving as allomones against herbivores and pathogens and as signals for insect–parasite recognition. These compounds also exhibit various pharmacological effects. The biosynthesis of cucurbitacins is largely regulated by the bitter (Bi) gene, encoding an oxidosqualene cyclase, which catalyzes the conversion of 2,3-oxidosqualene into cucurbitadienol, a common precursor for cucurbitacin synthesis. Previous studies focused on uncovering the Bi gene clusters in Cucurbitaceae, but their presence in other cucurbitacin-producing plants remained unexplored. Here, the evolutionary history of Bi genes and their clusters were investigated in twenty-one plant genomes spanning three families based on chemotaxonomy. Nineteen Bi genes were identified in fourteen Cucurbitaceae, four Begoniaceae, and one Aquilaria species. Phylogenetic analysis suggested that the genome of Aquilaria sinensis contained the earliest Bi gene clusters in this dataset. Moreover, the genomic analysis revealed a conserved microsynteny of pivotal genes for cucurbitacin biosynthesis in Cucurbitaceae, while interspersed Bi gene clusters were observed in Begoniaceae, indicating rearrangements during plant Bi gene cluster formation. The bitter gene in A. sinensis was found to promote cucurbitadienol biosynthesis in the leaves of Nicotiana benthamiana. This comprehensive exploration of plant Bi genes and their clusters provides valuable insights into the genetic and evolutionary underpinnings of cucurbitacin biosynthesis. These findings offer prospects for a deeper understanding of cucurbitacin production and potential genetic resources for their enhancement in various plants.
Keywords:
cucurbitacin; Aquilaria sinensis; Begonia; Bi gene; bitter gene cluster; origin; evolution 1. Introduction
Cucurbitacins (Cu) are characteristic oxygenated tetracyclic triterpenoids that are mainly present in the Cucurbitaceae family and are arbitrarily divided into 12 categories (cucurbitacins A–T), among which Cu B is ubiquitously distributed in this family of plants [1]. Being allomone in nature, cucurbitacins have a protective function in plants against the attacks of herbivores and pathogens. For example, the content of Cu C in cucumber was significantly correlated with resistance against Tetranychus urticae [2]. Cucurbitacins are also exploited as signals by insects to recognize parasites. Their contents can increase quickly once the plants are foraged by herbivores to induce an antifeedant effect [3]. Previous studies have also indicated that Cu B acts as a potential growth regulator for insects by antagonizing 20-hydroxyecdysone activity [4]. Cucurbitacins also show pharmacological effects, as they could have cytotoxic, purgative, anti-inflammatory, and anti-fertility activity [1,5].
The bitter taste of Cucumis sativus is from cucurbitacins and their biosynthesis is majorly rate-limited by the bitter (Bi) gene, which encodes a type of oxidosqualene cyclase (OSC). OSC catalyzes 2,3-oxidosqualene to generate cucurbitadienol, which acts as a common precursor for the biosynthesis of various cucurbitacins in plants (Figure 1a) [6]. The whole genome sequence of cucumber revealed a Bi gene cluster involved in cucurbitacin biosynthesis [6]. Further, nine genes involved in the biosynthesis of cucurbitacin C were identified in cucumber (Figure 1a) [7]. Subsequently, Bi gene clusters were also found in melon and watermelon. These functional genes, especially OSC (Bi), P450 encoding C25 or 2β hydroxylase, and acyltransferase (ACT), in Bi gene clusters are distributed in a collinear manner in different species of Cucurbitaceae, even showing the same order in their arrangements and direction of transcription across various species (Figure 1b) [8]. These observations suggest that Bi gene clusters might be highly conserved during the process of plant evolution. This hypothesis could not be investigated fully as the previous studies were focused on the species from Cucurbitaceae, whereas the Bi genes and their clusters in other species that produce cucurbitacins have not been investigated.
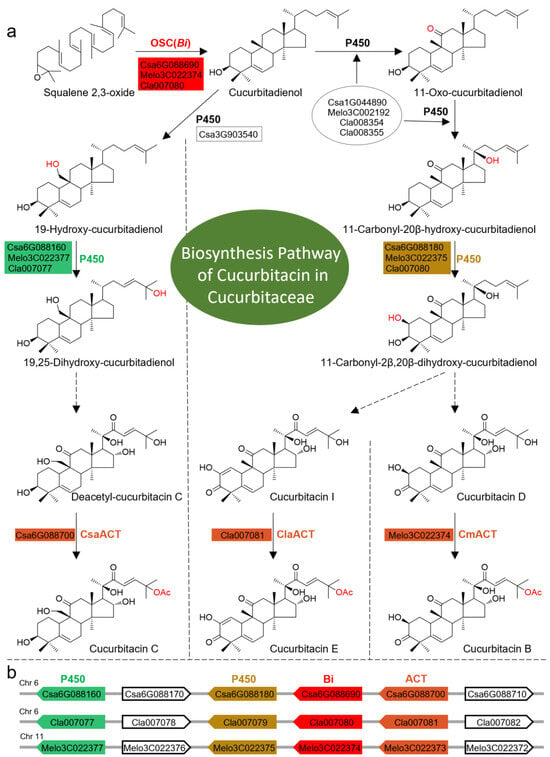
Figure 1.
The biosynthesis pathway of cucurbitacins in Cucurbitaceae and an illustration of the Bi gene cluster in the cucumber genome. (a) The biosynthesis pathway of cucurbitacins in Cucurbitaceae. The enzyme names, colored with red, green, brown, and mustard, are distributed in the Bi gene cluster and the gene IDs to the left of the enzymes are those corresponding to the IDs in the genomes of cucumber, watermelon, or melon. OSC: oxidosqualene cyclase; Bi: bitter gene; P450: cytochrome P450; ACT: acyltransferase; Csa: Cucumis sativus; Melo/Cm: Cucumis melo; Cla: Citrullus lanatus; Ac: acetyl. (b) A schematic diagram of Bi gene clusters and gene orders distributed on chr 6 in the cucumber genome, chr 6 in the watermelon genome, and chr 11 in the melon genome. The genes with the same color background are the syntenic genes in these three species from the Cucumis genus. The gray line in the background represents the chromosome and the arrow represents the transcriptomic direction of each gene. Chr: chromosome.
Plant chemotaxonomy provided the key information about the distribution of cucurbitacins, which can also be detected or extracted from the species belonging to Begoniaceae, Brassicaceae, Celastraceae, Elaeocarpaceae, Euphorbiaceae, Polemoniaceae, Rubiaceae, Scrophulariaceae, Sterculiaceae, Tetramelaceae, or Thymelaeaceae [1]. This implies that such species might also contain a Bi gene or Bi gene clusters in their genomes. Our previous study first reported that Cu D and Cu I were isolated from the fruits of Aquilaria sinensis [9]. Also, Cu E and Cu I were detected from callus, shoot, and in vitro plants of A. agallocha, and even the content of Cu E reached 1.235 mg/g in leaves after treatment with 0.5 mM methyl jasmonate [10]. These studies suggested that Aquilaria species might be an upstart natural resource for cucurbitacin production. In the last 10 years, an increasing number of plant genomes, including that of A. sinensis, have been sequenced [11]. They provide a useful resource for exploring the distribution of Bi genes and their clusters in the plant kingdom.
In this study, the Bi gene was first identified in the A. sinenesis genome with CLEAN [12], an annotation script of a machine learning algorithm. Then, plantiSMASH was employed to analyze a short scaffold containing this Bi gene belonging to the A. sinensis genome, resulting in this scaffold including a typical terpene gene cluster, which presents a characterization of the classical Bi gene clusters from the species of Cucurbitaceae. Subsequently, 20 other genomes from 2 plant families, of which 16 were from Cucurbitaceae and 4 were from Begoniaceae, based on the chemotaxonomy that they all produce cucurbitacin, were selected to present the evolution of plant Bi genes and Bi clusters together with the A. sinensis genome. The evolutionary history of these species suggested that plant Bi gene clusters may have originated from A. sinensis in this dataset. Finally, the Bi genes from the A. sinensis genome were demonstrated to promote cucurbitadienol biosynthesis. Understanding the evolutionary and genetic basis of plant Bi genes and their clusters could contribute to the biosynthetic pathway resolution of multifarious cucurbitacins thoroughly and provide crucial genetic or germplasm resources for cucurbitacin production.
2. Results and Discussion
2.1. Characterization of Plant Bitter Genes
To identify the bitter genes, a machine learning algorithm of CLEAN [12] was employed to screen the protein sequences to identify the Bi genes in 21 plant genome datasets. These species were selected from three families on the basis of their chemotaxonomy (cucurbitacin production). The results of CLEAN were verified by Blastp [13] and HmmScan [14]. Finally, a total of 19 Bi genes, annotated to code for a cucurbitadienol synthase (EC: 5.4.99.33; p-value > 0.95), were retained from 14 species of Cucurbitaceae, 4 from Begoniaceae, and 1 from Aquilaria. Generally, these genes in the Pfam database [15] synchronously contained both the domains of PF13249.9 (SQHop_cyclase_N) and PF13243.9 (SQHop_cyclase_C). In the A. sinensis genome dataset, As03G2784 was identified and annotated as EC: 5.4.99.33 (0.97) with CLEAN, and it simultaneously contained two conserved domains from Pfam. No bitter genes were identified in Benincasa hispida or Lagenaria siceraria.
To characterize the evolutionary relationships of the plant Bi genes, a maximum likelihood tree was created with IQ-tree2 by the best model of JTT+I+I+R2. Nineteen Bi genes were classified into three subfamilies, which was consistent with the plant taxonomy; the Bi genes from the Begoniaceae family were located in the root section of the phylogenetic tree (Figure 2a). But in a macroevolutionary tree of the 23 species, the origin time of Cucurbitaceae and Begoniaceae was around 37.72 million years ago (Mya) and 20.83 Mya, respectively. These are later than the origin time of A. sinensis, which is around 108.89 Mya (Figure 2b). These divergence times of the species are similar to those in previous reports [11,16,17]. As the outgroups, Amborella trichopoda and Vitis vinifera did not contain the Bi gene in their genomes.
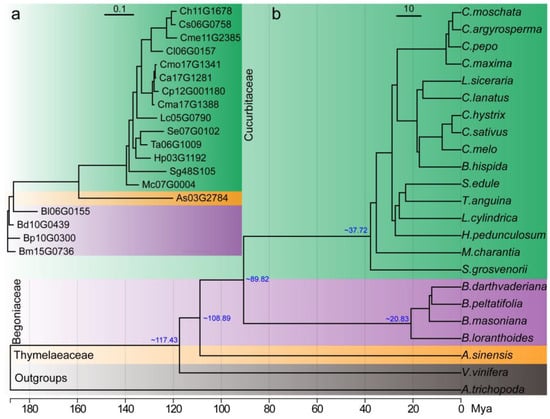
Figure 2.
Phylogenetic analysis of the plant bitter genes and their species. (a) Phylogenetic analysis of the plant bitter genes. The evolutionary trees were constructed by IQ-tree2 with the best model of JTT+I+I+R2. (b) Single copy gene tree of 21 plant species that produce cucurbitacin and two model species of Amborella trichopoda and Vitis vinifera as the outgroups. The blue numbers around the nodes are the approximate divergence times of the species. Mya: million years ago.
2.2. Identification and Microsynteny of Bitter Gene Clusters
To investigate the occurrence of a Bi gene cluster in the A. sinensis genome, the genes distributed on a single scaffold containing As03G2784 (Scaffod63), before the Hi-C anchoring, were re-analyzed. The total length of Scaffod63 was 2.15 Mb and it included 22 tactic genes (As03G2769-As03G2790), of which 15 had a homolog in a public database. Nine other genes besides As03G2784 might be associated with secondary metabolite biosynthesis or transport. As03G2774, As03G2778, As03G2780, As03G2786, and As03G2788 were classified as cytochrome P450s (P450), As03G2777 was identified as BAHD acyltransferase (ACT), and As03G2781 was annotated as an oxidoreductase (OR). As03G2770 and As03G2772 were identified as an ABC and a MATE transporter, respectively.
Then, Scaffold63 and its gff3 files were screened by plantiSMASH 1.0 and the results indicated that 1.38 Mb of Scaffold63 was a typical plant terpene biosynthesis cluster that contained 16 genes, from As03G2773 to As03G2788 (Figure 3a). The microsynteny analysis demonstrated that the six pivotal genes for cucurbitacin biosynthesis in Cucurbitaceae species showed collinear relationships with the Bi gene cluster from A. sinensis, including a Bi gene, four P450s, and an ACT (Figure 3b). In Momordica charantia, only the Bi gene presented collinearity with other species, whereas the conserved synteny gene pairs as P450s or ACT homologs were not similarly detected in this species [18]. A potential Bi gene in B. hispida was interrupted by an insertion of 864 bp non-coding sequences between Bh12G2682 (SQHop_cyclase_N) and Bh12G2683 (SQHop_cyclase_C), which would have formed two domains of the Bi protein. Now, this insertion has split the potential Bi gene in B. hispida into two separate genes that might have resulted in the Bi gene being lost in the wax gourd. The same characteristics were also found in the L. siceraria genome, which had a 3043 bp insertion of non-coding sequences between Ls09G1383 (SQHop_cyclase_N) and Ls09G1784 (SQHop_cyclase_C) (Figure 3b). The core biosynthesis clusters of cucurbitacins in the Cucurbita genus were separated into two chromosomes and lost the homolog of Cs06G0755 (Csa6G0088170) except in C. maxima, but there were two homologous genes of Cs06G0760 (Csa6G0088710) in the Cucurbita genus (Figure 3b). A syntenic pair of a MATE gene (Csa1G044870/Cs01G0775) contributing to cucurbitacin transport was also found in the nearby region of the Bi gene cluster from A. sinensis [19], although As03G2772 was not previously identified as a member of a typical plant biosynthesis gene cluster (Figure 3b).
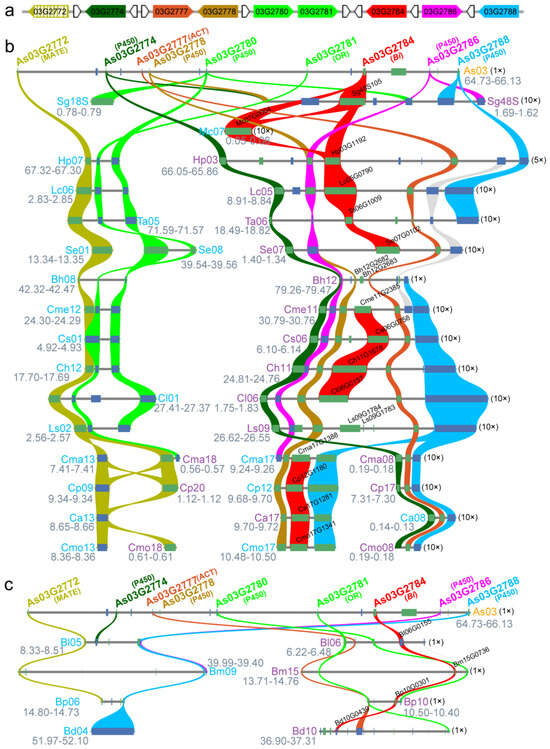
Figure 3.
A bitter gene (Bi) cluster on chr 3 of the A. sinensis genome and its microsynteny with clusters of species from Cucurbitaceae and Begoniaceae. (a) Schematic diagram of bitter gene (Bi) clusters on chr 3 of the A. sinensis genome. The gene named as As03G2772, in hatched grass yellow, is not in the typical gene cluster from the predication by plantiSMASH; the genes uncolored were annotated as uncharacterized proteins. (b) Microsynteny of bitter gene clusters between A. sinensis and the species from Cucurbitaceae. MATE: multidrug and toxic compound extrusion transporter; P450: cytochrome P450; ACT: acyltransferase; OR: oxidoreductase; Bi: bitter gene. Sg: Siraitia grosvenorii; Mc: Momordica charantia; Hp: Herpetospermum pedunculosum; Lc: Luffa cylindrica; Ta: Trichosanthes anguina; Se: Sechium edule; Bh: Benincasa hispida; Cme: Cucumis melo; Cs: Cucumis sativus; Ch: Cucumis hystrix; Cl: Citrullus lanatus; Ls: Lagenaria siceraria; Cma: Cucurbita maxima; Cp: Cucurbita pepo; Ca: Cucurbita argyrosperma; Cmo: Cucurbita moschata. (c) Microsynteny of bitter gene clusters between A. sinensis and the species from Begoniaceae. Bl: Begonia loranthoides; Bm: B. masoniana; Bp: B. peltatifolia; Bd: B. darthvaderiana. The number in the parentheses is the magnification for gene cluster visualization.
We inferred that a total of eight syntenic genes are involved in cucurbitacin biosynthesis and transport, which are present on Scaffold63 of the A. sinensis genome, except for As03G2772 (Figure 3a). It is interesting that a P450 gene (Csa1G044890/Cs01G0777), involved in cucurbitacin C biosynthesis [7], was found around this MATE gene in the cucumber genome. But the collinear pair of Csa1G044890/Cs01G0777, a syntenic gene of As03G2780, could not be detected around a MATE transporter in the four species of Cucurbita (Figure 3b).
Unlike the compact Bi gene clusters in Cucurbitaceae, the Bi gene clusters in Begoniaceae were interspersed, especially for the collinear gene pairs of six key biosynthesis genes for cucurbitacin C biosynthesis from cucumber. The syntenic pairs of two key P450s named Cs06G0759 (Csa6G0088700) and Cs06G0760 (Csa6G088710) were not distributed around the Bi genes in Begonia species, but were near a MATE transporter gene from another chromosome (Figure 3c). All four Begonia species do not contain a synteny pair of Cs06G0754 (Csa6G088160). In addition, the collinear pairs of P450 named Cs06G0759 (Csa6G0088700) were also lost in B. peltatifolia and B. darthvaderiana (Figure 3c). These results suggested that rearrangements by crossover or insertion between different chromosomes and gene loss have occurred during plant Bi gene cluster formation in different species.
2.3. Bitter Gene in A. sinensis Promotes Cucurbitadienol Biosynthesis in the Leaves of N. benthamiana
To explore the function of the Bi gene in A. sinensis, the full-length ORF of As03G2784 was cloned and inserted into the pCambia1300-35S vector to generate the overexpression plasmid of pCambia1300-35S-As03G2784. Then, a strain of Agrobacterium tumefaciens containing pCambia1300-35S-As03G2784 was infiltrated into the leaves of Nicotiana benthamiana for transient expression. One new peak at 52.655 min was detected in re-suspension liquid from an extract of the transfected N. benthamiana leaves (Figure 4a) by using the gas chromatography–mass spectrometry (GC-MS) system of Agilent 7820A-5977E compared with another peak named peak 2 at 52.899 min in all treatments except the cucurbitadienol standard (Figure 4 and Figure S1). The retention time of this new peak was nearer the retention time of 52.774 min of the cucurbitadienol standard (Figure 4b). Furthermore, the MS spectrum of peak 1 at 52.655 min from the transfection treatment (Figure 4a) was similar to the MS spectrum of peak 1 at 52.774 min of the cucurbitadienol standard (Figure 4b), and both presented the same electron ionization (EI) spectrum (m/z 426 [M]+, 411 [M−CH3]+, 393 [M−CH3−H2O]+, 109 [C8H13]+, 69 [C5H9]+, etc.) (Figure 4a,b). Peak 2 at 52.899 min in all treatments except the cucurbitadienol standard also presented the same electron ionization (Figure 4 and Figures S1–S5). In contrast, the characteristic fragment ions of the cucurbitadienol standard were not found in the spectra of two control treatments or in peak 2 (Figure 4c and Figures S1–S5). This result indicated that peak 1 at 52.655 min in Figure 1a might represent the compound cucurbitadienol and suggested that the Bi gene in A. sinensis could also promote cucurbitadienol biosynthesis.
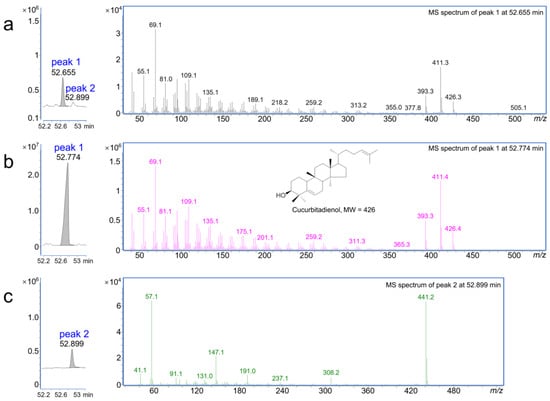
Figure 4.
GC-MS determined the function of As03G2784 by transient expression in the leaves of Nicotiana benthamiana. (a) MS chromatograms (EIC) of N. benthamiana transiently expressing As03G2784 (Bi) (left) and the EI–MS spectrum of peak 1 (right). (b) MS chromatogram (EIC) of the cucurbitadienol standard (left) and its EI–MS spectrum (right). (c) MS chromatogram of N. benthamiana transiently transfected with empty vector only (left) and the EI–MS spectrum of peak 2 (right).
3. Materials and Methods
3.1. Datasets and Plant Materials
The genome files of Amborella trichopoda and Vitis vinifera were collected from JGI [20]. The genome files of four Begonia species (B. loranthoides, B. masoniana, B. peltarifolia, and B. peltarifolia) were downloaded from CNGdb (https://db.cngb.org/, accessed on 21 August 2023), and the genome files of 16 gourd species were selected from CuGenDB [21], whereas the genome file of A. sinensis was from our previous study [11]. All genes were renamed by their order in pseudo chromosomes or scaffolds based on the bed format.
The functional gene transformation was performed on tobacco (Nicotiana benthamiana) plants that were grown in a growth chamber for 40 days. The plants were grown in pots filled with a soil and vermiculite mixture (2:1) under a photoperiod cycle of 16 h lightness and 8 h darkness at 25 °C and 70% relative humidity [17]. The seeds of N. benthamiana were obtained from the laboratory of Dr. Yuan Yao in our institute.
3.2. Identification and Evolution of Bitter Genes and Gene Clusters
A machine learning model, CLEAN (Contrastive Learning-enabled Enzyme Annotation) [12], was used to capture molecular features of proteins from amino acid sequences with the default parameters from 23 species at the genome-wide level. The target bitter genes were simultaneously annotated to two enzyme commission (EC) categories of EC: 5.4.99.8 and EC: 5.4.99.33, which were respectively classified as cycloartenol synthase and cucurbitadienol synthase in the EC number database. These sequences are presented in Table S1. Then, IQ-tree2 was employed to construct a phylogenetic tree of all bitter genes identified above with the following parameters: -m MFP -B 1000 --bnni -T AUTO [22]. The collinearity pairs of Bi and its nearby genes between cucumber and other species were also detected and visualized by JCVI v 0.8.4 on the basis of the species phylogenetic tree, which was initially constructed with 77 single copy genes from 23 species by using OrthoFinder v 2.5.4. The divergence time in each node was calibrated by r8s v 1.7.0 with the divergence time of A. trichopoda and V. vinifera at around 188 million years ago [23,24]. A. trichopoda and V. vinifera were set as the outgroup. The gene clusters were verified with plantiSMASH v 1.0 by using the default parameters [25].
3.3. Transient Transfection of Bi Gene of A. sinensis in N. benthamiana Leaves
The open reading frame (ORF) of As03G2784 was amplified and then inserted into the binary vector of pCambia1300-35S, resulting in a plasmid named pCambia1300-35S-As03G2784. A positive colony was inoculated in LB media supplemented with kanamycin (50 ng/µL) and rifampicin (30 ng/µL), and then cultured at 28 °C overnight after the plasmid pCambia1300-35S-As03G2784 was transformed into the strain EHA105 of Agrobacterium tumefaciens. The culture was incubated at 28 °C for 3 h until the OD600 of the culture reached 0.5, using induction media containing MgCl2 (10 mmol/L), MES (10 mmol/L), and acetosyringone (150 mmol/L) [26].
Subsequently, the culture was injected into 5-week-old N. benthamiana leaves. The Agrobacterium-transformed leaves of N. benthamiana were harvested after 5 days. Nine leaves of N. benthamiana from the control and treatments were harvested, with fresh weights ranging from 7.12 g to 7.45 g. These leaves were crushed into fine powder in liquid nitrogen. The powder was extracted with 30 mL of methanol for 5 min under vigorous vortexing and was then subjected to 30 min of ultrasonic extraction. This extraction procedure was repeated three times. Finally, the subsequent extraction was carried out in a chamber with analytical-grade methanol.
Chlorophyll was removed using a cartridge of solid phase extract and the re-suspension liquid of the metabolites from the extraction was analyzed by using gas chromatography–mass spectrometry (GC-MS, Agilent 7820A/5977E, Santa Clara, CA, USA). Separation of the samples by gas chromatography was carried out using an HP-5MS 5% phenyl methyl siloxane column (30 m × 0.25 mm × 0.25 µm) (Phenomenex, Torrence, CA, USA). The parameters were set as the following: injection volume, 1.0 µL; the front inlet temperature, 250 °C; no split ratio; the flow rate of the helium (carrier gas), 1.0 mL/min; the interface temperature, 280 °C; ionization of the compounds, electron impact (EI); emission current, 70 eV; the ion source temperature, 230 °C. The oven program commenced at 50 °C and increased to 310 °C at a rate of 5 °C/min, then held for 10 min. The spectra were obtained over the mass range of m/z 30 to 550 and the relative contents of the compounds were determined by normalization [27]. The plasmid containing no coding sequence (the empty vector) was also similarly transfected into leaves of N. benthamiana and served as one control check. Further, the extract from healthy leaves of N. benthamiana was also set as another control check. Due to the high similarity of the MS chromatograms (EIC) between these two control checks, only the EIC of the treatment with the empty vector is shown in the main text (Figure 4c) and the EIC of the extract from heathy N. benthamiana leaves is presented in the Supplementary Materials (Figure S1). Each treatment contained three biological replicates.
4. Conclusions
In summary, this study provided valuable insights into the evolution and distribution of bitter (Bi) genes and their clusters in cucurbitacin-producing plants. The identification of 19 Bi genes in various species and a macroevolutionary tree of 23 species indicated that Aquilaria sinensis is the earliest origin of plant Bi gene clusters among them, which sheds light on the genetic basis of cucurbitacin biosynthesis. Additionally, the functional validation of the Bi gene in A. sinensis further supports its role in promoting cucurbitadienol biosynthesis. The distinct arrangements of Bi gene clusters in Cucurbitaceae, Begoniaceae, and A. sinensis suggest complex evolutionary dynamics in plant Bi gene cluster formation. These findings shed light on the genetic basis and evolutionary dynamics of plant Bi gene clusters and contribute to understanding cucurbitacin production, even offering genetic resources for potential enhancements in various plant species.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants13020260/s1, Table S1: The protein sequences of Bi genes used in this study; Figure S1: The raw images of MS chromatograms (EIC) of N. benthamiana transiently transfected with As03G2784; Figure S2: The raw images of MS chromatograms (EIC) of N. benthamiana transiently transfected with As03G2784 in Figure 4a and Figure S1a; Figure S3: The raw images of MS chromatograms (EIC) of the cucurbitadienol standard in Figure 4b and Figure S1b; Figure S4: MS chromatogram of N. benthamiana transiently transfected with empty vector only in Figure 4c and Figure S1c; Figure S5: MS chromatogram of the extract from the healthy leaves of N. benthamiana in Figure S1d.
Author Contributions
Conceptualization, X.D. and W.M.; Methodology and Software, X.D. and Z.Y.; Writing—original draft, X.D.; Writing—review and editing, H.W. and J.Z.; Supervision, H.D. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (32171824 and 31870668) and the Earmarked Fund for CARS–Chinese Materia Medica (CARS-21).
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Chen, J.; Chiu, M.; Nie, R.; Cordell, G.; Qiu, S. Cucurbitacins and cucurbitane glycosides: Structures and biological activities. Nat. Prod. Rep. 2005, 22, 386–399. [Google Scholar] [CrossRef] [PubMed]
- Balkema-Boomstra, A.G.; Zijlstra, S.; Verstappen, F.W.A.; Inggamer, H.; Mercke, P.E.; Jongsma, M.A.; Bouwmeester, H.J. Role of cucurbitacin C in resistance to spider mite (Tetranychus urticae) in cucumber (Cucumis sativus L.). J. Chem. Ecol. 2023, 29, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Liang, W.; Yang, X.; Zhang, M.; Hu, F. Mechanism of Aulacophora femoralis chinensis Weise feeding behavior and chemical response of host Cucumis sativus L. Chin. Sci. Bull. 2004, 49, 1485–1489. [Google Scholar] [CrossRef]
- Zou, C.; Liu, G.; Liu, S.; Liu, S.; Song, Q.; Wang, J.; Feng, Q.; Su, Y.; Li, S. Cucurbitacin B acts a potential insect growth regulator by antagonizing 20-hydroxyecdysone activity. Pest Manag. Sci. 2018, 74, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Singha, S.; Kar, A.; Chanda, J.; Banerjee, S.; Dasgupta, B.; Haldar, P.; Sharma, N. Therapeutic importance of Cucurbitaceae: A medicinally important family. J. Ethnopharmacol. 2022, 282, 114599. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.I.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Ma, Y.; Zhou, Y.; Zhang, H.; Duan, L.; Chen, H.; Zeng, J.; Zhou, Q.; Wang, S.; Gu, W.; et al. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 2014, 346, 1084–1088. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, Y.; Zeng, J.; Duan, L.; Xue, X.; Wang, H.; Lin, T.; Liu, Z.; Zeng, K.; Zhong, Y.; et al. Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nat. Plants 2016, 2, 16183. [Google Scholar] [CrossRef] [PubMed]
- Mei, W.; Lin, F.; Zuo, W.; Wang, H.; Dai, H. Cucurbitacins from fruits of Aquilaria sinensis. Chin. J. Nat. Med. 2012, 10, 234–237. [Google Scholar] [CrossRef]
- Chen, C.H.; Kuo, T.C.Y.; Yang, M.H.; Chien, T.Y.; Chu, M.J.; Huang, L.C.; Lo, H.F.; Jeng, S.T.; Chen, L.F.O. Identification of cucurbitacins and assembly of a draft genome for Aquilaria agallocha. BMC Genom. 2014, 15, 578. [Google Scholar] [CrossRef][Green Version]
- Ding, X.; Mei, W.; Lin, Q.; Wang, H.; Wang, J.; Peng, S.; Li, H.; Zhu, J.; Li, W.; Wang, P.; et al. Genome sequence of the agarwood tree Aquilaria sinensis (Lour.) Spreng: The first chromosome-level draft genome in the Thymelaeceae family. GigaScience 2020, 9, giaa013. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Cui, H.; Li, J.; Luo, Y.; Jiang, G.; Zhao, H. Enzyme function prediction using contrastive learning. Science 2023, 379, 1358–1363. [Google Scholar] [CrossRef]
- Mahram, A.; Herbordt, M.C. NCBI BLASTP on high-performance reconfigurable computing systems. ACM T. Reconfig. Technol. 2015, 7, 1–20. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Li, L.; Chen, X.; Fang, D.; Dong, S.; Guo, X.; Li, N.; Campos-Dominguez, L.; Wang, W.; Liu, Y.; Lang, X.; et al. Genomes shed light on the evolution of Begonia, a mega-diverse genus. N. Phytol. 2022, 234, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ding, X.; Wang, H.; Chen, H.; Dong, W.; Zhu, J.; Wang, J.; Peng, S.; Dai, H.; Mei, W. Systematic evolution of bZIP transcription factors in Malvales and functional exploration of AsbZIP14 and AsbZIP41 in Aquilaria sinensis. Front. Plant Sci. 2023, 14, 1243323. [Google Scholar] [CrossRef]
- Cui, J.; Yang, Y.; Luo, S.; Wang, L.; Huang, R.; Wen, Q.; Han, X.; Miao, N.; Cheng, J.; Liu, Z.; et al. Whole-genome sequencing provides insights into the genetic diversity and domestication of bitter gourd (Momordica spp.). Hortic. Res. 2020, 7, 85. [Google Scholar] [CrossRef]
- Ma, Y.; Li, D.; Zhong, Y.; Zhong, Y.; Wang, X.; Li, L.; Osbourn, A.; Lucas, W.J.; Huang, S.; Shang, Y. Vacuolar MATE/DTX protein-mediated cucurbitacin C transport is co-regulated with bitterness biosynthesis in cucumber. N. Phytol. 2023, 238, 995–1003. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Yu, J.; Wu, S.; Sun, H.; Wang, X.; Tang, X.; Guo, S.; Zhang, Z.; Huang, S.; Xu, Y.; Weng, Y.; et al. CuGenDBv2: An updated database for cucurbit genomics. Nucleic Acids Res. 2023, 51, D1457–D1464. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Yang, Z.; Mei, W.; Wang, H.; Zeng, J.; Dai, H.; Ding, X. Comprehensive Analysis of NAC Transcription Factors Reveals Their Evolution in Malvales and Functional Characterization of AsNAC019 and AsNAC098 in Aquilaria sinensis. Int. J. Mol. Sci. 2023, 24, 17384. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef] [PubMed]
- Kautsar, S.A.; Suarez Duran, H.G.; Blin, K.; Osbourn, A.; Medema, M.H. plantiSMASH: Automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. 2017, 45, W55–W63. [Google Scholar] [CrossRef]
- Wang, X.H.; Gao, B.W.; Nakashima, Y.; Mori, T.; Zhang, Z.X.; Kodama, T.; Lee, Y.-E.; Zhang, Z.-K.; Wong, C.-P.; Liu, Q.-Q.; et al. Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl) chromones in agarwood. Nat. Commun. 2020, 13, 348. [Google Scholar] [CrossRef]
- Yu, Z.; Dong, W.; Wang, Y.; Li, W.; Guo, Z.; Mei, W.; Dai, H. Identification of aroma-active components from cultivated agarwood ‘Qi-Nan’ based on GC-O-MS combined with aroma extract dilution analysis. Flavour Fragr. J. 2023, 38, 392–403. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).