Twin Embryos in Arabidopsis thaliana KATANIN 1 Mutants
Abstract
1. Introduction
2. Results
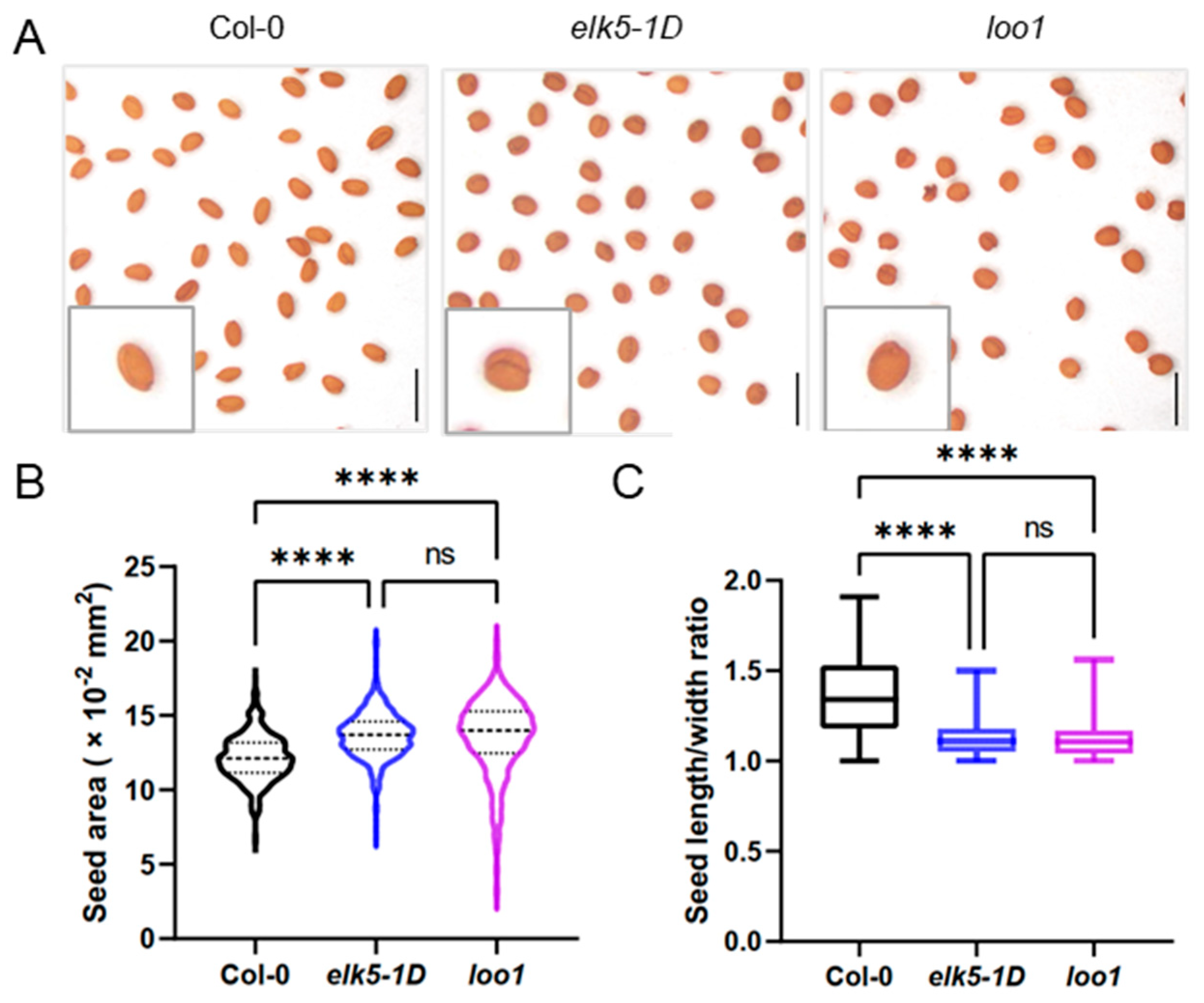
2.1. Identification and Phenotypic Characterization of Two Round Seed Mutants
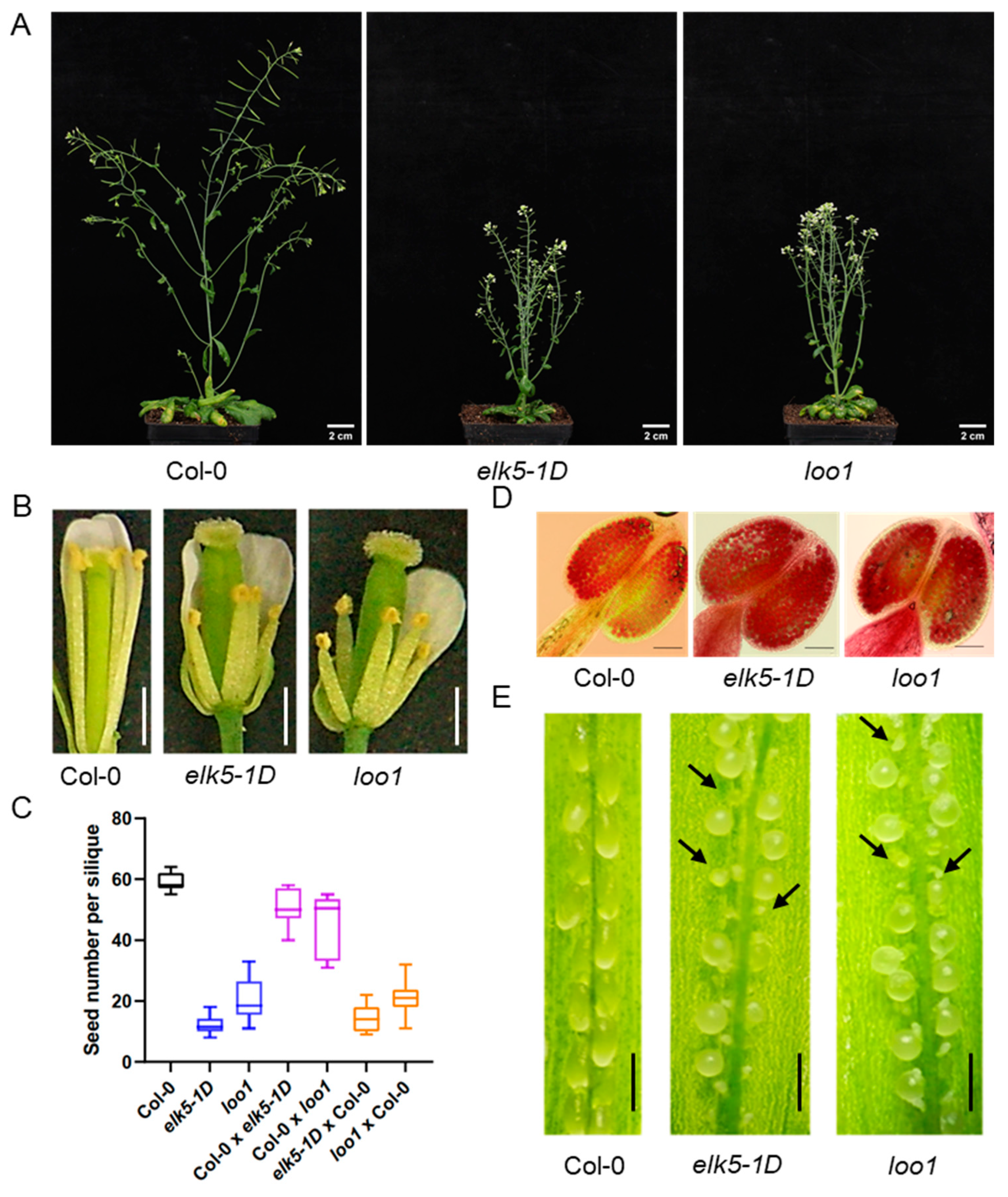
2.2. Decline in Fertility in elk5-1D and loo1 Mutants
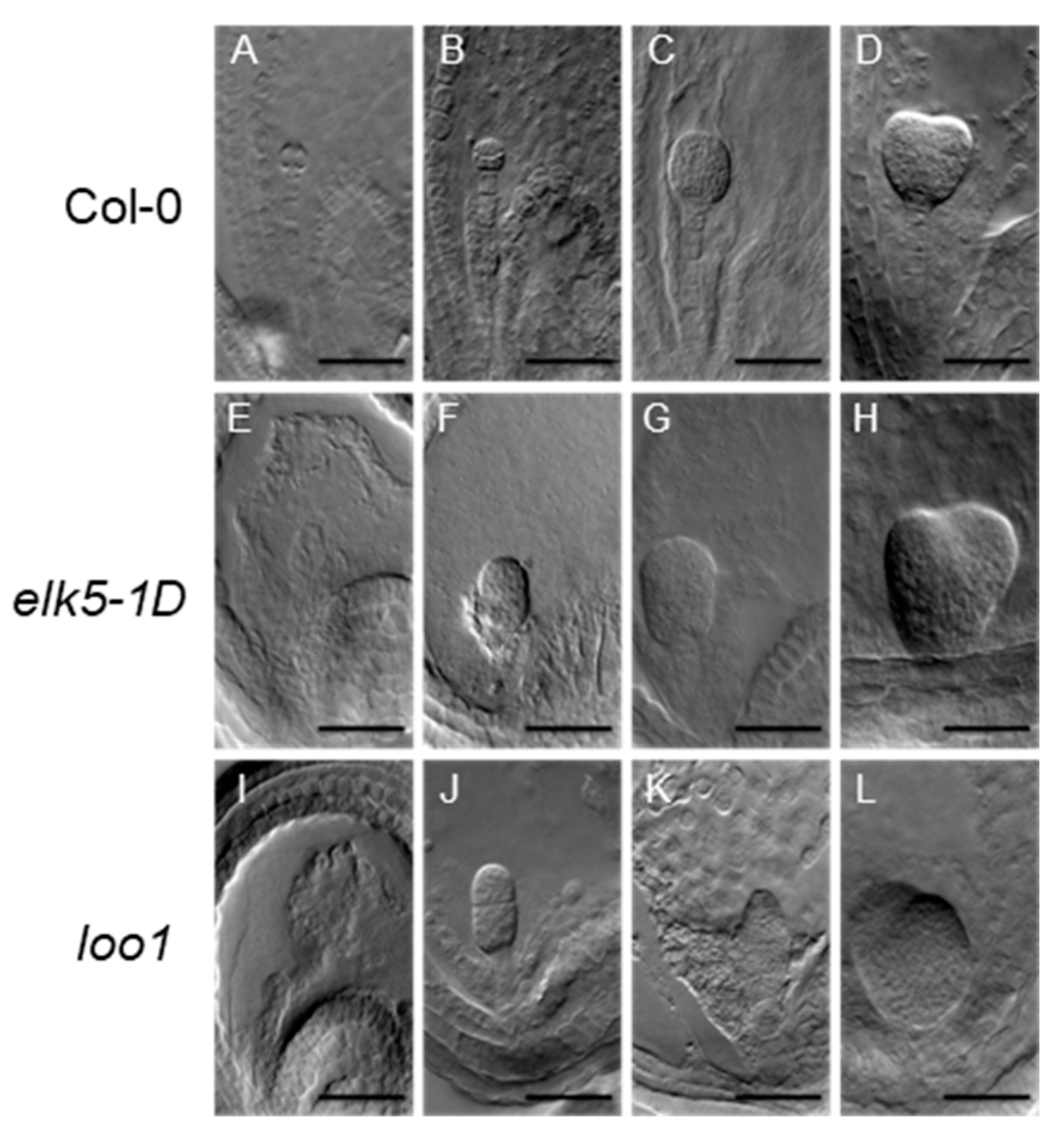
2.3. Phenotypic Alterations in the Morphology of elk5-1D and loo1 Mutants
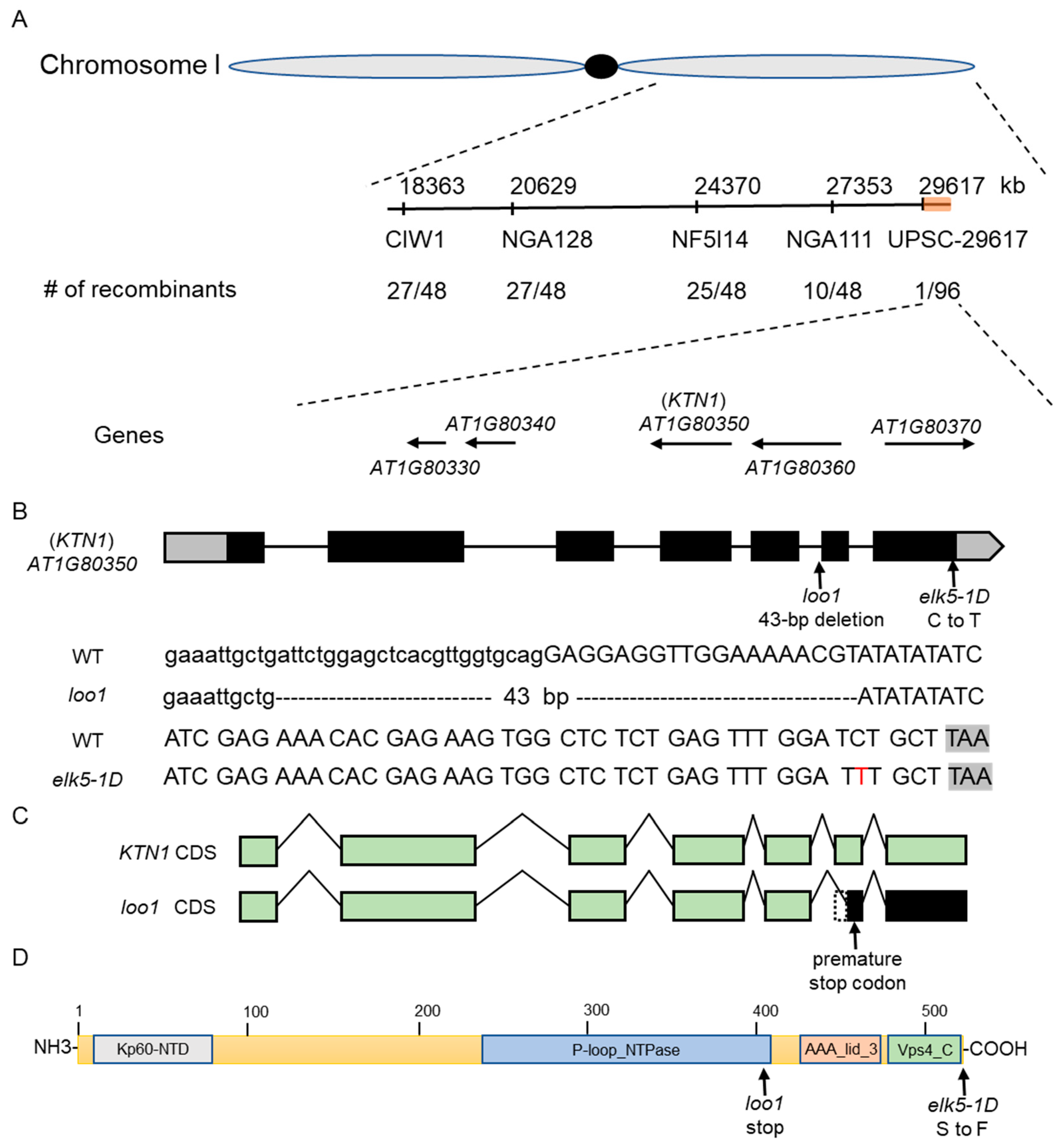
2.4. Positional Cloning of the elk5-1D and loo1 Gene
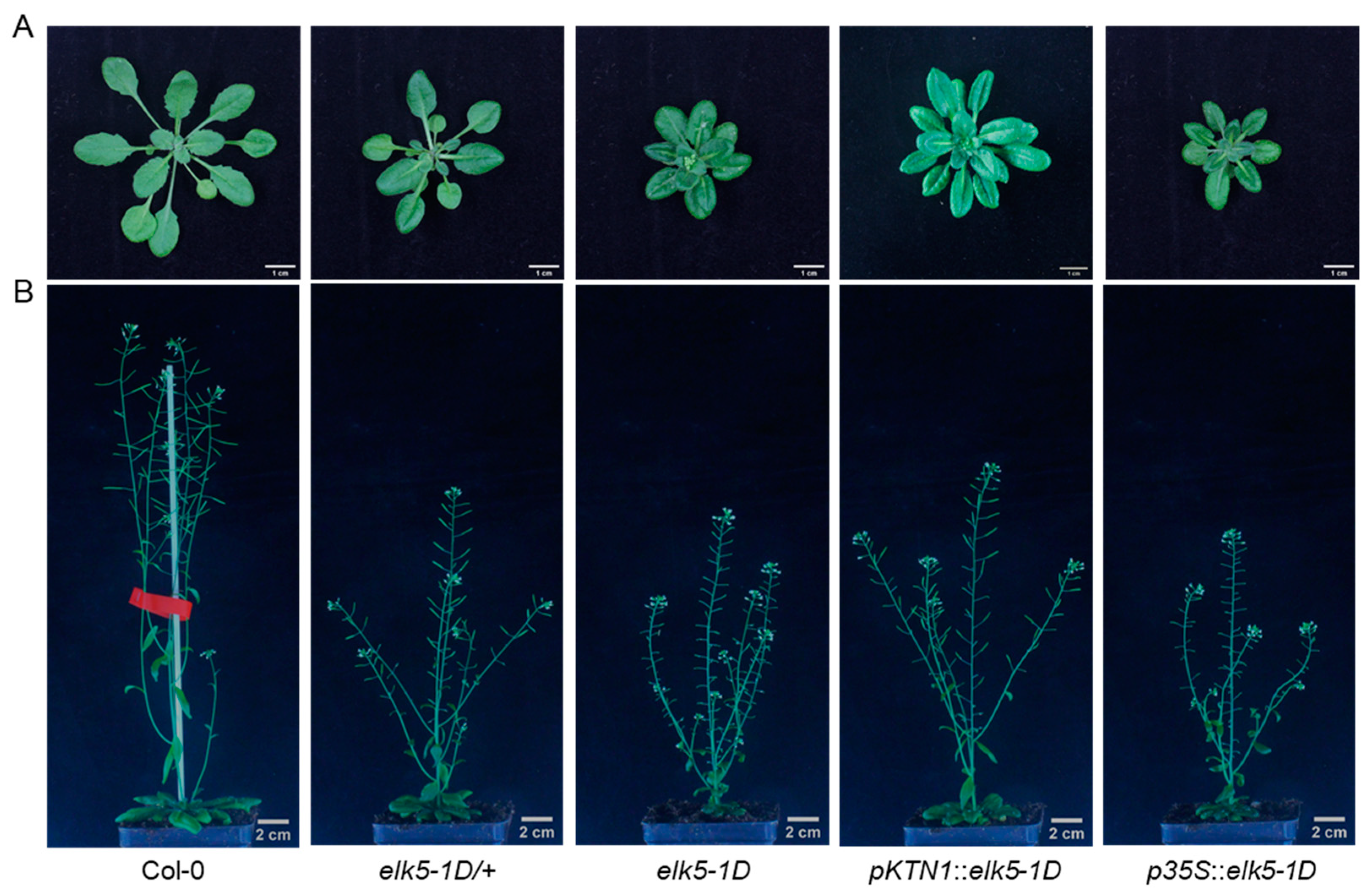
2.5. elk5-1D Is a Dominant Mutant
2.6. Aberrant Embryo Development and Twin Embryos in ktn1 Mutants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Alexander Staining
4.3. Phenotyping of the Mutant Lines
4.4. Genetic Mapping
4.5. Plasmid Construction and Generation of Transgenic Plant Generation
4.6. Reverse-Transcription PCR (RT-PCR) Analysis
4.7. Light Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sprunck, S. Twice the fun, double the trouble: Gamete interactions in flowering plants. Curr. Opin. Plant Biol. 2020, 53, 106–116. [Google Scholar] [CrossRef]
- Ramalho, J.J.; Jones, V.A.S.; Mutte, S.; Weijers, D. Pole position: How plant cells polarize along the axes. Plant Cell 2022, 34, 174–192. [Google Scholar] [CrossRef]
- Ueda, M.; Zhang, Z.; Laux, T. Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev. Cell 2011, 20, 264–270. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, T.; Dong, J. Asymmetric cell division in plant development. J. Integr. Plant Biol. 2023, 65, 343–370. [Google Scholar] [CrossRef]
- Dante, R.A.; Larkins, B.A.; Sabelli, P.A. Cell cycle control and seed development. Front. Plant Sci 2014, 5, 493. [Google Scholar] [CrossRef]
- Rasmussen, C.G.; Humphries, J.A.; Smith, L.G. Determination of Symmetric and Asymmetric Division Planes in Plant Cells. Annu. Rev. Plant Biol. 2011, 62, 387–409. [Google Scholar] [CrossRef]
- Van Damme, D.; Vanstraelen, M.; Geelen, D. Cortical division zone establishment in plant cells. Trends Plant Sci. 2007, 12, 458–464. [Google Scholar] [CrossRef]
- Forth, S.; Kapoor, T.M. The mechanics of microtubule networks in cell division. J. Cell Biol. 2017, 216, 1525–1531. [Google Scholar] [CrossRef]
- Nicklas, R.B. Measurements of the force produced by the mitotic spindle in anaphase. J. Cell Biol. 1983, 97, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Hamada, T. Microtubule organization and microtubule-associated proteins in plant cells. Int. Rev. Cell Mol. Biol. 2014, 312, 1–52. [Google Scholar] [CrossRef] [PubMed]
- Paredez, A.R.; Somerville, C.R.; Ehrhardt, D.W. Visualization of cellulose synthase demonstrates functional association with microtubules. Science 2006, 312, 1491–1495. [Google Scholar] [CrossRef]
- Lei, L.; Singh, A.; Bashline, L.; Li, S.; Yingling, Y.G.; Gu, Y. Cellulose Synthase Interactive1 Is Required for Fast Recycling of Cellulose Synthase Complexes to the Plasma Membrane in Arabidopsis. Plant Cell 2015, 27, 2926–2940. [Google Scholar] [CrossRef]
- Bouquin, T.; Mattsson, O.; Naested, H.; Foster, R.; Mundy, J. The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J. Cell Sci. 2003, 116, 791–801. [Google Scholar] [CrossRef]
- McNally, F.J.; Vale, R.D. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 1993, 75, 419–429. [Google Scholar] [CrossRef]
- Roll-Mecak, A.; Vale, R.D. The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr. Biol. 2005, 15, 650–655. [Google Scholar] [CrossRef]
- Zhang, D.; Rogers, G.C.; Buster, D.W.; Sharp, D.J. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J. Cell Biol. 2007, 177, 231–242. [Google Scholar] [CrossRef]
- Frickey, T.; Lupas, A.N. Phylogenetic analysis of AAA proteins. J. Struct. Biol. 2004, 146, 2–10. [Google Scholar] [CrossRef]
- McClinton, R.S.; Chandler, J.S.; Callis, J. cDNA isolation, characterization, and protein intracellular localization of a katanin-like p60 subunit from Arabidopsis thaliana. Protoplasma 2001, 216, 181–190. [Google Scholar] [CrossRef]
- Burk, D.H.; Liu, B.; Zhong, R.; Morrison, W.H.; Ye, Z.H. A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 2001, 13, 807–827. [Google Scholar] [CrossRef] [PubMed]
- McNally, K.P.; Bazirgan, O.A.; McNally, F.J. Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. J. Cell Sci. 2000, 113 Pt 9, 1623–1633. [Google Scholar] [CrossRef] [PubMed]
- Hartman, J.J.; Vale, R.D. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science 1999, 286, 782–785. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, W.; Wang, G.; Li, J.; Dong, L.; Han, L.; Wang, Q.; Tian, J.; Yu, Y.; Gao, C.; et al. KTN80 confers precision to microtubule severing by specific targeting of katanin complexes in plant cells. EMBO J. 2017, 36, 3435–3447. [Google Scholar] [CrossRef] [PubMed]
- Webb, M.; Jouannic, S.; Foreman, J.; Linstead, P.; Dolan, L. Cell specification in the Arabidopsis root epidermis requires the activity of ECTOPIC ROOT HAIR 3--a katanin-p60 protein. Development 2002, 129, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Stoppin-Mellet, V.; Gaillard, J.; Vantard, M. Functional evidence for in vitro microtubule severing by the plant katanin homologue. Biochem. J. 2002, 365, 337–342. [Google Scholar] [CrossRef]
- Nakamura, M.; Ehrhardt, D.W.; Hashimoto, T. Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 2010, 12, 1064–1070. [Google Scholar] [CrossRef] [PubMed]
- Wightman, R.; Turner, S.R. Severing at sites of microtubule crossover contributes to microtubule alignment in cortical arrays. Plant J. 2007, 52, 742–751. [Google Scholar] [CrossRef]
- Zhang, Q.; Fishel, E.; Bertroche, T.; Dixit, R. Microtubule severing at crossover sites by katanin generates ordered cortical microtubule arrays in Arabidopsis. Curr. Biol. 2013, 23, 2191–2195. [Google Scholar] [CrossRef]
- Bichet, A.; Desnos, T.; Turner, S.; Grandjean, O.; Höfte, H. BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 2001, 25, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Burk, D.H.; Ye, Z.H. Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule-severing protein. Plant Cell 2002, 14, 2145–2160. [Google Scholar] [CrossRef]
- Luptovciak, I.; Samakovli, D.; Komis, G.; Samaj, J. KATANIN 1 Is Essential for Embryogenesis and Seed Formation in Arabidopsis. Front. Plant Sci. 2017, 8, 728. [Google Scholar] [CrossRef]
- Hernandez-Lagana, E.; Mosca, G.; Mendocilla-Sato, E.; Pires, N.; Frey, A.; Giraldo-Fonseca, A.; Michaud, C.; Grossniklaus, U.; Hamant, O.; Godin, C.; et al. Organ geometry channels reproductive cell fate in the Arabidopsis ovule primordium. Elife 2021, 10, e66031. [Google Scholar] [CrossRef] [PubMed]
- Gudimchuk, N.B.; McIntosh, J.R. Regulation of microtubule dynamics, mechanics and function through the growing tip. Nat. Rev. Mol. Cell Biol. 2021, 22, 777–795. [Google Scholar] [CrossRef] [PubMed]
- Tang, E.I.; Lee, W.M.; Cheng, C.Y. Coordination of Actin- and Microtubule-Based Cytoskeletons Supports Transport of Spermatids and Residual Bodies/Phagosomes During Spermatogenesis in the Rat Testis. Endocrinology 2016, 157, 1644–1659. [Google Scholar] [CrossRef]
- Joly, N.; Martino, L.; Gigant, E.; Dumont, J.; Pintard, L. Microtubule-severing activity of the AAA+ ATPase Katanin is essential for female meiotic spindle assembly. Development 2016, 143, 3604–3614. [Google Scholar] [CrossRef]
- O’Donnell, L.; Rhodes, D.; Smith, S.J.; Merriner, D.J.; Clark, B.J.; Borg, C.; Whittle, B.; O’Connor, A.E.; Smith, L.B.; McNally, F.J.; et al. An essential role for katanin p80 and microtubule severing in male gamete production. PLoS Genet. 2012, 8, e1002698. [Google Scholar] [CrossRef]
- Schmidt, A.; Schmid, M.W.; Grossniklaus, U. Plant germline formation: Common concepts and developmental flexibility in sexual and asexual reproduction. Development 2015, 142, 229–241. [Google Scholar] [CrossRef]
- Yang, W.-C.; Shi, D.-Q.; Chen, Y.-H. Female gametophyte development in flowering plants. Annu. Rev. Plant Biol. 2010, 61, 89–108. [Google Scholar] [CrossRef]
- Drews, G.N.; Koltunow, A.M.G. The female gametophyte. Arab. Book 2011, 9, e0155. [Google Scholar] [CrossRef]
- Li, J.; Berger, F. Endosperm: Food for humankind and fodder for scientific discoveries. New Phytol. 2012, 195, 290–305. [Google Scholar] [CrossRef]
- Pagnussat, G.C.; Yu, H.-J.; Sundaresan, V. Cell-Fate Switch of Synergid to Egg Cell in Arabidopsis eostre Mutant Embryo Sacs Arises from Misexpression of the BEL1-Like Homeodomain GeneBLH1. Plant Cell 2007, 19, 3578–3592. [Google Scholar] [CrossRef] [PubMed]
- Zehr, E.; Szyk, A.; Piszczek, G.; Szczesna, E.; Zuo, X.; Roll-Mecak, A. Katanin spiral and ring structures shed light on power stroke for microtubule severing. Nat. Struct. Mol. Biol. 2017, 24, 717–725. [Google Scholar] [CrossRef]
- Mains, P.E.; Kemphues, K.J.; Sprunger, S.A.; Sulston, I.A.; Wood, W.B. Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics 1990, 126, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Clark-Maguire, S.; Mains, P.E. mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases. Genetics 1994, 136, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Mergner, J.; Frejno, M.; List, M.; Papacek, M.; Chen, X.; Chaudhary, A.; Samaras, P.; Richter, S.; Shikata, H.; Messerer, M.; et al. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 2020, 579, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Gao, X.; Cao, J.; Liu, Z.; Jin, C.; Wen, L.; Yao, X.; Ren, J. A summary of computational resources for protein phosphorylation. Curr. Protein Peptide Sci. 2010, 11, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, X.; Pan, L.; Xie, F.; Dai, B.; Sun, M.; Peng, X. Plant egg cell fate determination depends on its exact position in female gametophyte. Proc. Natl. Acad. Sci. USA 2021, 118, e2017488118. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, X.; Zhao, J.; Tang, X.; Tian, S.; Chen, J.; Shi, C.; Wang, W.; Zhang, L.; Feng, X.; et al. Direct evidence that suspensor cells have embryogenic potential that is suppressed by the embryo proper during normal embryogenesis. Proc. Natl. Acad. Sci. USA 2015, 112, 12432–12437. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D.; Ahn, J.H.; Blázquez, M.A.; Borevitz, J.O.; Christensen, S.K.; Fankhauser, C.; Ferrándiz, C.; Kardailsky, I.; Malancharuvil, E.J.; Neff, M.M.; et al. Activation tagging in Arabidopsis. Plant Physiol. 2000, 122, 1003–1013. [Google Scholar] [CrossRef]
- Cao, L.; Wang, S.; Venglat, P.; Zhao, L.; Cheng, Y.; Ye, S.; Qin, Y.; Datla, R.; Zhou, Y.; Wang, H. Arabidopsis ICK/KRP cyclin-dependent kinase inhibitors function to ensure the formation of one megaspore mother cell and one functional megaspore per ovule. PLoS Genet. 2018, 14, e1007230. [Google Scholar] [CrossRef]


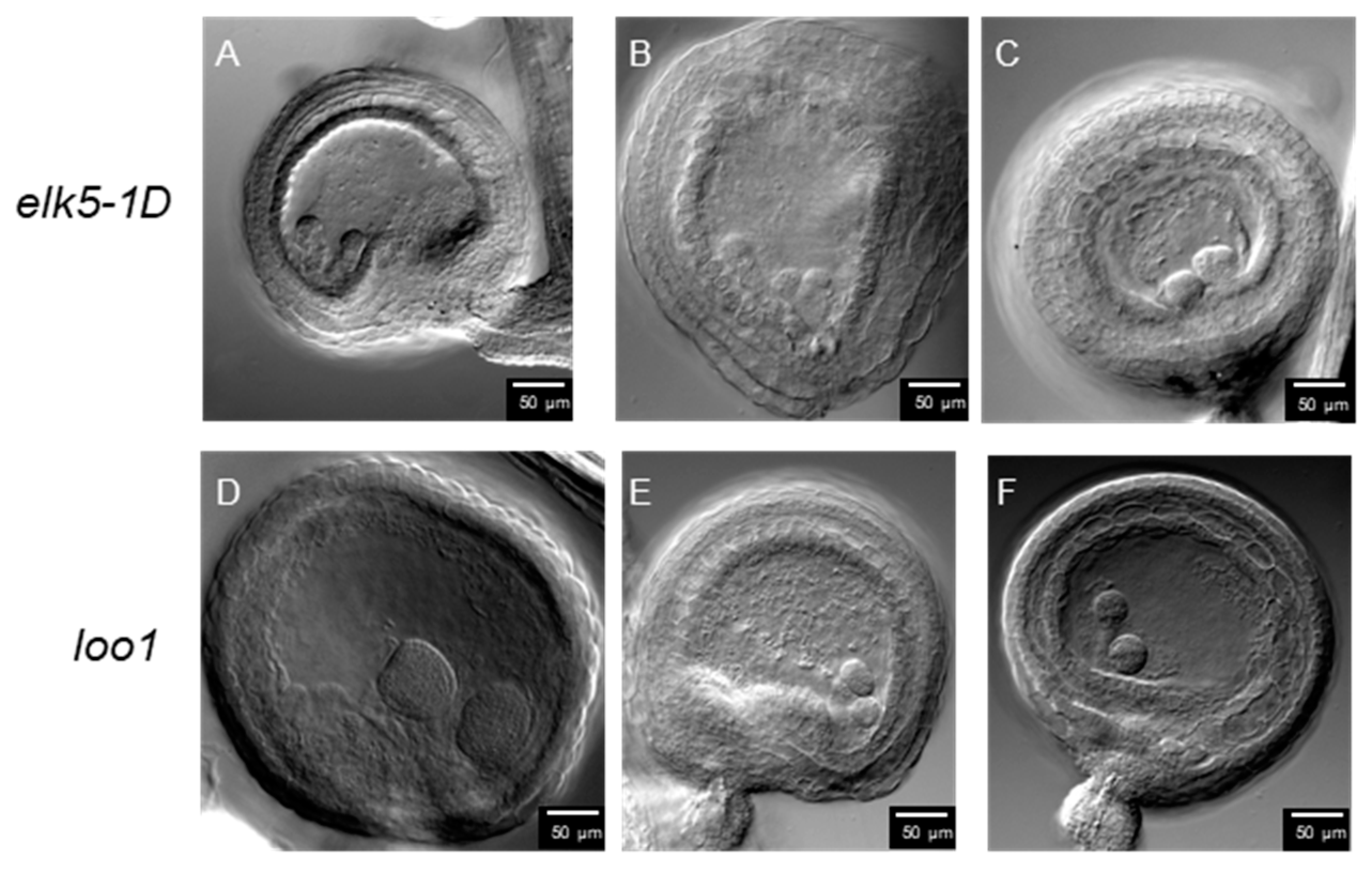
| No. of Fertilized Ovules | No. of Twin Embryos | Percentage of Twin Embryos | |
|---|---|---|---|
| Col-0 | 1035 | 0 | 0 |
| elk5-1D | 1126 | 7 | 0.62% |
| loo1 | 1382 | 8 | 0.58% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.; Zhu, R.; Xu, H.; Enugutti, B.; Schneitz, K.; Wang, X.; Li, J. Twin Embryos in Arabidopsis thaliana KATANIN 1 Mutants. Plants 2024, 13, 1824. https://doi.org/10.3390/plants13131824
Yu Y, Zhu R, Xu H, Enugutti B, Schneitz K, Wang X, Li J. Twin Embryos in Arabidopsis thaliana KATANIN 1 Mutants. Plants. 2024; 13(13):1824. https://doi.org/10.3390/plants13131824
Chicago/Turabian StyleYu, Youfeng, Rui Zhu, Hao Xu, Balaji Enugutti, Kay Schneitz, Xuanpeng Wang, and Jing Li. 2024. "Twin Embryos in Arabidopsis thaliana KATANIN 1 Mutants" Plants 13, no. 13: 1824. https://doi.org/10.3390/plants13131824
APA StyleYu, Y., Zhu, R., Xu, H., Enugutti, B., Schneitz, K., Wang, X., & Li, J. (2024). Twin Embryos in Arabidopsis thaliana KATANIN 1 Mutants. Plants, 13(13), 1824. https://doi.org/10.3390/plants13131824







