Class III Peroxidases in the Peach (Prunus persica): Genome-Wide Identification and Functional Analysis
Abstract
1. Introduction
2. Results
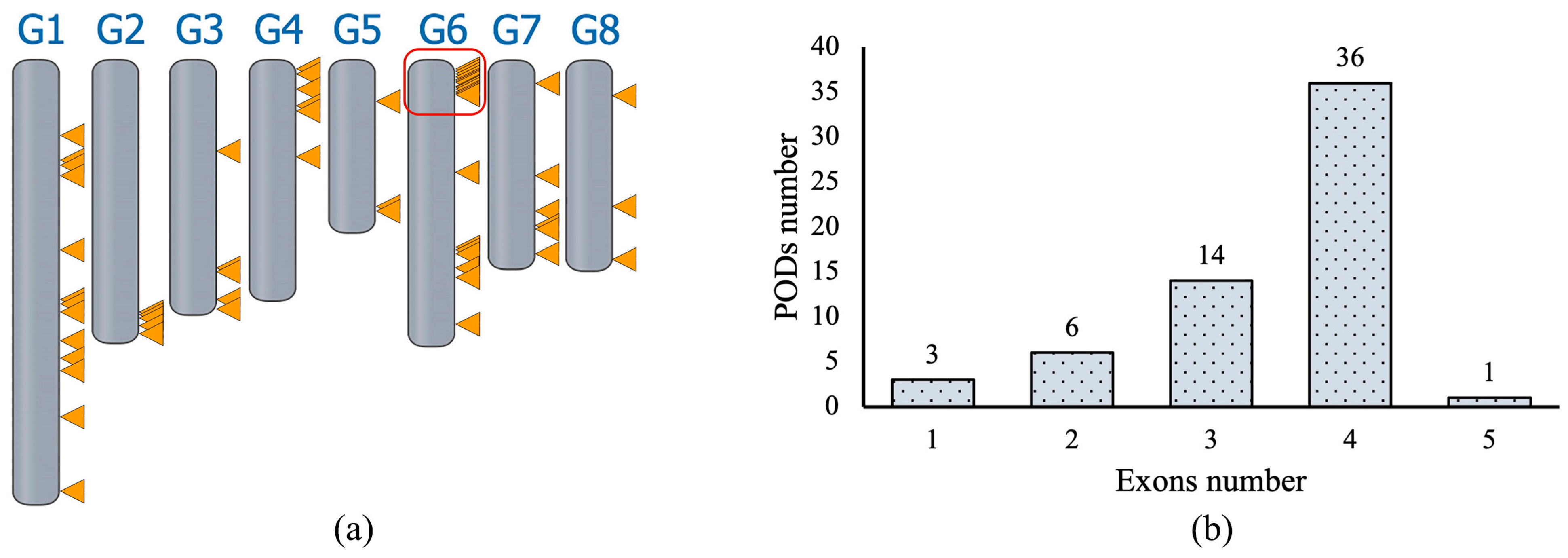
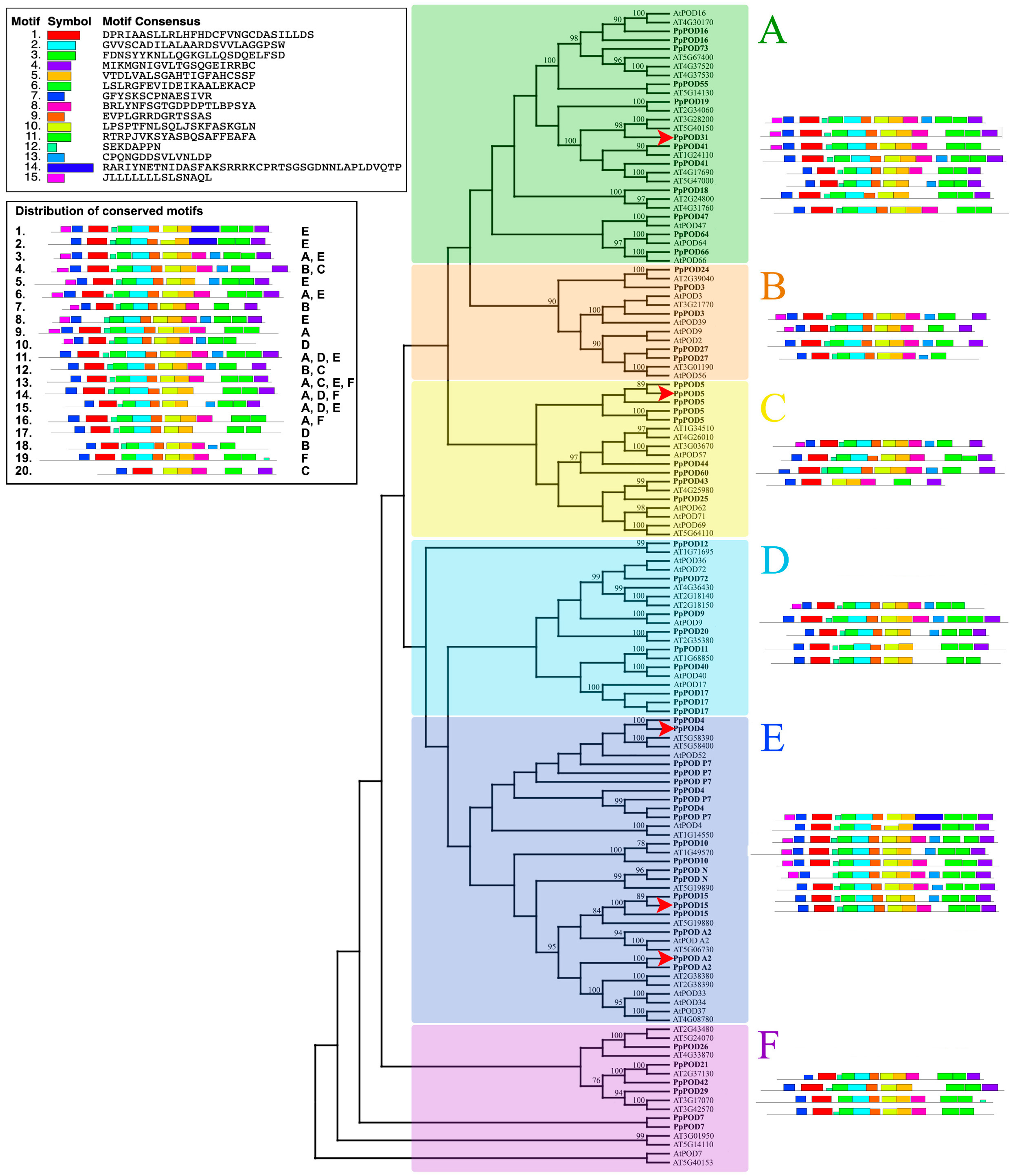
2.1. Peroxidase Genes in P. persica
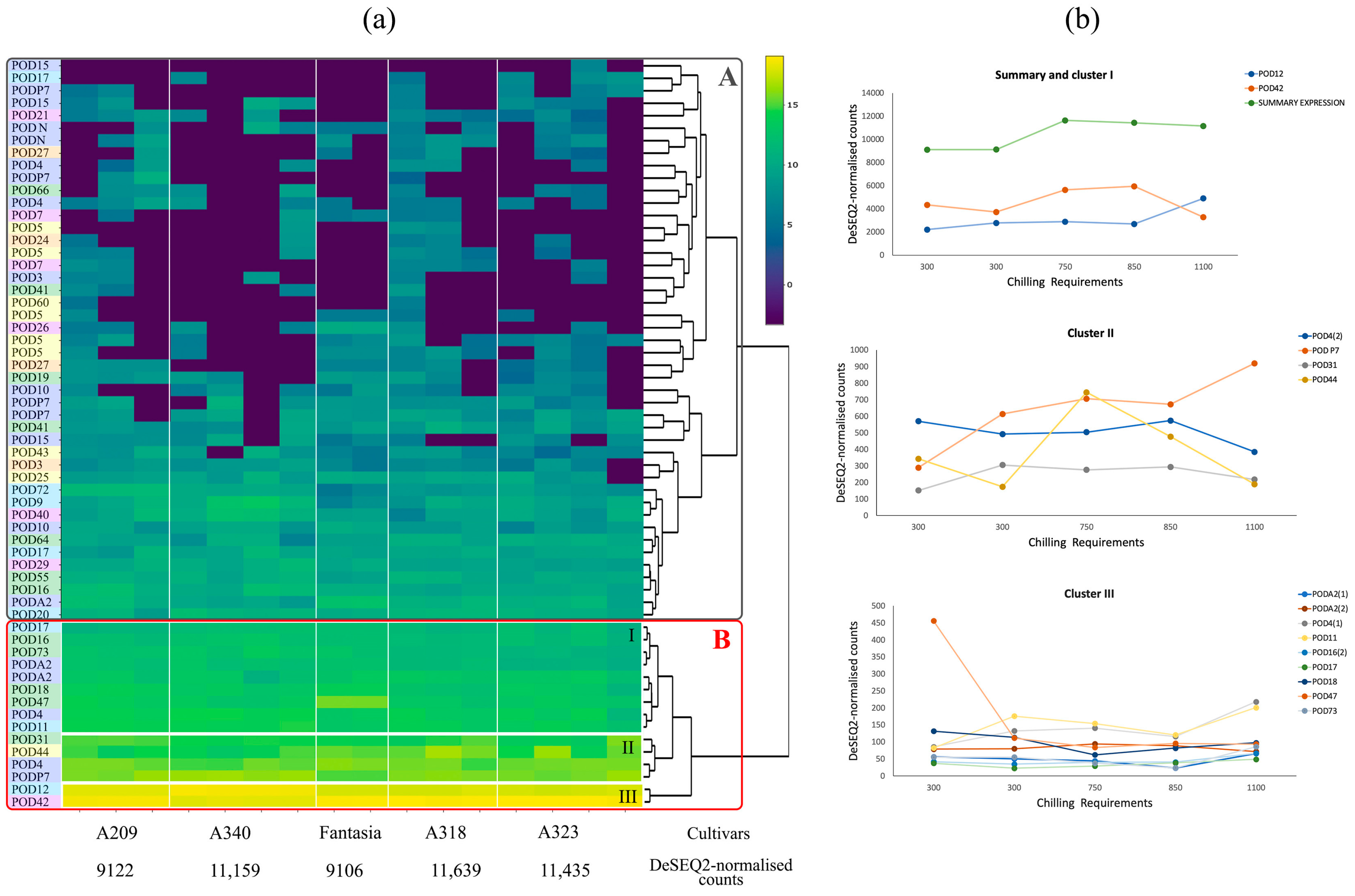
2.2. Expression of Peroxidases in Different Cultivars
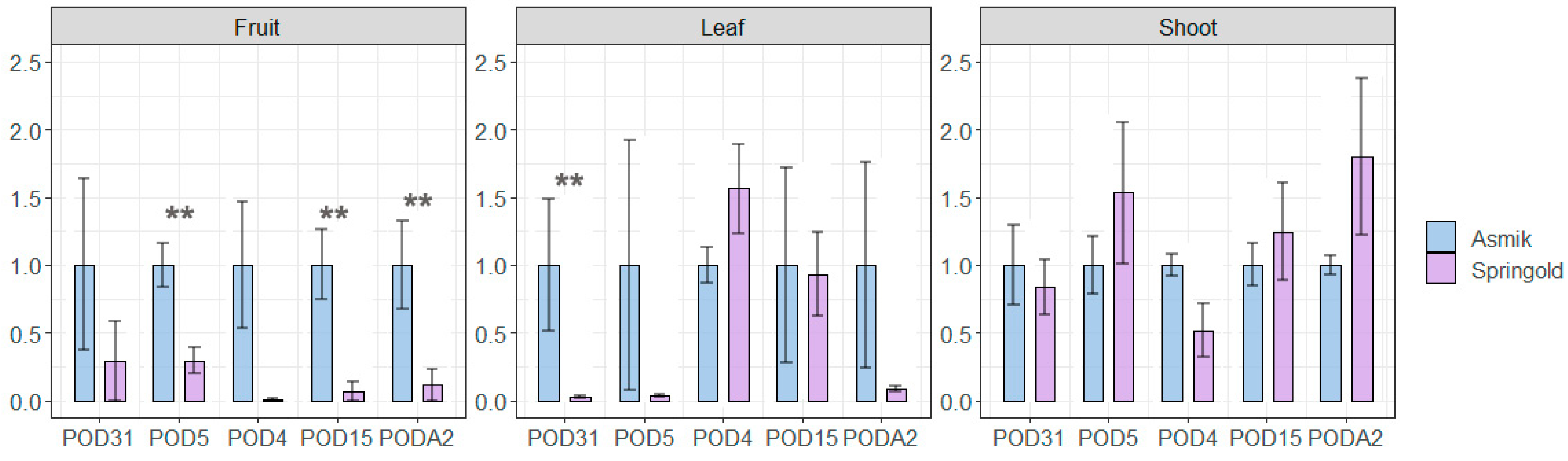
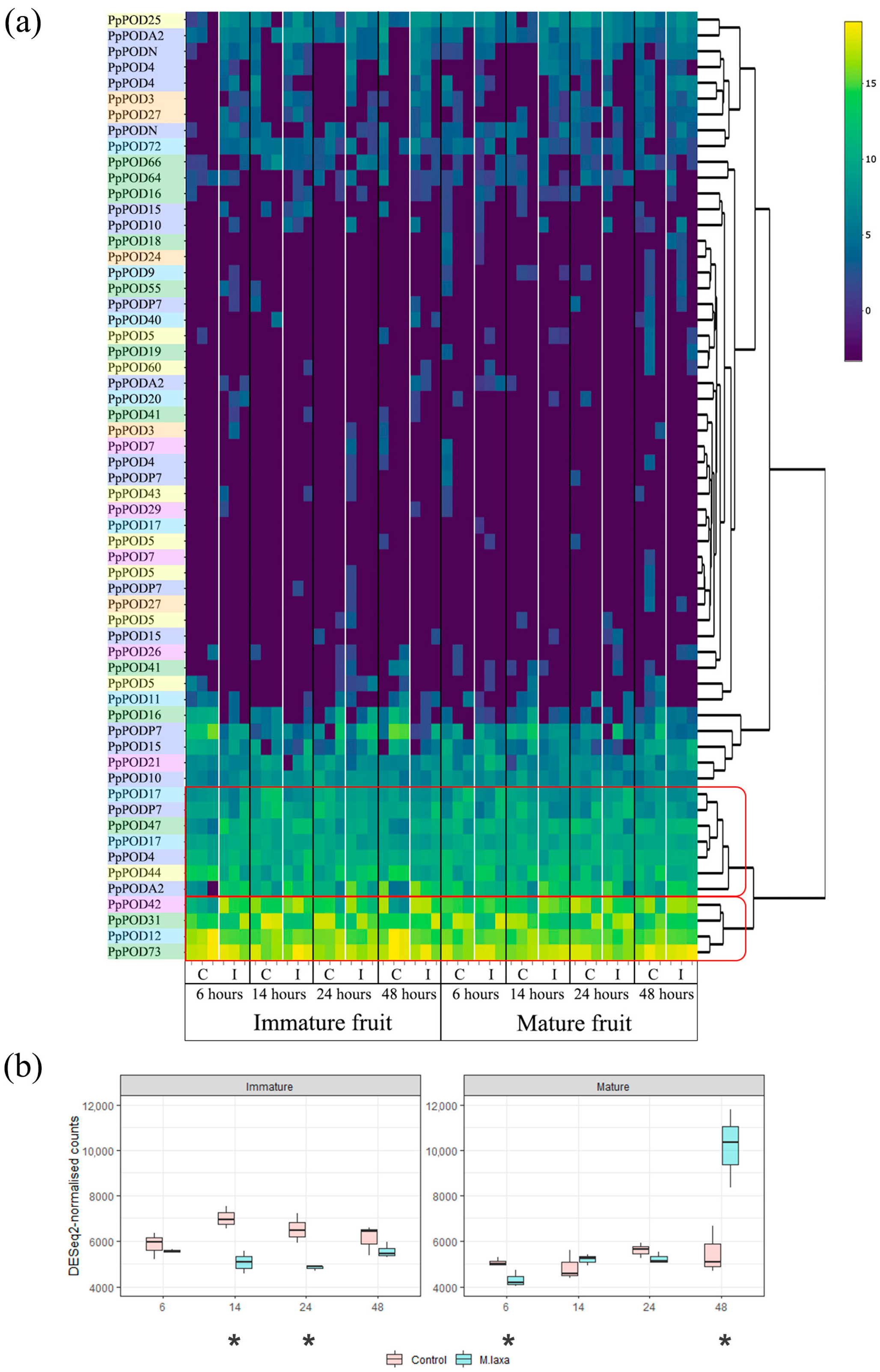
2.3. The Expression of Peroxidases under Stress Factors
3. Discussion
4. Materials and Methods
4.1. Description and Phylogeny of Peroxidases in P. persica
4.2. Expression of Some Peroxidases Based on RT-PCR
4.2.1. Sample Collection
4.2.2. Molecular Analyses
4.3. Expression of Peroxidases Based on Transcriptomic Data
4.3.1. Different Cultivars
4.3.2. Stress Conditions
4.3.3. Bioinformatic Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Passardi, F.; Cosio, C.; Penel, C.; Dunand, C. Peroxidases have more functions than a swiss army knife. Plant Cell Rep. 2005, 24, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Kidwai, M.; Ahmad, I.Z.; Chakrabarty, D. Class III peroxidase: An indispensable enzyme for biotic/abiotic stress tolerance and a potent candidate for crop improvement. Plant Cell Rep. 2020, 39, 1381–1393. [Google Scholar] [CrossRef] [PubMed]
- Hiraga, S.; Sasaki, K.; Ito, H.; Ohashi, Y.; Matsui, H. A Large Family of Class III Plant Peroxidases. Plant Cell Physiol. 2001, 42, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Bindschedler, L.V.; Dewdney, J.; Blee, K.A.; Stone, J.M.; Asai, T.; Plotnikov, J.; Denoux, C.; Hayes, T.; Gerrish, C.; Davies, D.R. Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 2006, 47, 851–863. [Google Scholar] [CrossRef] [PubMed]
- Almagro, L.; Ros, L.G.; Belchi-Navarro, S.; Bru, R.; Barceló, A.R.; Pedreño, M. Class III peroxidases in plant defence reactions. J. Exp. Bot. 2009, 60, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Ritonga, F.N.; Chen, S. Physiological and Molecular Mechanism Involved in Cold Stress Tolerance in Plants. Plants 2020, 9, 560. [Google Scholar] [CrossRef]
- Hou, X.; Lu, Z.; Hong, K.; Song, K.; Gu, H.; Hu, W.; Yao, Q. The class iii peroxidase gene family is involved in ascorbic acid induced delay of internal browning in pineapple. Front. Plant Sci. 2022, 13, 953623. [Google Scholar] [CrossRef]
- Tognolli, M.; Penel, C.; Greppin, H.; Simon, P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 2002, 288, 129–138. [Google Scholar] [CrossRef]
- Wu, C.; Ding, X.; Ding, Z.; Tie, W.; Yan, Y.; Wang, Y.; Yang, H.; Hu, W. The Class III Peroxidase (POD) Gene Family in Cassava: Identification, Phylogeny, Duplication, and Expression. Int. J. Mol. Sci. 2019, 20, 2730. [Google Scholar] [CrossRef]
- Cai, K.W.; Liu, H.; Chen, S.; Zhao, X.Y.; Chen, S. Genome-wide identification and analysis of Class III peroxidases in Betula pendula. BMC Genom. 2021, 22, 314. [Google Scholar] [CrossRef]
- Passardi, F.; Longet, D.; Penel, C.; Dunand, C. The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry 2004, 65, 1879–1893. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.L.; Liu, Y.J.; Liu, H.J.; Qian, T.T.; Qi, L.W.; Wang, X.R.; Zeng, Q.Y. Subcellular relocalization and positive selection play key roles in the retention of duplicate genes of populus Class III peroxidase family. Plant Cell 2014, 26, 2404–2419. [Google Scholar] [CrossRef] [PubMed]
- Behr, M.; Legay, S.; Hausman, J.F.; Guerriero, G. Analysis of cell wall-related genes in organs of Medicago sativa L. under different abiotic stresses. Int. J. Mol. Sci. 2015, 16, 16104–16124. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jin, Q.; Lin, Y.; Cai, Y. Structural, Evolutionary, and Functional Analysis of the Class III Peroxidase Gene Family in Chinese Pear (Pyrus bretschneideri). Front. Plant Sci. 2016, 7, 1874. [Google Scholar] [CrossRef]
- Yang, T.; Zhang, P.; Pan, J.; Amanullah, S.; Luan, F.; Han, W.; Liu, H.; Wang, X. Genome-Wide Analysis of the Peroxidase Gene Family and Verification of Lignin Synthesis-Related Genes in Watermelon. Int. J. Mol. Sci. 2022, 23, 642. [Google Scholar] [CrossRef]
- Aleem, M.; Riaz, A.; Raza, Q.; Aleem, M.; Aslam, M.; Kong, K.; Atif, R.M.; Kashif, M.; Bhat, J.A.; Zhao, T. Genome-wide characterization and functional analysis of class III peroxidase gene family in soybean reveal regulatory roles of GsPOD40 in drought tolerance. Genomics 2022, 114, 45–60. [Google Scholar] [CrossRef]
- Meng, G.; Fan, W.; Rasmussen, S.K. Characterisation of the class III peroxidase gene family in carrot taproots and its role in anthocyanin and lignin accumulation. Plant Physiol. Biochem. 2021, 167, 245–256. [Google Scholar] [CrossRef]
- Su, P.S.; Yan, J.; Li, W.; Wang, L. A member of wheat class III peroxidase gene family, Ta PRX-2A, enhanced the tolerance of salt stress. BMC Plant Biol. 2020, 20, 15. [Google Scholar] [CrossRef]
- Flurkey, W.H.; Jen, J.J. Peroxidase and PPO activites in developing peaches. J. Food Sci. 1978, 43, 1826–1828. [Google Scholar] [CrossRef]
- Abeles, F.B.; Biles, C.L. Characterisation of peroxidases in lignifying peach fruit endocarp. Plant Physiol. 1991, 95, 269–273. [Google Scholar] [CrossRef]
- Jiménez-Atiénzar, M.; Pedreño, M.A.; Caballero, N.; Cabanes, J.; García-Carmona, F. Characterization of polyphenol oxidase and peroxidase from peach mesocarp (Prunus persica L. cv. Babygold). J. Sci. Food Agric. 2007, 87, 1682–1690. [Google Scholar] [CrossRef]
- An, X.; Xu, Y.; Jiang, L.; Huan, C.; Yu, Z. Effects of postharvest temperature on apoptosis-related enzyme activity and gene expression in peach fruits (Prunus persica L. cv. Xiahui 8). Sci. Hortic. 2019, 245, 178–184. [Google Scholar] [CrossRef]
- Oldonil, C.M.; Nienow, A.A.; Schons, J.; Mayer, A.N. Peroxidase activity and initial growth of ‘Barbosa’ peach on clonal rootstocks. Rev. Bras. Frutic. 2019, 2, 1–10. [Google Scholar] [CrossRef]
- Haider, M.S.; Kurjogi, M.M.; Khalil-ur-Rehman, M.; Pervez, T.; Songtao, J.; Fiaz, M.; Fang, J. Drought stress revealed physiological, biochemical and gene-expressional variations in ‘Yoshihime’peach (Prunus persica L.) cultivar. J. Plant Interact. 2018, 13, 83–90. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Z.; Tian, Q.; Zhou, Y.; Xu, J.; Chang, R.; Chen, H.; Liu, G. Quantitative Proteomic Analyses on the Mechanisms of Cold Tolerance in Two Peach Cultivars (Prunus persica L. Batsch) Based on ITRAQ. Eur. J. Hortic. Sci. 2021, 86, 308–319. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Zhao, Y.; Han, G.; Zhu, S. Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 2015, 566, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Xin, F.; Wei, S.; Liu, Y.; Han, Y.; Xie, J.; Ding, Q.; Ma, L. Genome-wide identification, phylogeny and expression profiling of class III peroxidases gene family in Brachypodium distachyon. Gene 2019, 700, 149–162. [Google Scholar] [CrossRef]
- Yang, X.; Yuan, J.; Luo, W.; Qin, M.; Yang, J.; Wu, W.; Xie, X. Genome-Wide Identification and Expression Analysis of the Class III Peroxidase Gene Family in Potato (Solanum tuberosum L.). Front. Genet. 2020, 11, 593577. [Google Scholar] [CrossRef]
- Chen, Y.; Feng, J.; Qu, Y.; Zhang, J.; Zhang, L.; Liang, D.; Yang, Y.; Huang, J. Genome-wide identification and functional analysis of class III peroxidases in Gossypium hirsutum. PeerJ 2022, 10, e13635. [Google Scholar] [CrossRef]
- Cheng, L.; Ma, L.; Meng, L.; Shang, H.; Cao, P.; Jin, J. Genome-Wide Identification and Analysis of the Class III Peroxidase Gene Family in Tobacco (Nicotiana tabacum). Front. Genet. 2022, 13, 916867. [Google Scholar] [CrossRef]
- González-Gordo, S.; Muñoz-Vargas, M.A.; Palma, J.M.; Corpas, F.J. Class III Peroxidases (POD) in Pepper (Capsicum annuum L.): Genome-Wide Identification and Regulation during Nitric Oxide (NO)-Influenced Fruit Ripening. Antioxidants 2023, 12, 1013. [Google Scholar] [CrossRef] [PubMed]
- Shang, H.; Fang, L.; Qin, L.; Jiang, H.; Duan, Z.; Zhang, H.; Yang, Z.; Cheng, G.; Bao, Y.; Xu, J.; et al. Genome-Wide Identification of the Class III Peroxidase Gene Family of Sugarcane and Its Expression Profiles under Stresses. Front. Plant Sci. 2023, 14, 1101665. [Google Scholar] [CrossRef] [PubMed]
- Muthamilarasan, M.; Khandelwal, R.; Yadav, C.B.; Bonthala, V.S.; Khan, Y.; Prasad, M. Identification and Molecular Characterization of MYB Transcription Factor Superfamily in C4Model Plant Foxtail Millet (Setaria italica L.). PLoS ONE 2014, 9, e109920. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Ding, T.; Su, B.; Jiang, H. Genome-Wide Identification, Characterization and Expression Analysis of the Chalcone Synthase Family in Maize. Int. J. Mol. Sci. 2016, 17, 161. [Google Scholar] [CrossRef] [PubMed]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, E.; Veronico, P.; Melillo, M.T.; Delibes, A.; Andrés, M.F.; López-Braña, I. Analysis of class III peroxidase genes expressed in roots of resistant and susceptible wheat lines infected by Heterodera avenae. Mol. Plant–Microbe Interact. 2009, 22, 1081–1092. [Google Scholar] [CrossRef]
- Chassot, C.; Nawrath, C.; Métraux, J.P. Cuticular defects lead to full immunity to a major plant pathogen. Plant J. 2007, 49, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Cheong, Y.H.; Chang, H.-S.; Gupta, R.; Wang, X.; Zhu, T.; Luan, S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002, 129, 661–677. [Google Scholar] [CrossRef]
- Cosio, C.; Dunand, C. Specific Functions of Individual Class III Peroxidase Genes. J. Exp. Bot. 2009, 60, 391–408. [Google Scholar] [CrossRef]
- Herrero, J.; Fernándezpérez, F.; Yebra, T.; Novouzal, E.; Pomar, F.; Pedreño, M.Á.; Cuello, J.; Guéra, A.; Estebancarrasco, A.; Zapata, J.M. Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 2013, 237, 1599–1612. [Google Scholar] [CrossRef]
- Wellmer, F.; Riechmann, J.L.; Alves-Ferreira, M.; Meyerowitz, E.M. Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 2004, 16, 1314–1326. [Google Scholar] [CrossRef] [PubMed]
- Balsells-Llauradó, M.; Silva, C.J.; Usall, J.; Vall-Llaura, N.; Serrano-Prieto, S.; Teixidó, N.; Mesquida-Pesci, S.D.; de Cal, A.; Blanco-Ulate, B.; Torres, R. Depicting the battle between nectarine and Monilinia laxa: The fruit developmental stage dictates the effectiveness of the host defenses and the pathogen’s infection strategies. Hortic. Res. 2020, 7, 167. [Google Scholar] [CrossRef] [PubMed]
- Fanucchi, F.; Alpi, E.; Olivieri, S.; Cannistraci, C.V.; Bachi, A.; Alpi, A.; Alessio, M. Acclimation increases freezing stress response of Arabidopsis thaliana at proteome level. Biochim. Biophys. Acta 2012, 1824, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-G.; Ahsan, N.; Lee, S.-H.; Kang, K.Y.; Lee, J.J.; Lee, B.H. An approach to identify cold-induced low-abundant proteins in rice leaf. Comptes Rendus Biol. 2007, 330, 215–225. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Szalay, L.; Hegedűs, A.; Stefanovits-Bányai, É. Presumable protective role of peroxidase and polyphenol oxidase enzymes against freezing stress in peach (Prunus persica L. Batsch). Acta Biol. Szeged. 2005, 49, 121–122. Available online: https://abs.bibl.u-szeged.hu/index.php/abs/article/view/2441 (accessed on 12 December 2023).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Sychov, A.I. Flower buds and wood tissues hardiness in the dormant phase of some peach varieties and interspecific hybrids. Breed. Var. Cultiv. Fruit Berry Crops 2018, 5, 131–135. Available online: https://s3-eu-west-1.amazonaws.com/vniispk-storage/ckeditor_assets/attachments/330/SSSK._2018._T.5__%E2%84%961.pdf (accessed on 12 December 2023).
- Fedorova, O.S.; Smykov, A.V.; Ivashchenko, I.A.; Ivashchenko, Y.V.; Tsiupka, S.Y. Drought resistance and photoactivity of leaf apparatus of peach cultivars under conditions of the southern coast of Crimea. Acta Hortic. 2021, 1324, 407–412. [Google Scholar] [CrossRef]
- Tsiupka, S.; Mesyats, N.; Smykov, A.; Tsiupka, V. Assessment of drought tolerance in some peach cultivars by chlorophyll fluorescence induction method. Acta Hortic. 2022, 1339, 353–361. [Google Scholar] [CrossRef]
- Smykov, A.; Komar-Tyomnaya, L.; Gorina, V.; Shoferistov, E.; Fedorova, O.; Shishova, T.; Korzin, V.; Tsiupka, S.; Ivashchenko, I. Gene pool collections of stone fruit crops of the Nikitsky Botanical Garden. Sci. Notes “Cape Martyan” Nat. Reserve 2015, 6, 164–228. Available online: https://scnote.nbgnsc.ru/vypuski-sbornika/2015-%e2%84%96-6/ (accessed on 12 December 2023).
- Moale, C. Influence of Winter Frosts on Some Peach Cultivars of Dobrogea. Sci. Pap. Ser. B Hortic. 2013, 57, 213–217. Available online: https://horticulturejournal.usamv.ro/pdf/vol.LVII/Art42.pdf (accessed on 12 December 2023).
- Moale, C.; Șeptar, L.; Gavăț, C.; Petrișor, C. The effect of frost and hale on the peach tree cultivars from R.S.F.G. Constanța. Sci. Pap. Ser. B Hortic. 2016, 60, 37–44. Available online: https://horticulturejournal.usamv.ro/pdf/2016/Art6.pdf (accessed on 12 December 2023).
- Steinberg, S.L. Leaf and Tree Water Relations of Peach (Prunus persica (L.) Batsch). Ph.D. Thesis, Texas A&M University, College Station, TX, USA, 1988. Available online: https://www.proquest.com/openview/8681a793e851acaf1591db98f9e109e8/1?pq-origsite=gscholar&cbl=18750&diss=y (accessed on 12 December 2023).
- Ivashchenko, I.A.; Smykov, A.V.; Fedorova, O.S.; Pilkevich, R.A. The drought resistance on peach varieties of foreign breeding in conditions of the South coast of Nikitsky botanical garden. Pomic. Small Fruits Cult. Russ. 2017, 51, 140–145. Available online: https://www.plodovodstvo.com/jour/article/view/298/298 (accessed on 12 December 2023).
- Tsiupka, V.; Tsiupka, S.; Plugatar, Y.; Bulavin, I.; Komar-Tyomnaya, L. Assessment of the Drought-Tolerance Criteria for Screening Peach Cultivars. Horticulturae 2023, 9, 1045. [Google Scholar] [CrossRef]
- Moale, C.; Septar, L.; Gavat, C.; Caplan, I.; Oprita, A.; Lamureanu, G.; Oancea, F.; Ghiurea, M.; Desliu-Avram, M. The resistance of Peach to the attack of some pathogenes in climate change conditions. Sci. Pap. Ser. B Hortic. 2022, 66. Available online: https://horticulturejournal.usamv.ro/pdf/2022/issue_1/Art22.pdf (accessed on 12 December 2023).
- Okie, W.R.; Beckman, T.G.; Reighard, G.L.; Newall, W.C.; Graham, C.J.; Werner, D.J.; Powell, A.; Krewer, G. Spring freeze damage to the 1996 peach and nectarine crop in the southeastern United States. HortTechnology Horttech 1998, 8, 381–386. [Google Scholar] [CrossRef]
- Gavat, C.; Dumitru, L.M.; Dima, V.; Șeptar, L.; Moale, C.; Oprita, V.A. Study regarding the productivity of some peach varieties. Peach 1990, 4, 30. Available online: https://horticulturejournal.usamv.ro/pdf/2016/Art7.pdf (accessed on 12 December 2023).
- Horsáková, J.; Krška, B. Evaluation of dormancy break in some selected peach (Prunus persica) cultivars. Hort. Sci. 2016, 43, 181–187. [Google Scholar] [CrossRef]
- Scorza, R.; Okie, W.R. Peaches (Prunus). Acta Hortic. 1991, 290, 177–234. [Google Scholar] [CrossRef]
- Melnichenko, L.A. Assessment of the nectarine gene pool for resistance to major diseases. In Problems of Agroecology and Adaptability of Varieties in Modern Horticulture in Russia; Kuznetsov, M.N., Ed.; VNIISPK: Orel, Russia, 2008; pp. 189–192. [Google Scholar]
- Dirlewanger, E.; Cosson, P.; Boudehri, K.; Renaud, C.; Capdeville, G.; Tauzin, Y.; Laigret, F.; Moing, A. Development of a second-generation genetic linkage map for peach [Prunus persica (L.) Batsch] and characterization of morphological traits affecting flower and fruit. Tree Genet. Genomes 2006, 3, 1–13. [Google Scholar] [CrossRef]
- You, S.; Cao, K.; Chen, C.; Li, Y.; Wu, J.; Zhu, G.; Fang, W.; Wang, X.; Wang, L. Selection and validation reference genes for qRT-PCR normalization in different cultivars during fruit ripening and softening of peach (Prunus persica). Sci. Rep. 2021, 11, 7302. [Google Scholar] [CrossRef]
- Szalay, L.; Papp, J.; Szaobo, Z. Evaluation of frost tolerance of peach varieties in artificial freezing tests. Acta Hortic. 1999, 538, 407–410. [Google Scholar] [CrossRef]
- Carrasco-Valenzuela, T.; Munoz-Espinoza, C.; Riveros, A.; Pedreschi, R.; Arus, P.; Campos-Vargas, R.; Meneses, C. Expression QTL (eQTLs) Analyses Reveal Candidate Genes Associated With Fruit Flesh Softening Rate in Peach [Prunus persica (L.) Batsch]. Front. Plant Sci. 2019, 10, 1581. [Google Scholar] [CrossRef]
- Balsells-Llauradó, M.; Vall-Llaura, N.; Usall, J.; Silva, C.J.; Blanco-Ulate, B.; Teixidó, N.; Caballol, M.; Torres, R. Transcriptional profiling of the terpenoid biosynthesis pathway and in vitro tests reveal putative roles of linalool and farnesal in nectarine resistance against brown rot. Plant Sci. 2023, 327, 111558. [Google Scholar] [CrossRef]
- Li, P.; Zhang, Q.; Shi, B.; Liu, L.; Zhang, X.; Wang, J.; Yi, H. Integration of genome and transcriptome reveal molecular regulation mechanism of early flowering trait in Prunus genus (Prunus mume and Prunus persica). Front. Plant Sci. 2022, 13, 1036221. [Google Scholar] [CrossRef]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhebentyayeva, T.N.; Fan, S.; Chandra, A.; Bielenberg, D.G.; Reighard, G.L.; Okie, W.R.; Abbott, A.G. Dissection of chilling requirement and bloom date QTLs in peach using a whole genome sequencing of sibling trees from an F2 mapping population. Tree Genet. Genomes 2014, 10, 35–51. [Google Scholar] [CrossRef]
- Fan, S.; Bielenberg, D.G.; Zhebentyayeva, T.N.; Reighard, G.L.; Okie, W.R.; Holland, D.; Abbott, A.G. Mapping quantitative trait loci associated with chilling requirement, heat requirement and bloom date in peach (Prunus persica L.). New Phytol. 2010, 185, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Fadón, E.; Herrera, S.; Guerrero, B.; Guerra, M.; Rodrigo, J. Chilling and heat requirements of temperate stone fruit trees (Prunus sp.). Agronomy 2020, 10, 409. [Google Scholar] [CrossRef]
- Canton, M.; Forestan, C.; Marconi, G.; Carrera, E.; Bonghi, C.; Varotto, S. Evidence of chromatin and transcriptional dynamics for cold development in peach flower bud. New Phytol. 2022, 236, 974–988. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Anders, S.; McCarthy, D.J.; Chen, Y.; Okoniewski, M.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, A.P.; Qvist, P.; Lazarus, R.; Lescai, F.; Ju, J.; Nyegaard, M.; Mors, O.; Børglum, A.D.; Li, Q.; Christensen, J.H. Experimental validation of methods for differential gene expression analysis and sample pooling in RNA-seq. BMC Genom. 2015, 16, 548. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Reference |
|---|---|---|---|
| ACT | GTTATTCTTCATCGGCGTCTTCG | CTTCACCATTCCAGTTCCATTGTC | [73] |
| GAPDH | ATTTGGAATCGTTGAGGGTCTTATG | AATGATGTTGAAGGAAGCAGCAC | [73] |
| PODA2 | ACTTAGACCCCACAACTCCG | CTCCCCATTACTTCCCACCA | [25] |
| POD4 | CATTGCTGCTCGAGACTCCGTT | AGCTGGCTGAGGGTAGAAGTGG | Present study |
| POD5 | CAAGGGSTGCGATGCCTCAATT | CTGKCCTTGATCTCATCAATGA | Present study |
| POD15 | TCCAGGGTTGTGATGGTTCG | AGACAACACCAGGGCAAACA | [24] |
| POD31 | CCCTACTACAACGTACCGCT | ACCTGGATGAGCTGAGACAC | [25] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vodiasova, E.; Meger, Y.; Uppe, V.; Tsiupka, V.; Chelebieva, E.; Smykov, A. Class III Peroxidases in the Peach (Prunus persica): Genome-Wide Identification and Functional Analysis. Plants 2024, 13, 127. https://doi.org/10.3390/plants13010127
Vodiasova E, Meger Y, Uppe V, Tsiupka V, Chelebieva E, Smykov A. Class III Peroxidases in the Peach (Prunus persica): Genome-Wide Identification and Functional Analysis. Plants. 2024; 13(1):127. https://doi.org/10.3390/plants13010127
Chicago/Turabian StyleVodiasova, Ekaterina, Yakov Meger, Victoria Uppe, Valentina Tsiupka, Elina Chelebieva, and Anatoly Smykov. 2024. "Class III Peroxidases in the Peach (Prunus persica): Genome-Wide Identification and Functional Analysis" Plants 13, no. 1: 127. https://doi.org/10.3390/plants13010127
APA StyleVodiasova, E., Meger, Y., Uppe, V., Tsiupka, V., Chelebieva, E., & Smykov, A. (2024). Class III Peroxidases in the Peach (Prunus persica): Genome-Wide Identification and Functional Analysis. Plants, 13(1), 127. https://doi.org/10.3390/plants13010127





